Abstract
Key message
The altered rice leaf color based on the knockout of CAO1 gene generated using CRISPR/Cas9 technology plays important roles in chlorophyll degradation and ROS scavenging to regulate both natural and induced senescence in rice.
Abstract
Rice chlorophyllide a oxygenase (OsCAO1), identified as the chlorophyll b synthesis under light condition, plays a critical role in regulating rice plant photosynthesis. In this study, the development of edited lines with pale green leaves by knockout of OsCAO1 gene known as a chlorophyll synthesis process is reported. Eighty-one genetically edited lines out of 181 T0 plants were generated through CRISPR/Cas9 system. The edited lines have short narrow flag leaves and pale green leaves compared with wild-type ‘Dongjin’ plants (WT). Additionally, edited lines have lower chlorophyll b and carotenoid contents both at seedling and mature stages. A transcriptome analysis identified 580 up-regulated and 206 downregulated genes in the edited lines. The differentially expressed genes (DEGs) involved in chlorophyll biosynthesis, magnesium chelatase subunit (CHLH), and glutamate-1-semialdehyde2, 1-aminomutase (GSA) metabolism decreased significantly. Meanwhile, the gel consistency (GC) levels of rice grains, chalkiness ratios and chalkiness degrees (CD) decreased in the edited lines. Thus, knockout of OsCAO1 influenced growth period, leaf development and grain quality characters of rice. Overall, the result suggests that OsCAO1 also plays important roles in chlorophyll degradation and ROS scavenging to regulate both natural and induced rice senescence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chloroplasts include chlorophyll and many other protein-binding pigments, which are responsible for capturing light energy needed for photosynthesis. Green chlorophyll is the most abundant pigment and is most important in capturing the light required for photosynthesis (Stenbaek and Jensen 2010). These processes are critical for photosynthetic organisms. There are several types of chlorophyll with different substituents, such as chlorophyll a and chlorophyll b. The chlorophyll synthesis process starts from glutamate through a series of processes to produce chlorophyll a, and then catalyzed by chlorophyllide a oxygenase (CAO) to form chlorophyll b (Wettstein 1995). Chlorophyll a not only acts as a photosynthetic reaction center but also an important factor of the light-harvesting chlorophyll–protein complex (Green and Durnford 1996). However, chlorophyll b is present only in chlorophyll–protein complex (Liu et al. 2004). Chlorophyll b also controls the photosynthetic antenna size, binds to LHC (light-harvesting complex) proteins and stabilizes on the thylakoid membrane. However, LHC proteins that do not bind to chlorophyll b are degraded by proteases (Paulsen et al. 1993; Lindahl et al. 1995; Hoober and Eggink 2001). The gene involved in chlorophyll b synthesis was isolated from the knockout mutants of Chlamydomonas reinhardtii (Tanaka et al. 1998; Mueller et al. 2012). The CAO protein contains three domains known as A, B, and C at the N terminus (Nagata et al. 2004). Also, the mutations in the CAO genes result in a total loss of chlorophyll b, indicating that CAO is exclusively responsible for chlorophyll b synthesis in Arabidopsis plant (Espineda et al. 1999). In an in vitro experiment, the methyl group of chlorophyll a was converted to a formyl group without other proteins in recombinant Arabidopsis as a CAO (Oster et al. 2000). Therefore, CAO is believed to be the sole enzyme responsible for chlorophyll b synthesis. The all green plants tested as of this study, including Streptophyta and Chlorophyta, contains a single copy of a CAO gene (Kunugi et al. 2016), but rice has two CAO genes (OsCAO1 and OsCAO2) (Lee et al. 2005). The two rice genes are tandemly positioned in rice genome, inferring that they were modified by a recent gene duplication event. Chlorophyll b deficiency was showed in the T-DNA mutant inserted in the first intron site of the OsCAO1 gene. Therefore, it was reported that the CAO1 gene plays a direct role in chlorophyll b production (Lee et al. 2005). In addition, these mutants have pale green leaves, which are induced by light (Lee et al. 2005; Yang et al. 2016).
The CRISPR/Cas9 system as a powerful and highly efficient genome editing tool for breeding programs has been utilized to modify the genomes of main crops. For instance, the Osvp1 rice mutant generated by the CRISPR/Cas9 system produced the acquisition of seed dormancy breaking than wild-type plants (Jung et al. 2019). Moreover, knockout of EIF4E increased the immunity of cucumber to a number of viruses, including zucchini yellow mosaic virus, cucumber vein yellowing virus, and papaya ringspot mosaic virus (Bastet et al. 2019). Gene editing of VvWRKY52 via CRISPR/Cas9 in grape increased its resistance to Botrytis cinerea infection (Wang et al. 2018). In this study, we aim (i) to identify whether homozygous mutant lines in rice can be efficiently generated using CRISPR/Cas9 technology, (ii) to determine the phenotype of OsCAO1 knockout mutant, and (iii) to quantify the extent of changes in gene expression through RNA-sequencing to knockout (KO) mutants of OsCAO1 in rice. These KO mutant lines, as previously reported, confirmed that chlorophyll biosynthesis was significantly suppressed and phenotype of a pale green leaf was observed.
Materials and methods
Plant materials and growth conditions
Korean rice cultivar Dongjin (Oryza sativa L., ssp. Japonica) has used as a wild-type in the experiment. Plants have grown under a greenhouse and in the field of Hankyong National University in Korea. Seeds were harvested and dried to have about 14% moisture content and kept at 4 °C.
CRISPR/Cas9 target site selection and CRISPR/Cas9 vector construction
Two target sites were selected from the OsCAO1 sequence for designing sgRNA (single-guide RNA) based on their location in the target gene using the CRISPR RGEN tool program (https://www.rgenome.net/) (Table S1, Fig. S1) (Park et al. 2015). The validation test for sgRNA selection was performed through T7E1 assay using protoplast isolated from rice seedling leaves (Kim et al. 2013; Zhu et al. 2015) (Fig. S2). The CRISPR/Cas9 construct was generated according to the previously established method (Jung et al. 2019). Briefly, the sgRNA expression cassette sequences with XhoI and EcoRI sites between sgRNA scaffold and OsU3 promoter sequence were constructed by Bioneer co., LTD (Dajeon, Korea). The sgRNA expression cassettes were then cloned into AarI-digested OsU3/pBOsC binary vector (Jung et al. 2019). Construction of the sgRNA expression vector and its flanking sequences were investigated by Sanger sequencing and next-generation (deep) sequencing (NGS) (Fig. S3a). The constructed plasmid was inserted into the Agrobacterium tumefaciens strain, EHA105, with the electroporation method (Fig. S3b) (Lee et al. 2011).
Rice transformations and generation of edited lines
Rice transformation was performed as described by Lee et al. (2011). Transgenic plants were regenerated from inoculated calli using a medium containing phosphinothricin (6 mg/L) and cefotaxime (250 mg/L) before transplanting to greenhouse. After 4 weeks of root development, transgenic rice plants were transplanted to pots in a greenhouse maintained at 30 °C during the day and at 23 °C during night. All transgenic plants were carefully cultivated in greenhouse and fields condition (Fig. S3b). Genomic DNA was extracted from approximately 100 mg leaf tissue using DNA Quick Plant Kit (Inclone, Korea). To determine whether T-DNA is present in transgenic plants, PCR analysis was conducted with PrimeSTAR HS DNA Polymerase (TAKARA, Japan) to amplify the Bar gene and the fragments surrounding the target sites (Fig. S3c). PCR amplicons were used to sequence the gene-edited sites using MiniSeq (Illumina, San Diego, CA, USA). In addition, the selection of T-DNA free lines was carried out by the method previously described by Jung et al. (2019). Briefly, a 1% (v/v) Basta solution was embedded in gauze on a part of the seedling leaves grown to the three-leaf stage, and observed 3 days later to distinguish between resistance and sensitivity. Also, these plants were used to reconfirm whether they were resistant or sensitive by the bar-strip test. The NGS data obtained with MiniSeq was performed using Cas-Analyzer previously reported by Park et al (2017) (https://www.rgenome.net/cas-analyzer). To reduce errors in experiments, reads that appear only once were excluded. The insertion and deletion mutations induced by Cas9 were considered to occur only around the cleavage site 3 bp upstream of the protospacer. Also, stably inherited edited plants possessing single-copy insertion of the T-DNA region were selected and used in this study.
Morphological, physiological, and cellular analysis
To estimate pigment contents, photosynthetic and chlorophyll fluorescence parameters for leaves, experiment methods were performed as described by Akhter et al. (2018). TEM analysis (Hitachi, H-600, Tokyo, Japan) was used to examine the structure of chloroplasts in plant cells according to the method previously described by Nakajima et al. (2012). Agronomic traits containing grain length, width, and 1000-grain weight were investigated using conventional methods. In addition, the accumulation of peroxide anions and peroxidase was also performed using 3,3′-diaminobenzidine (DAB), as previously reported by Wang et al. (2013b). The data were analyzed using the GraphPad Prism 7.0 (https://www.graphpad.com/, San Diego, CA, USA) for mean values and SD or SEM. The significance of statistic data was identified using Student’s t test and Duncan’s test.
Transcriptome analysis by RNA-seq
Total RNA was extracted from 4-week-old WT, cao1-1–2 (−32/−32) and cao1-2–16 (−4/−4) seedlings using TRIzol reagent for RNA-seq analysis. The construction of cDNA library and sequencing analysis were carried out by Macrogen Co (Seoul, Korea). Clean reads were produced by removing low-quality reads and mapped to Nipponbare (IRGSP-1.0 pseudomolecule/MSU7) reference genome (https://plants.ensembl.org/) using TopHat (V 2.0.13) with default parameters (https://ccb.jhu.edu/software/tophat). According to information of mapped reads, gene expression levels were normalized to RPKM (Reads per kilobase per million mapped reads). DEGs analysis between edited plants (cao1-1–2 (−32/−32), cao1-2–16 (−4/−4)) and WT was performed to screening standard of FC (Fold Change) ≥ 2 and FDR (False Discovery Rate) < 0.01. GO (Gene Ontology) analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) database were performed as previously reported by Marjohn et al. (2020).
Quantitative RT-PCR analysis
To confirm the accuracy of the RNA-seq data, qRT-PCR was conducted on selected genes. The cao1-1–2 and cao1-2–16 lines were assessed according to WT, and relative gene expression levels were normalized by the Actin gene. All assays for each gene were performed in triplicate with the same conditions. Primers list to amplify the genes used in this experiment are shown in Table S2. And the RNA-seq data have been deposited into NCBI database.
Results
Production of OsCAO1 knockout mutants using CRISPR/Cas9 system
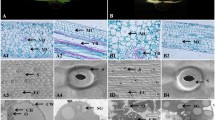
To develop the edited plants, we generated OsCAO1 loss-of-function lines using CRISPR/Cas9 system. A total of 98 and 83 positive transgenic plants for sgRNA:cao1-1 and sgRNA:cao1-2 were obtained in T0 generation, respectively (Fig. S3c, Table 1). Subsequently, we sequenced the target regions using all positive transgenic plants to analyze the mutations at the target sites. The mutation rate was 37.5 and 36.7% in the sgRNA:cao1-1 and sgRNA:cao1-2, respectively (Table 1). These results may reveal very high editing efficiency in the same genetic backgrounds. Sequence results of the mutated region revealed that there were various mutations that occurred, including insertion (1–2 bp) and deletion (1–61 bp) of different nucleotides (Fig. 1b-c). These mutants could be divided into edited plants with homo-, hetero- and bi-alleles (Fig. 1b-c). The most common type of mutations were 1 bp insertions and deletions (Indels) (Fig. S4). Among allele mutation types, there were 69.8% (56/81) for deletions, 29.6% (24/81) for insertions, and 1.2% (1/81) for insertions/deletions, respectively (Fig. S4). Also, we investigated the sgRNA using Cas-OFFinder (https://www.rgenome.net/cas-offinder/) (Bae et al. 2014) and chose two potential off-target sites. However, we could not find any off-target mutations after sequencing these loci (data not shown). To eliminate the bialaphos resistance gene (bar) as a selection marker and T–DNA sequences, T0 edited plants identified by NGS analysis were self-pollinated to generate T1 progeny seeds. T1 progeny plants that were either homo-alleles or T-DNA free plants were analyzed by Bar-strip test, phenotyping and genotyping (Fig. S5a-d). The mutations found can lead to functionally altered proteins, and could have different mutation sites compared with previously reported Oscao1 alleles (Herbert et al. 2020). In this experiment, we edited the other sites of OsCAO1 gene, and the edited lines showed phenotype of a pale green leaf at the seedling stages (Fig. 2a). Also, we measured the photosynthetic pigment contents to explore the pale green leaf phenotype of cao1-1–2 and cao1-2–16 lines. The results showed that cao1-1–2 and cao1-2–16 lines had significant reduction of chlorophyll a, total chlorophyll’s and carotenoids, whereas the chlorophyll b was not detected in two edited lines (Fig. 2b). Also, the ratio of chlorophyll a/chlorophyll b of these mutant lines was dramatically increased compared with that of WT at the seedling (21 day) and mature stages (90 day) (Fig. 2b). Therefore, these data suggested that pale green leaf of cao1-1–2 and cao1-2–16 lines was caused by the reduced chlorophyll b contents. To investigate the chloroplast development of pale green leaf phenotype in edited lines, a TEM (transmission electron microscope) analysis was carried out to compare to the leaf of WT for ultrastructure of chloroplasts. The mesophyll cells appear fully developed chloroplasts in WT, but a decrease in the number of thylakoid lamellar layers was observed in cao1-1–2 and cao1-2–16, and these thylakoid lamellar layers were not well arranged compared to WT (Fig. 2c). These results suggested that cao1-1–2 and cao1-2–16 did not develop chloroplast normally due to the impairment of pale green color and chlorophyll content, and poor arrangement of thylakoid lamellar layers. Thus, the expression levels of genes related to photosynthesis were assessed by quantitative real-time PCR (qRT-PCR). The results revealed that the transcription levels of a psaA gene were upregulated slightly in cao1-1–2 and cao1-2–16, whereas psbA, rbcS and rbcL were apparently downregulated in cao1-1–2 and cao1-2–16 compared with those of the WT (Fig. 2d). Thus, the knockout of OsCAO1 influences the expression of other genes related to plant photosynthesis as well.
Pigment contents in leaves between cao1-1–2 and cao1-2–16 and WT. a Phenotypes of 21 days seedling stage and 90 days mature stage for wild-type (WT) and OsCAO1 mutant lines. b Contents of chlorophyll a, chlorophyll b, total chlorophyll, chlorophyll a/b ratio and carotenoid ratio on leaf mass basis, in different periods of 21-day seedling stage and 90-day mature stage. c Transmission electron microscopy (TEM) analysis of leaves of cao1-1–2, cao1-2–16 and WT lines. d Relative expression levels of psaA, psbA, rbcS and rbcL genes by qRT-PCR. As a loading control, the cao1-1–2, cao1-2–16 and WT were also amplified with actin-specific primers for qRT-PCR analysis. CT values were calculated by actin expression levels, and error bars represent from the values obtained through three replications
Agronomic traits of edited lines
Several agronomic traits of edited lines were compared to the WT control. The cao1-1–2 and cao1-2–16 lines remarkably reduced plant height, number of tillers per plant, and number of grains per panicle, grain size, grain weight and seed-setting rate than that of the WT (Table 2). Grain qualities including chalkiness degree (CD), gel consistency (GC), amylose content (AC) and gelatinization temperature (GT) in edited lines and WT were further investigated. The CD, GC and AC in cao1-1–2 and cao1-2–16 lines were lower than that in WT (Fig. 3, Table S3). The GT showed no significant difference between edited lines and WT (Table S3). Therefore, the cao1-1–2 and cao1-2–16 lines demonstrated decreased photosynthetic efficiency, biomass, grain yield and grain quality due to lack of chlorophyll b compared to the WT. It is also known that leaf variegation is induced by ROS accumulation because it usually leads to cell apoptosis (Jiang et al. 2011). Therefore, we investigated whether ROS accumulation in the flag leaves of edited lines and WT was detected using DAB solution (Fig. S6). More ROS was accumulated in leaves of cao1-1–2 and cao1-2–16 lines compared to the WT, suggesting that the edited plants lost more membrane integrity during development compared with the WT (Fig. S6 b).
Comparative analysis of transcriptome profiling
To understand the impact of OsCAO1 knockout on the rice transcriptome, we performed RNA-seq experiments on the cao1-1–2 and cao1-2–16 and WT plants (Fig. 4). The RNA-seq results showed that gene expression was altered significantly in cao1-1–2, cao1-2–16 and WT plants (Fig. 4b-c). The differentially expressed genes (DEGs) were identified using the screening standards FC ≥ 2. A total of 786 DEGs, containing 580 up- and 206 down-regulated genes in cao1-1–2 vs. WT and cao1-2–16 vs. WT were identified, respectively (https://210.115.36.99:8099/R-seq_results-jung.xlsx) (Fig. 4b-d, Tables S4, S5). Gene ontology (GO) was used to classify the DEG functions. DEGs were grouped into three major GO categories, cellular component, biological process, and molecular function, that totally contained 49 functional groups (Fig. 4e). In addition, the determination of the most enriched GO (gene ontology) terms revealed that the DEGs were grouped into 13 different GO terms, including 2 terms for cellular component, 3 terms for biological process, and 9 terms for molecular function (Fig. 4e). The results underlined that the DEGs annotated with GO terms were mostly involved in oxidation–reduction process, protein phosphorylation, carbohydrate metabolic process, ATP-binding, protein kinase activity, protein binding, oxidoreductase activity, hydrolase activity, hydrolyzing O-glycosyl compounds and integral component of membrane. Moreover, to further understand the molecular functions of the DEGs, we mapped the DEGs and obtained 58 genes related to chlorophyll biosynthesis pathways that were the most enriched (Table 3). This analysis revealed that most of the DEGs in the pathways were downregulated (Fig. S7, Table 3). In particular, five DEGs in “chlorophyll biosynthesis” were all down-regulated. Among them, “Os03g0323200”, encoding magnesium chelatase subunit (CHLH), and “Os08g0532200”, encoding glutamate-1-semialdehyde2, 1-aminomutase (GSA), are important genes involved in chlorophyll biosynthesis (Fig. S7, Table 3). Because glutamic acid and magnesium chelatase is the precursor of chlorophyll b, it is demonstrated that the cao1-1–2 and cao1-2–16 plants influenced the expression of genes upstream of the chlorophyll cycle.
Global gene expression changes in knockout OsCAO1 in rice. a Phenotypes of WT, cao1-1–2 and cao1-2–16 leaves. b Hierarchical cluster analysis of the 554 differentially expressed genes (DEGs) between WT vs. cao1-1–2 and WT vs. cao1-2–16 lines. c Comparison the number of DEGs in WT vs cao1-1–2 and WT vs cao1-2–16. d Volcano plot of DEGs. Axis of abscissas indicates standards FC ≥ 2 and FDR < 0.01. Each point represents a gene. e Gene ontology (GO) classification of DEGs shared by WT vs cao1-1–2 and WT vs cao1-2–16 lines
Gene expression analysis
To confirm the DEGs identified from the transcriptome analysis, we selected 14 genes that were significantly up- or down-regulated in the cao1-1–2 vs WT and cao1-2–16 vs WT from RNA-seq results. The expression levels of six up-regulated DEGs showed higher in cao1-1–2 and cao1-2–16 than in WT (Fig. 5a), while the expression levels of eight down-regulated DEGs revealed lower in cao1-1–2 and cao1-2–16 than in WT (Fig. 5b). Therefore, the expression profiles of DEGs analyzed by RT-PCR were consistent with the transcriptome analysis. Also, to understand the molecular basis of CAO that encodes chlorophyll a, which catalyze the synthesis of chlorophyll b from chlorophyll a oxygenase (Wang et al. 2010; 2013a), the temporal expression profiles of several chlorophyll synthesis-associated genes were also examined using leaves of cao1-1–2 and cao1-2–16 and WT plants. As a result of qRT-PCR, the expression level of OsGSA, OsCHLH, OsHCAR, OsNYC and OsCRD genes in cao1-1–2 and cao1-2–16 seedlings was significantly decreased compared to WT (Fig. S7).
Relative expression levels of cao1-1–2 and cao1-2–16 compared to that of WT and actin detected by quantitative RT-PCR. a Six up-regulated genes (Os10g0502400, HEMA; Os03g0351200, DVR; Os05g0349700, CHLG; Os06g0354700, PPH; Os04g0671300, PAO4; Os10g0389200, RCCR)-associated chlorophyll biosynthesis pathway. b Eight down-regulated genes (Os04g0320100, HCAR; Os01g0227100, NYC1; Os08g0532200, GSA; Os03g0186100, HEM4; Os03g0323200, CHLH; Os01g0279100, CRD1; Os03g0146400, PAO; Os04g0623300, PAO2)-associated chlorophyll biosynthesis pathway
Discussion
Photosynthesis is an important element of the metabolic system and is a process that converts light energy into chemical energy, which has a great influence on plant growth and development. The chlorophyll is existing in chloroplast, consists of chlorophyll a and chlorophyll b, and has almost the same structure except for the side chain. According to Tanaka et al. (1998), chlorophyll a has a methyl group and chlorophyll b has a formyl group at the position of C7, and absorption spectra show that chlorophyll a absorbs mainly at 662 nm and 430 nm and chlorophyll b absorbs at 453 nm and 642 nm. Such difference broadens the light absorption spectrum so that plants can utilize light optimally. So far, rice leaf color mutations have been subdivided into eight types: albino, yellow, light green, yellow-green, green-white, white-green, green-yellow and striped (Nori et al. 2005). These mutants have been used to identify mechanisms that regulate leaf color, such as chlorophyll synthesis and metabolism, plant photosynthesis and photochemical reactions (Li et al. 2018). Until now, many genes in rice have been reported to function in chlorophyll biogenesis such as OsCAO1, OsCHLD, OsCHLH, OsPORA, OsPORB, DVR, FGL, LYL1, YGL1, YGL2, and YLC1 (Masuda et al. 2003). Leaf chlorophyll content has a great influence on photosynthesis and causes many changes in grain formation in rice (Zhou et al. 2006). In this work, we generated rice knockout mutants, cao1-1–2 and cao1-2–16 lines displayed pale green leaves with a significant reduction of chlorophyll content and impaired chloroplast development and photosynthesis (Fig. 2a). The mutation involved in the metabolic pathway of the chlorophyll synthesis usually results in the accumulation of their respective substrates in plants (Mock and Grimm 1997; Wu et al. 2007). In cao1-1–2 and cao1-2–16 lines, chlorophyll a was decreased to 75% that of the WT while chlorophyll b was not detected. These results showed almost the same results to T-DNA insertional mutants and RNAi transformants in the previous reports (Abe et al. 2012; Lee et al. 2005). To investigate the chloroplast development in cao1-1–2 and cao1-2–16 lines, transmission electron microscope (TEM) analysis for ultrastructure of chloroplasts was performed. The mesophyll cells appear fully developed chloroplasts in WT, but a decrease in the number of thylakoid lamellar layers was observed in cao1-1–2 and cao1-2–16, and these thylakoid lamellar layers were not well arranged compared to WT (Fig. 2c). These results suggested that edited lines did not develop chloroplast normally due to the impairment of pale green color and chlorophyll content, and poorly arrangement of thylakoid lamellar layers. Also, the expression levels of genes related to photosynthesis were assessed by qRT-PCR analysis. As a result, the transcript levels of the psaA gene was upregulated slightly in cao1-1–2 and cao1-2–16, whereas psbA, rbcS and rbcL apparently downregulated in cao1-1–2 and cao1-2–16 compared with those of the WT (Fig. 2d). Thus, the knockout of OsCAO1 could really influence the expression of genes related to photosynthesis. Also Wu et al. (2007), reported that leaves are the main photosynthetic apparatus, accounting for 90–95% of dry matter in rice. Several agronomic traits of cao1-1–2 and cao1-2–16 lines were compared to the WT. The cao1-1–2 and cao1-2–16 lines remarkably reduced plant height, number of tillers per plant, and number of grains per panicle, grain weight and seed-setting rate than that of the WT (Table 2). Grain qualities including chalkiness degree (CD), gel consistency (GC) and amylose content (AC) in cao1-1–2 line was lower than that in WT (Fig. 3, Table S3). The GC is one of the key chemical properties of starch in the endosperm that play an important role in eating and cooking quality of rice (Fan et al. 2005). Therefore, the cao1-1–2 and cao1-2–16 lines were demonstrated to have a tendency to decrease in photosynthetic efficiency, biomass, grain yield and grain quality due to lack of chlorophyll b compared to the WT which can lead to an inadequate supply of grain nutrients at grain filling stage (Long et al. 2013). It is also known that leaf variegation can be caused by ROS accumulation because it usually leads to cell apoptosis (Jiang et al. 2011). Thus, we investigated whether ROS accumulation in the flag leaves of cao1-1–2, cao1-2–16 and WT using DAB solution (Fig. S6). There was more ROS accumulation in cao1-1–2 and cao1-2–16 compared to the WT leaves, suggesting that edited plants lost more membrane integrity during growth and development potentially by affecting the ROS scavenging system (Morita et al. 2009; Yamatani et al. 2013) (Fig. S6). Furthermore, RNA-seq was used to analyze a widespread of transcriptome changes according to chlorophyll reduction from the seeding stage to the cao1-1–2, cao1-2–16 oscao1-2 lines. For RNA-seq analysis of cao1-1–2, cao1-2–16 plants, we focused on reactions to related genes such as photosynthesis-related enzymes, redox activity, cofactor binding (Tables S4, S5). To further understand the molecular functions of the DEGs, we mapped the DEGs and obtained 26 genes related to chlorophyll biosynthesis pathways that were the most enriched (Table 3). This analysis showed that the five genes (OsGSA, OsCHLH, OsHCAR, OsNYC and OsCRD) in “chlorophyll biosynthesis” were all down-regulated. Among them, “Os03g0323200”, encoding magnesium chelatase subunit (CHLH), and “Os08g0532200”, encoding glutamate-1-semialdehyde2, 1-aminomutase (GSA), are important genes involved in chlorophyll biosynthesis (Fig S7). Because glutamic acid and magnesium chelatase are the precursors of chlorophyll b, it is demonstrated that the cao1-1–2, cao1-2–16 plants influenced the expression of genes upstream of chlorophyll cycle. Photosynthesis is the main driving force for plant growth, and provides the necessary energy for synthesizing organic compounds (Katz et al. 1978; Krause and Weis 1991). Aside from providing green color, chlorophyll is also highly important in photosynthesis because it plays essential roles in harvesting light energy and converting it to chemical energy (Fromme et al. 2003). Increasing the chlorophyll content in rice is regarded as an important approach to enhancing the photosynthesis rate (Huang et al. 2013) to drive the accumulation of more photoassimilates and ultimately increase crop yield (Mitchell and Sheehy, 2006). The assembly of organelles involves thousands of genes that encode a complex network of metabolic, signaling, and biosynthetic functions (Wang et al. 2020). Heatmap results expand our understanding of the gene mechanisms in chlorophyll biosynthesis pathway. In conclusion, our transcriptomic data provide evidence that OsCAO1 is essential for chlorophyll b biosynthesis in rice. OsCAO1 is induced by various factors. OsCAO1 regulates the expression of genes related to responses to oxidation–reduction, protein phosphorylation, carbohydrate metabolic process, oxidoreductase activity. Overall, the result suggests that OsCAO1 also plays important roles in chlorophyll degradation and ROS scavenging to regulation of many developmental processes including senescence, and responses to environment stresses in rice.
Abbreviations
- LHC:
-
Chlorophyll–protein complex
- CAO :
-
Chlorophyllide a oxygenase
- KO:
-
Knockout
- GSA:
-
Glutamate-1-semialdehyde2, 1-aminomutase
- CHLH:
-
Magnesium-chelatase subunit
- GC:
-
Gel consistency
- CD:
-
Chalkiness degrees
- T-DNA:
-
Transfer, DNA
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Innan H (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178. https://doi.org/10.1038/nbt.2095
Akhter D, Qin R, Nath UK, Alamin M, Jin X, Shi C (2018) The brown midrib leaf (bml) mutation in rice (Oryza sativa L.) causes premature leaf senescence and the induction of defense responses. Genes 9:203. https://doi.org/10.3390/genes9040203
Bae S, Park J, Kim JS (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. J Bioinform 30:1473–1475. https://doi.org/10.1093/bioinformatics/btu048
Bastet A, Zafirov D, Giovinazzo N, Guyon-Debast A, Nogué F, Robaglia C, Gallois JL (2019) Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol J 17:1736–1750. https://doi.org/10.1111/pbi.13096
Espineda CE, Linford AS, Devine D, Brusslan JA (1999) The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 96:10507–10511. https://doi.org/10.1073/pnas.96.18.10507
Fan CC, Yu XQ, Xing YZ, Xu CG, Luo LJ, Zhang Q (2005) The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theor Appl Genet 110:1445–1452. https://doi.org/10.1007/s00122-005-1975-y
Fromme P, Melkozernov A, Jordan P, Krauss N (2003) Structure and function of photosystem I: interaction with its soluble electron carriers and external antenna systems. FEBS Lett 555:40–44. https://doi.org/10.1016/S0014-5793(03)01124-4
Green BR, Durnford DG (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Biol 47:685–714. https://doi.org/10.1146/annurev.arplant.47.1.685
Herbert L, Meunier AC, Bes M, Vernet A, Portefaix M, Durandet F, Périn C (2020) Beyond seek and destroy: how to generate allelic series using genome editing tools. Rice 13:1–9. https://doi.org/10.1186/s12284-020-0366-y
Hoober JK, Eggink LL (2001) A potential role of chlorophylls b and c in assembly of light-harvesting complexes. FEBS Lett 489:1–3. https://doi.org/10.1016/S0014-5793(00)02410-8
Huang J, Qin F, Zang G, Kang Z, Zou H, Hu F, Wang G (2013) Mutation of OsDET1 increases chlorophyll content in rice. Plant Sci 210:241–249. https://doi.org/10.1016/j.plantsci.2013.06.003
Jiang H, Chen Y, Li M, Xu X, Wu G (2011) Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J Integr Plant Biol 53:375–387. https://doi.org/10.1111/j.1744-7909.2011.01037.x
Jung YJ, Lee HJ, Kim JH, Kim DH, Kim HK, Cho YG, Kang KK (2019) CRISPR/Cas9-targeted mutagenesis of F3′ H, DFR and LDOX, genes related to anthocyanin biosynthesis in black rice (Oryza sativa L.). Plant Biotechnol Rep 13:521–531. https://doi.org/10.1007/s11816-019-00579-4
Katz JJ, Norris JR, Shipman LL, Thurnauer MC, Wasielewski MR (1978) Chlorophyll function in the photosynthetic reaction center. Annu Rev Biophys Bioeng 7:393–434
Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim D (2013) A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31:251. https://doi.org/10.1038/nbt.2517
Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Biol 42:313–349. https://doi.org/10.1146/annurev.pp.42.060191.001525
Kunugi M, Satoh S, Ihara K, Shibata K, Yamagishi Y, Kogame K, Tanaka A (2016) Evolution of green plants accompanied changes in light-harvesting systems. Plant Cell Physiol 57:1231–1243. https://doi.org/10.1093/pcp/pcw071
Kurata N, Miyoshi K, Nonomura KI, Yamazaki Y, Ito Y (2005) Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol 46:48–62. https://doi.org/10.1093/pcp/pci506
Lee S, Kim JH, Yoo ES, Lee CH, Hirochika H, An G (2005) Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol Biol 57:805–818. https://doi.org/10.1007/s11103-005-2066-9
Lee HJ, Abdula SE, Jee MG, Jang DW, Cho Y-G (2011) High-efficiency and Rapid Agrobacterium-mediated genetic transformation method using germinating rice seeds. Plant Biotech J 38:251–257. https://doi.org/10.5010/JPB.2011.38.4.251
Li WX, Yang SB, Lu Z, He ZC, Ye YL, Zhao BB, Jin B (2018) Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic Res 5:1–14. https://doi.org/10.1038/s41438-018-0015-4
Lindahl M, Yang DH, Andersson B (1995) Regulatory proteolysis of the major light-harvesting chlorophyll a/b protein of photosystem II by a light-induced membrane-associated enzymic system. Eur J Biochem 231:503–509. https://doi.org/10.1111/j.1432-1033.1995.0503e.x
Liu XD, Shen YG (2004) NaCl-induced phosphorylation of light harvesting chlorophyll a/b proteins in thylakoid membranes from the halotolerant green alga, Dunaliella salina. FEBS Lett 569:337–340. https://doi.org/10.1016/j.febslet.2004.05.065
Long JR, Yz W, Cf S, Sun J, Rj Q (2013) Effects of nitrogen fertilizer level on chlorophyll fluorescence characteristics in flag leaf of super hybrid rice at late growth stage. Rice Sci 20:220–228. https://doi.org/10.1016/S1672-6308(13)60138-9
Marjohn CN, Kang KK, Cho Y-G (2020) Genome-wide transcriptional response of papain-like cysteine protease-mediated resistance against Xanthomonas oryzae pv. oryzae in rice. Plant Cell Rep 39:457–472. https://doi.org/10.1007/s00299-019-02502-1
Masuda T, Fusada N, Oosawa N, Takamatsu KI, Yamamoto YY, Ohto M, Hayashi H (2003) Functional analysis of isoforms of NADPH: protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol 44:963–974. https://doi.org/10.1093/pcp/pcg128
Mitchell PL, Sheehy JE (2006) Supercharging rice photosynthesis to increase yield. New Phytol 171:688–693. https://doi.org/10.1111/j.1469-8137.2006.01855.x
Mock HP, Grimm B (1997) Reduction of uroporphyrinogen decarboxylase by antisense RNA expression affects activities of other enzymes involved in tetrapyrrole biosynthesis and leads to light-dependent necrosis. Plant Physiol 113:1101–1112. https://doi.org/10.1104/pp.113.4.1101
Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59:940–952. https://doi.org/10.1111/j.1365-313X.2009.03919.x
Mueller AH, Dockter C, Gough SP, Lundqvist U, von Wettstein D, Hansson M (2012) Characterization of mutations in barley fch2 encoding chlorophyllide a oxygenase. Plant Cell Physiol 53:1232–1246. https://doi.org/10.1093/pcp/pcs062
Nagata N, Satoh S, Tanaka R, Tanaka A (2004) Domain structures of chlorophyllide a oxygenase of green plants and Prochlorothrix hollandica in relation to catalytic functions. Planta 218:1019–1025. https://doi.org/10.1007/s00425-003-1181-6
Nakajima HH, Pisano DV, Roosli C, Hamade MA, Merchant GR, Mahfoud L, Merchant SN (2012) Comparison of ear-canal reflectance and umbo velocity in patients with conductive hearing loss: a preliminary study. Ear Hear 33:35. https://doi.org/10.1063/1.3658147
Oster U, Tanaka R, Tanaka A, Rüdiger W (2000) Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J 21:305–310. https://doi.org/10.1046/j.1365-313x.2000.00672.x
Park J, Bae S, Kim JS (2015) Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31:4014–4016. https://doi.org/10.1093/bioinformatics/btv537
Park J, Lim K, Kim JS, Bae S (2017) Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33:286–288. https://doi.org/10.1093/bioinformatics/btw561
Paulsen H, Finkenzeller B, KÜHLEIN N, (1993) Pigments induce folding of light-harvesting chlorophyll a/b-binding protein. Eur J Biochem 215:809–816. https://doi.org/10.1111/j.1432-1033.1993.tb18096.x
Stenbaek A, Jensen PE (2010) Redox regulation of chlorophyll biosynthesis. Phytochemistry 71:853–859. https://doi.org/10.1016/j.phytochem.2010.03.022
Tanaka A, Ito H, Tanaka R, Tanaka NK, Yoshida K, Okada K (1998) Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc Natl Acad Sci 95:12719–12723. https://doi.org/10.1073/pnas.95.21.12719
Von Wettstein D, Gough S, Kannangara CG (1995) Chlorophyll biosynthesis. Plant Cell 7:1039
Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Deng X (2010) Divinyl chlorophyll (ide) a can be converted to monovinyl chlorophyll (ide) a by a divinyl reductase in rice. Plant Physiol 153:994–1003. https://doi.org/10.1104/pp.110.158477
Wang P, Wan C, Xu Z, Wang P, Wang W, Sun C, Deng X (2013a) One divinyl reductase reduces the 8-vinyl groups in various intermediates of chlorophyll biosynthesis in a given higher plant species, but the isozyme differs between species. Plant Physiol 161:521–534. https://doi.org/10.1104/pp.112.208421
Wang YH, Zhang LR, Zhang LL, Xing T, Peng JZ, Sun SL, Chen G, Wang XJ (2013b) A novel stress-associated protein SbSAP14 from Sorghum bicolor confers tolerance to salt stress in transgenic rice. Mol Breeding 32:437–449. https://doi.org/10.1007/s11032-013-9882-4
Wang X, Tu M, Wang D, Liu J, Li Y, Li Z, Wang X (2018) CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol J 16:844–855. https://doi.org/10.1111/pbi.12832
Wang B, Zhong Z, Wang X, Han X, Yu D, Wang C, Zhang Y (2020) Knockout of the OsNAC006 transcription factor causes drought and heat sensitivity in rice. Int J Mol Sci 21:2288. https://doi.org/10.3390/ijms21072288
Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Wang C (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant physiol 145:29–40. https://doi.org/10.1104/pp.107.100321
Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Kusaba M (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll–protein complexes during leaf senescence. Plant J 74:652–662. https://doi.org/10.1111/tpj.12154
Yang CJ, Yang YT, Song MA, Song JH (2016) Pest biodiversity and their characteristic damage caused to greenhouse strawberries in Jeju. Korean J Appl Entomol 55:431–437. https://doi.org/10.5656/KSAE.2016.10.0.059
Zhou XS, Wu DX, Shen SQ, Sun JW, Shu QY (2006) High photosynthetic efficiency of a rice (Oryza sativa L.) xantha mutant. Photosynthetica 44:316–319. https://doi.org/10.1007/s11099-006-0025-6
Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, Song G, Gao X, Yao L, Fan D, Meng S, Zhang X, Hu S, Tian Y (2015) An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci Rep 4:1–8. https://doi.org/10.1038/srep06420
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01477604; PJ01319302)” Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
Formal analysis, JY, LH and YJ; investigation, CY-G; resources, BS and KK; software, LH, YJ and BS; supervision, KK; writing-original draft, JY; writing-review and editing, KK. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jung, Y.J., Lee, H.J., Yu, J. et al. Transcriptomic and physiological analysis of OsCAO1 knockout lines using the CRISPR/Cas9 system in rice. Plant Cell Rep 40, 1013–1024 (2021). https://doi.org/10.1007/s00299-020-02607-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02607-y