Abstract
Key message
Benzoate-Coenzyme A ligase enzyme activity catalyzing the conversion of free benzoic acid to benzoyl-CoA was detected and biochemically characterized in the elicitor-treated pear cell cultures.
Abstract
Asian pear (Pyrus pyrifolia) is an economically and nutritionally important fruit-bearing tree of the subtribe Malinae. Upon pathogen attack, pears produce unique benzoate-derived biphenyl phytoalexins. The upstream biosynthesis of the biphenyl in Malinae is still incomplete. Previously, protein preparations from yeast extract-treated pear cultures were able to convert l-phenylalanine to cinnamic acid catalyzed by the activity of the phenylalanine ammonia lyase. The same extract was able to perform a C2 side-chain cleavage of cinnamic acid to benzaldehyde followed by oxidation of the latter to benzoic acid owing to the molecularly-undefined benzaldehyde synthase and benzaldehyde dehydrogenase activities, respectively. The biosynthesis of biphenyls starts with benzoate-Coenzyme A ligase (BZL), which converts benzoic acid to benzoyl-CoA. Subsequently, the previously-defined biphenyl synthase uses benzoyl-CoA to form the biphenyls. The current study reports the first time detection and characterization of BZL activity in elicitor-treated pear cell cultures. The preferred substrate was benzoic acid (Km = 62 ± 4 µM). Magnesium or manganese was prerequisite for the activity, which was enhanced by ~ 70% in the presence of potassium. Maximum BZL activity was observed 18 h post elicitation, which is in agreement with the coordinate induction reported for the enzymes in the same pathway. The induced BZL activity preceded the accumulation of biphenyls supporting its involvement in their biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Pyrus, popularly known as pear, is an economically important fruit-bearing crop plant, which belongs to the subtribe Malinae of the Rosaceae family. This genus comprises about 24 species and had arisen in course of evolution mainly due to interspecific hybridization (Fotirić Akšić et al. 2015; Jiang et al. 2016). Based on geographical distribution and fruit size, pears are further divided into occidental and oriental pears, out of which the latter group is economically more important in terms of fruit yield (Jiang et al. 2016). Oriental pear is represented by approximately 15 species, out of which P. pyrifolia is the most widely grown species and popularly known as the Asian pear (Teng et al. 2002).
Pear fruits are nutritionally important because of the high contents of dietary fibers, polyphenols, flavonoids and antioxidant metabolites (Sarkate et al. 2017; Saini et al. 2019). The production of pear is threatened by two serious diseases, scab and fire blight. Considering the high economic importance of pears, there is an increasing need to develop new disease-resistant cultivars. To resist the pathogen infection, pear and some other members of the subtribe Malinae are known to produce two unique but related classes of phytoalexin, biphenyls and dibenzofurans (Chizzali et al. 2016; Sarkate et al. 2018). These biphenyl phytoalexins showed significant antibacterial activity against the fire blight bacterium Erwinia amylovora in vitro (Chizzali et al. 2012).
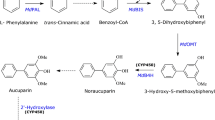
The formation of biphenyl skeleton initiates with benzoyl-CoA as the starter substrate, which is condensed with three molecules of malonyl-CoA in a reaction catalyzed by the enzyme biphenyl synthase (BIS, Fig. 1) (Liu et al. 2004) to form the first biphenyl backbone 3,5-dihydroxybiphenyl. BIS gene function has been recently characterized from the European pear (P. communis) (Chizzali et al. 2016). Then, 3,5-dihydroxybiphenyl undergoes hydroxylation, followed by O-methylation to produce other substituted biphenyls (Khalil et al. 2015; Sircar et al. 2015; Sarkate et al. 2019).
Proposed biosynthesis of benzoate-derived biphenyls in pear. PAL, phenylalanine ammonia lyase; BS, benzaldehyde synthase; BD, benzaldehyde dehydrogenase; BZL, benzoate-CoA ligase; BIS, biphenyl synthase; B4H, biphenyl 4-hydroxylase; OMT, O-methyltransferase. Dashed arrows indicate yet unidentified reactions in pear
The early enzymatic steps of benzoic acid formation have been recently characterized from the elicitor-treated cell cultures of P. pyrifolia (Saini et al. 2017, 2019). However, neither biochemical investigations nor molecular cloning have been explored for the formation of the benzoyl-CoA precursor molecule.
Recently, we have shown that benzoic acid formation in pear cell cultures proceed through the CoA-independent and non-β-oxidative pathway and results in the formation of free benzoic acid. This route involves a C2 side-chain cleavage of cinnamic acid to yield benzaldehyde catalyzed by benzaldehyde synthase (BS) (Fig. 1; Saini et al. 2019). The next enzyme in the biosynthetic pathway is benzaldehyde dehydrogenase (BD) (Fig. 1; Saini et al. 2017), which converts benzaldehyde into benzoic acid.
During benzoate-derived biphenyl formation, the activation of benzoic acid is catalyzed by an adenylate-forming enzyme generally referred to as acyl-CoA ligases (benzoate-CoA ligase; BZL), which is a two-step-mediated activation (Schmelz and Naismith 2009). In this two-step mechanism of CoA ester activation, first an acyl-adenosine monophosphate (AMP) is formed, which is converted in the next step to acyl-CoA ester with the release of one AMP molecule. A well-characterized example of plant aromatic-CoA ligases is the enzyme 4-hydroxycinnamoyl-CoA ligase (4CL) (Bjorklund and Leete 1992; Stuible et al. 2000), which is predominantly involved in the biosynthesis of flavonoids, anthocyanins and lignins. Another example of characterized plant CoA ligases is cinnamte-CoA ligase (CNL), which converts cinnamic acid into cinnamoyl-CoA. CNL gene function has been well-studied in Petunia hybrida (Colquhoun et al. 2012; Klempien et al. 2012), which is responsible for producing benzenoid volatiles, and was shown to initiate xanthone biosynthesis in Hypericum calycinum (Gaid et al. 2012). In Petunia, cinnamoyl-CoA is further metabolized into 3-oxo-3-phenylpropionoyl-CoA (OPP-COA) catalyzed by cinnamoyl-CoA hydratase/dehydrogenase (PhCHD) (Qualley et al. 2012; Bussell et al. 2014). Further, CHD-catalyzed OPP–COA is then converted to benzoyl-CoA by the enzyme 3-ketoacyl CoA thiolase from P. hybrida (PhKAT) (Van Moerkercke et al. 2009). Interestingly, 4CLs and CNLs failed to show any activity with benzoic acid (Gaid et al. 2011; Teotia et al. 2019). BZL activity is scantly reported in plants. It has been detected in crude protein extracts prepared from Clarkia breweri the benzenoid volatile producer (Beuerle and Pichersky 2002b) and xanthone producing H. androsaemum (Abd El-Mawla and Beerhues 2002). Another example of plant aromatic-CoA ligase is the 3-hydroxybenzoyl-CoA ligase. It was detected in the xanthone-producing cell cultures of Centaurium erythraea (Barillas and Beerhues 1997; 2000). Although the protein preparations were found to prefer 3-hydroxybenzoic acid as substrate (100%) it could also accept benzoic acid to a certain level (18%) (Barillas and Beerhues 1997).
CoA-independent and non-β-oxidative route of benzoic acid formation is predominant in pear cell cultures during biphenyl formation. Thus, it is likely that the BD-catalyzed formation of benzoic acid in the pear cell cultures is followed by its thio-esterification into benzoyl-CoA in a reaction catalyzed by BZL. In our previous work, we have detected and characterized BD (Saini et al. 2017) and BS (Saini et al. 2019) activities from the biphenyl producing P. pyrifolia cell cultures. In this study, the same P. pyrifolia cell cultures were used to report the novel detection and biochemical characterization of BZL in Malinae. The detected BZL activity in pear completes the up-stream pathway in the biphenyl biosynthesis.
Materials and methods
Chemicals and reagents
Benzoic acid, benzoyl-CoA, luciferase and luciferin were purchased from Sigma-Aldrich (Bangalore, India). Yeast extract, Murashige and Skoog (MS) medium and plant growth regulators were purchased from HiMedia Laboratories Pvt Ltd (Mumbai, India). High-performance liquid chromatography (HPLC) grade solvents from Merck Life Science Pvt Ltd (Mumbai, India) were used for chromatography. Aucuparin and noraucuparin were synthesized according to Hüttner et al. (2010).
Cell cultures and elicitor treatment
Cell cultures of P. pyrifolia were grown in dark as described by Saini et al. (2017). Eight-day-old cell cultures were treated with yeast extract elicitor (YE, stock solution 150 mg/mL) as previously described (Saini et al. 2019). After treatment with 1 mL YE (150 mg/50 mL cultures), cells were harvested at particular time points [0, 4, 8, 12, 14, 16, 18, 20, 24, 28, 32 and 36 h post elicitation (hpe)] to prepare cell-free extracts for BZL assay or for the analysis of biphenyls. In control treatments, equal volume of sterile distilled water was added to the cell cultures in lieu of the YE. All experiments were carried out at least in triplicates.
Extraction and quantification of biphenyls
Biphenyl phytoalexins were extracted and analyzed by HPLC at different time points (0–36 hpe) after the YE treatments as described previously (Saini et al. 2017).
Preparation of partially-purified protein extract for BZL assay
Cell-free extracts were prepared at 4 °C according to Barillas and Beerhues (1997) except that YE-treated cell cultures (10 g collected through vacuum filtration) were gently homogenized in 20 mL of 200 mM potassium phosphate buffer (pH 7.5) containing 1 mM dithithreitol (DTT), 1% (w/w) polyvinylpyrrolidone and 1 mM ascorbic acid. The resulting homogenate was centrifuged for 30 min at 14,000g and the supernatant was collected. Ammonium sulfate (35–40% saturation) was then added stepwise to the crude extract and mixed gently to precipitate the target protein fraction by centrifugation for 15 min at 10,000g. The protein pellets were re-suspended in the aforementioned buffer and the precipitation step was repeated. The resuspended pellets were subsequently desalted using a PD10 column (GE Healthcare) pre-equilibrated with 100 mM potassium phosphate buffer (pH 7.0). The protein concentration was measured by the Bradford method using bovine serum albumin (BSA; Bradford 1976) as the standard.
BZL assay
To monitor BZL activity, in vitro conversion of benzoic acid to benzoyl-CoA was detected by HPLC. The standard BZL assay consisted of 100 µg partially-purified protein, 0.4 mM benzoic acid (substrate), 0.5 mM CoA, 2.5 mM ATP and 2.5 mM MgCl2. The final assay volume was adjusted to 250 µL using 100 mM potassium phosphate buffer (pH 7.0). The assay was incubated for 120 min at 30 °C. The reaction was terminated by adding 10 µL of 3 M trichloroacetic acid (ice cold). Assay mixture was then centrifuged at 12,000g for 10 min. An aliquot of the resulting supernatant was analyzed by HPLC for detection of BZL activity. In separate assays, a range of monovalent and divalent cations (2.5 mM) was supplemented in the assay to test their impact on BZL activity. Different biochemical parameters were tested for the enzyme characterization. The tested parameters were incubation temperature (20–60 °C), pH (from 6.5–10.5), incubation times (from 5–180 min), and protein concentrations from 12.5–200 µg. The stability of BZL activity was tested at room temperature (for 24 h), − 80 °C (for 2 months) and at 4 °C (for 24 h).
HPLC separation of product of the BZL assay (benzoyl-CoA) was carried out on a X-Bridge™ (Waters, Milford, USA) C18 column (5 µm, 150 × 4.6 mm) using a Waters HPLC system (Milford, USA) equipped with a binary pump (Waters; Binary pump 1525) and photo diode array (Waters; PDA 2998) detector. Chromatograms were analyzed on a Windows 10 platform with an EMPOWER™ software ver. 3 feature release 4 (Waters). BZL reaction product was separated using a gradient solvent system consisting of 5 mM ammonium acetate buffer pH 5.7 (solvent A) and acetonitrile (solvent B). Following gradient was used: 0% B for 2 min, 0–40% B in 23 min, 40–100% B in 26 min, 100% B continued till 30 min. Flow rate was 1.0 mL/min. Detection wavelength was set at 261 nm for benzoyl-CoA. Benzoyl-CoA, the BZL product was identified by comparing the retention time and UV spectrum with those of authentic benzoyl-CoA.
Analysis of BZL assay product by ESI–MS/MS
The chemical identity of benzoyl-CoA, the product of BZL assay was further confirmed by electrospray ionization mass spectrometry (ESI–MS) analysis. The BZL assay volume was up-scaled to 5 mL with proportionate increase in all the reaction components. After incubation at 30 °C for 120 min, the resulting CoA ester was purified and analyzed as described previously (Beuerle and Pichersky 2002a). ESI–MS analysis was carried out in negative ion mode on a 3200 QTrap mass spectrometer (Applied Biosystem/MDS SCIEX) equipped with an electrospray ionization interface (ESI, Turbo V). Analyst Software version 1.4.2 (Applied Biosystems/MDS SCIEX) was used for data acquisition and evaluation.
Substrate specificity test of BZL
Substrate specificity of BZL was tested by luciferase assay as described previously (Gaid et al. 2012). The reaction mixture (total volume 200 µL) consisted of potential carboxylic acid substrate (200 µM of benzoic acid, cinnamic acid, 4-coumaric acid, caffeic acid, or their structurally related acids), ATP (50 µM), MgCl2 (200 µM), CoA (100 µM), 20 µg partially-purified protein and 100 mM potassium phosphate buffer pH 7.0. This reaction mixture was incubated at 30 °C for 120 min and then an aliquot (2 µL) of the assay was taken out and diluted with 100 mM potassium phosphate buffer pH 7.0 to a final volume of 100 µL. A second reaction was started in a 96 microwell plate by adding 100 µL of this diluted mixture with 100 µL of luciferase–luciferin mixture consisting of 1 µg firefly luciferase, 4.6 µg luciferin prepared in potassium phosphate buffer pH 7.0. After 15 s time delay with gentle shaking, luminescence was measured by a luminometer (BMG Labtech Fluostar Omega, Germany). The result was expressed as relative luciferase activity, which inversely correlates with the left-over ATP in the assay. A control assay containing heat-denatured BZL protein and without any substrate was used to normalize the ATP quantification and resulting ATP content was considered as 100% (control).
Determination of apparent kinetic parameters of BZL
The kinetic properties of BZL were determined using Hyper 32 software (https://hyper32.software.informer.com/). For kinetic parameter determination, protein amount was reduced to 50 µg, and the incubation time to 20 min. A range of concentration of benzoic acid (10–500 µM) was used keeping a fixed concentration of CoA (0.5 mM), ATP (2.5 mM), MgCl2 (2.5 mM). Similarly, to determine the apparent Km value for ATP, its concentration was varied from 1–1000 μM keeping a fixed concentration of benzoic acid (0.4 mM), CoA (0.5 mM) and MgCl2 (2.5 mM). For CoA, the apparent Km value was calculated using CoA concentration ranges 1–500 μM keeping fixed concentration of benzoic acid (0.4 mM), ATP (2.5 mM) and MgCl2 (2.5 mM). Three experiments were done for each value with independent enzyme preparations and the mean values were taken for the calculation of apparent Km and Vmax values using Hanes plot algorithm.
Results
Yeast extract induced accumulation of biphenyl phytoalexins in pear cell cultures
When pear cell cultures were treated with YE elicitor, biphenyl phytoalexins were formed as detected by HPLC analyses. The contents of total biphenyl phytoalexins (sum of noraucuparin and aucuparin) increased till 28 hpe and thereafter accumulation decreased slightly. Highest total biphenyl phytoalexin accumulation was observed at 24 hpe (15.8 ± 0.6 µg/g FW) (Fig. 2). Biphenyl phytoalexins were not detected in the untreated control cells.
Detection of BZL activity from the yeast extract treated pear cell cultures
YE-treated (18 hpe) pear cell cultures were used to prepare partially-purified protein extract for incubation with benzoic acid along with CoA, ATP and MgCl2 to test the BZL activity. HPLC analysis of BZL assays showed the formation of benzoyl-CoA (Fig. 3a) suggesting the existence of BZL activity converting benzoic acid to benzoylCoA. The identity of BZL-catalyzed enzymatic product was confirmed by matching retention time and UV spectrum with authentic benzoyl-CoA standard (Fig. 3a). The benzoyl-CoA product was further confirmed by ESI–MS/MS analysis in negative ion mode (Fig. 3b). The obtained molecular ion [M − H]− at m/z 870.2 was an indicative peak for benzoyl-CoA formation, the resulting mass spectrum and the molecular ion peak matched the previously reported fragmentation pattern of benzoyl-CoA (Beuerle and Pichersky 2002a).
Analyses of BZL assay (a) HPLC analysis. Inserts show the UV-spectra of BZL product and benzoyl-CoA reference. b ESI–MS/MS analyses of BZL product (upper panel) and reference (lower panel). S substrate (benzoic acid), P BZL product (benzoyl-CoA), R1 authentic reference benzoic acid, R2 authentic reference benzoyl-CoA. HPLC chromatograms were monitored at 261 nm
Changes in BZL activity after yeast extract treatment
After YE treatment, pear cell cultures were harvested at defined time points to prepare the partially-purified protein for time course BZL assay. Time course analyses showed that initially after the elicitor treatment, BZL activity rapidly increased and attained peak at 18 hpe (7.6 ± 0.4 pkat/mg protein), thereafter, a sharp decrease in the activity was detected, which reached to basal level at 36 hpe (Fig. 2). No basal BZL activity was detected at 0 hpe. Untreated control cell cultures failed to show any BZL activity during the similar time course studied. Interestingly, the peak of biphenyl accumulation (Fig. 2) was preceded by the peak of BZL activity suggesting its involvement in the biosynthesis of the biphenyls.
Biochemical characterization of BZL
Maximum BZL activity was detected in protein preparations from treated cell cultures at 18 hpe, thus the same protein was used for the biochemical characterization of BZL. Protein fraction was obtained by partial purification of cell-free extracts using ammonium sulfate precipitation and desalting by PD10 column. This enzyme activity was found to be dependent on the presence of monovalent and divalent cations. Assay without cations failed to show any BZL activity (Table 1). Among cations, K+ and Mg2+ gave highest activity among monovalent and divalent cations, respectively. Negligible or no activity was detected in the assay supplemented with either Cu2+, Zn2+ or Ca2+. The highest BZL activity (set as 100%) was detected when a combination of divalent (Mg2+, 2.5 mM) and monovalent (K+, 100 mM) cations were used in the assay. The temperature and pH optima of the BZL were found to be 30 °C and 7.5, respectively. BZL activity was found to be linear with time up to 45 min and with the protein concentration up to 150 µg per assay. Significant loss (60%) in BZL activity was detected when stored (as 20% glycerol mix) at − 80 °C for 2 months. Around 40% and 78% losses in the BZL activity were observed after 24 h storage at 4 °C and room temperature, respectively.
BZL substrate specificity was tested using a luciferase-based assay. In this process, CoA ligase activity was measured indirectly by measuring the luciferase activity, which is proportional to the left-over ATP in the assay. More ATP suggests higher luciferase activity, which is indicative of lower CoA ligase activity. A range of structurally related substrates was tested (Fig. 4). The preferred substrate was benzoic acid, followed by 2-aminobenzoic acid, 4-hydroxybenzoic acid, 2-hydroxybenzopic acid and 3-hydroxybenzoic acid. Previously, it was shown that CNL and 4CL cannot activate benzoic acid but actively convert cinnamic and 4-coumaric acids to their thioesters. Partially-purified protein preparations showed residual 4CL and CNL activities (Fig. 4). Notably, protein preparations from untreated cells were able to activate 4-coumaric but not benzoic acid indicating the absence of BZL but the presence of 4CL activities in these control samples (data not shown). After YE treatment, BZL activity gradually increased to reach peak at 18 hpe, which preceded the accumulation of biphenyls.
Substrate specificity of BZL tested by luciferase-based assay. The left-over ATP in the assay was measured as luciferase-dependent bioluminescence, which is inversely correlated with BZL activity. Control reaction with heat-denatured protein extract was used as a negative control and was set at 100% luciferase activity
The BZL-catalyzed reaction followed Michaelis–Menten kinetics, where apparent Km values for benzoic acid, ATP and CoA were found to be 62 ± 4 µM, 108 ± 6 µM and 128 ± 8 µM, respectively.
Discussion
Despite its simple structure, benzoic acid serves as biosynthetic core of a number of economically important and complex plant natural products, such as paclitaxel, xanthones, and benzophenones (Abd El-Mawla and Beerhues 2002; Stewart et al. 2017; Gaid el al. 2012). Notably, benzoic acid serves as the structural building block of biphenyl phytoalexins in Malinae, which serve as the marker defense metabolites (Chizzali and Beerhues 2012). Despite tremendous economic importance of Malinae, biosynthesis of its defense specific marker metabolite, biphenyls, is partially confirmed (Chizzali et al. 2016). We have recently reported that elicitor-treated cell cultures of P. pyrifolia are promising systems for analyzing the biphenyl metabolism (Saini et al. 2017, 2019). It has been shown that benzoic acid biosynthesis in the pear cell cultures proceeds through a CoA-independent and non-β-oxidative pathway. In this process, first cinnamic acid is converted to benzaldehyde by BS activity (Saini et al. 2019) and subsequently benzaldehyde is converted to benzoic acid by BD activity (Saini et al. 2017). The biphenyl and the induction patterns of BZL activity confirm the activation of free benzoic acid to form benzoyl-CoA as intermediate during biphenyl biosynthesis in pear. Cinnamic acid is the product of phenylalanine ammonia lyase (PAL), which was confirmed in crude protein preparations from pear cultures (Saini et al. 2017). Consequently, in elicitor-treated pear cell cultures, PAL, BS, BD, and BIS activities attained peak at 9, 12, 16 and 18 hpe, respectively (Saini et al. 2017, 2019). The coordinated induction of the aforementioned pear enzymes together with the herein studied BZL indicates their involvement in the biphenyl biosynthetic sequence (Fig. 1).
During the last 2 decades, only three reports recorded the biochemical properties of benzoate-mediated BZL activation in planta, from crude protein extracts of C. breweri (Beuerle and Pichersky 2002b), H. androsaemum (Abd El-Mawla and Beerhues 2002) and C. erythraea (Barillas and Beerhues 2000). Similar to the detected BZL activity in crude protein preparations from C. breweri and H. androsaemum, the preferred substrate for pear BZL was benzoic acid. Likewise C. breweri, pear BZL also showed significant activity with 2-aminobenzoic acid. The protein preparations from pear cultures showed residual activities with both cinnamic- and 4-coumaric acids due to the remaining 4CL or CNL after ammonium sulphate precipitation. The sequential increase of the benzoate-activating property of the extract followed by the accumulation of the biphenyl provides evidence for the involvement of the detected BZL activity in phytoalexins formation. Previously reported Malinae 4CLs and CNL (Gaid et al. 2011; Teotia et al. 2019) and other plants CNLs (Colquhoun et al. 2012; Gaid et al. 2012; Klempien et al. 2012) failed to activate benzoic acid, which rules out the involvement of these enzymes in the induced BZL activity after YE treatment of pear cultures. The detected pear BZL activity showed apparent Km values of 62, 108 and 128 µM for benzoic acid, ATP and CoA, respectively. Pear BZL showed lower affinity towards benzoic acid than C. breweri BZL (45 µM). The same is true for 3-hydroxybenzoate-CoA ligase characterized in C. erythraea that showed Km value of 14.7 µM for 3-hydroxybenzoic acid (Barillas and Beerhues 2000). Interestingly, 3-hydroxybenzoate-CoA ligase could also accept benzoic acid as substrate to a certain extent when compared to 3-hydroxybenzoic acid (18%) (Barillas and Beerhues 1997).
In conclusion, the biphenyl formation in pear cell cultures is subsequent to the coordinated induction of PAL, BS, BD, BZL and BIS activities. These results ruled out the possible operation of CoA-dependent and β-oxidative pathway and confirmed the CoA-independent non-β-oxidative sequences of benzoic acid formation. Except for PAL and BIS, none of the above-mentioned coding sequences have been isolated from Malinae. Thus, these cultures can be the system of choice for targeting the needed cDNA-rich pool. Based on these data further research is warranted to involve molecular identification of pear BZL and the subsequent characterizations of the heterologously expressed protein which will support the gene function in vitro. Furthermore, gene silencing of the identified coding sequence would help to further understand the role of this enzyme in biphenyl phytoalexin biosynthesis in vivo.
References
Abd El-Mawla AM, Beerhues L (2002) Benzoic acid biosynthesis in cell cultures of Hypericum androsaemum. Planta 214:727–733. https://doi.org/10.1007/s004250100657
Barillas W, Beerhues L (1997) 3-Hydroxybenzoate:coenzyme A ligase and 4-coumarate: coenzyme A ligase from cultured cells of Centaurium erythraea. Planta 202:112–116
Barillas W, Beerhues L (2000) 3-Hydroxybenzoate:coenzyme A ligase from cell cultures of Centaurium erythraea: isolation and characterization. Biol Chem 381:155–160
Beuerle T, Pichersky E (2002a) Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem 302:305–312
Beuerle T, Pichersky E (2002b) Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri. Arch Biochem Biophys 400:258–264
Bjorklund JA, Leete E (1992) Biosynthesis of the benzoyl moiety of cocaine from cinnamic acid via (R)-(+)-3-hydroxy-3-phenylpropanoic acid. Phytochemistry 31:3883–3887. https://doi.org/10.1016/S0031-9422(00)97546-0
Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bussell JD, Reichelt M, Wiszniewski AAG et al (2014) Peroxisomal ATP-binding cassette transporter comatose and the multifunctional protein abnormal inflorescence meristem are required for the production of benzoylated metabolites in Arabidopsis seeds. Plant Physiol 164:48–54. https://doi.org/10.1104/pp.113.229807
Chizzali C, Beerhues L (2012) Phytoalexins of the pyrinae: biphenyls and dibenzofurans. Beilstein J Org Chem 8:613–620. https://doi.org/10.3762/bjoc.8.68
Chizzali C, Khalil MNA, Beuerle T et al (2012) Formation of biphenyl and dibenzofuran phytoalexins in the transition zones of fire blight-infected stems of Malus domestica cv. “Holsteiner Cox” and Pyrus communis cv. “Conference”. Phytochemistry 77:179–185. https://doi.org/10.1016/j.phytochem.2012.01.023
Chizzali C, Swiddan AK, Abdelaziz S et al (2016) Expression of biphenyl synthase genes and formation of phytoalexin compounds in three fire blight-infected Pyrus communis cultivars. PLoS One 11:e0158713. https://doi.org/10.1371/journal.pone.0158713
Colquhoun TA, Marciniak DM, Wedde AE et al (2012) A peroxisomally localized acyl-activating enzyme is required for volatile benzenoid formation in a Petunia × hybrida cv. “Mitchell Diploid” flower. J Exp Bot 63:4821–4833. https://doi.org/10.1093/jxb/ers153
Fotirić Akšić MM, Dabić DČ, Gašić UM et al (2015) Polyphenolic profile of pear leaves with different resistance to Pear Psylla (Cacopsylla pyri). J Agric Food Chem 63:7476–7486. https://doi.org/10.1021/acs.jafc.5b03394
Gaid MM, Scharnhop H, Ramadan H et al (2011) 4-Coumarate:coA ligase family members from elicitor-treated Sorbus aucuparia cell cultures. J Plant Physiol 168:944–951. https://doi.org/10.1016/j.jplph.2010.11.021
Gaid MM, Sircar D, Muller A et al (2012) Cinnamate:coA ligase initiates the biosynthesis of a benzoate-derived xanthone phytoalexin in Hypericum calycinum Cell Cultures. Plant Physiol 160:1267–1280. https://doi.org/10.1104/pp.112.204180
Hüttner C, Beuerle T, Scharnhop H, Ernst L, Beerhues L (2010) Differential effect of elicitors on biphenyl and dibenzofuran formation in sorbus aucuparia cell cultures. J Agric Food Chem 58:11977-11984.
Jiang S, Zheng X, Yu P et al (2016) Primitive genepools of asian pears and their complex hybrid origins inferred from fluorescent sequence-specific amplification polymorphism (SSAP) markers based on LTR retrotransposons. PLoS One 11:e0149192. https://doi.org/10.1371/journal.pone.0149192
Khalil MNA, Brandt W, Beuerle T et al (2015) O-Methyltransferases involved in biphenyl and dibenzofuran biosynthesis. Plant J 83:263–276. https://doi.org/10.1111/tpj.12885
Klempien A, Kaminaga Y, Qualley A et al (2012) Contribution of CoA ligases to benzenoid biosynthesis in Petunia flowers. Plant Cell 24:2015–2030. https://doi.org/10.1105/tpc.112.097519
Liu B, Beuerle T, Klundt T, Beerhues L (2004) Biphenyl synthase from yeast-extract-treated cell cultures of Sorbus aucuparia. Planta 218:492–496. https://doi.org/10.1007/s00425-003-1144-y
Qualley AV, Widhalm JR, Adebesin F et al (2012) Completion of the core -oxidative pathway of benzoic acid biosynthesis in plants. Proc Natl Acad Sci 109:16383–16388. https://doi.org/10.1073/pnas.1211001109
Saini SS, Teotia D, Gaid M et al (2017) Benzaldehyde dehydrogenase-driven phytoalexin biosynthesis in elicitor-treated Pyrus pyrifolia cell cultures. J Plant Physiol 215:154–162. https://doi.org/10.1016/j.jplph.2017.06.004
Saini SS, Teotia D, Gaid M, Sircar D (2019) A new enzymatic activity from elicitor-treated pear cell cultures converting trans-cinnamic acid to benzaldehyde. Physiol Plant 167:64–74. https://doi.org/10.1111/ppl.12871
Sarkate A, Banerjee S, Mir JI, Roy P, Sircar D (2017) Antioxidant and cytotoxic activity of bioactive phenolic metabolites isolated from the yeast-extract treated cell culture of apple. Plant Cell, Tissue Organ Cult 130:641–664
Sarkate A, Saini SS, Teotia D, Gaid M, Mir JI, Roy P, Agrawal PK, Sircar D (2018) Comparative metabolomics of scab-resistance and susceptible apple cell cultures in response to scab fungus elicitor treatment. Sci Reoprts 8:17844
Sarkate A, Saini SS, Gaid M, Teotia D, Mir JI, Agrawal PK, Beerhues L, Sircar D (2019) Molecular cloning and functional analysis of a biphenyl phytoalexin-specific O-methyltransferase from apple cell suspension cultures. Planta 249:677. https://doi.org/10.1007/s00425-018-3031-6
Schmelz S, Naismith JH (2009) Adenylate-forming enzymes. Curr Opin Struct Biol 19:666–671
Sircar D, Gaid M, Chizzali C et al (2015) Biphenyl 4-hydroxylases involved in aucuparin biosynthesis in Rowan and Apple are CYP736A proteins. Plant Physiol 168:428–442. https://doi.org/10.1104/pp.15.00074
Stewart CJ, Woods K, Macias G, Allan AC, Hellens RP, Noel JP (2017) Molecular architectures of benzoic acid-specific type III polyketide synthases. Acta Crystallogr D Struct Biol 73:1007–1019. https://doi.org/10.1107/S2059798317016618
Stuible HP, Büttner D, Ehlting J et al (2000) Mutational analysis of 4-coumarate:coA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett 467:117–122. https://doi.org/10.1016/S0014-5793(00)01133-9
Teng Y, Tanabe K, Tamura F, Itai A (2002) Genetic relationships of Pyrus species and cultivars native to east asia revealed by randomly amplified polymorphic DNA markers. J Am Soc Hortic Sci 127:262–270. https://doi.org/10.21273/jashs.127.2.262
Teotia D, Gaid M, Saini SS et al (2019) Cinnamate-CoA ligase is involved in biosynthesis of benzoate-derived biphenyl phytoalexin in Malus domestica ‘Golden Delicious’ cell cultures. Plant J. https://doi.org/10.1111/tpj.14506
Van Moerkercke A, Schauvinhold I, Pichersky E et al (2009) A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J 60:292–302. https://doi.org/10.1111/j.1365-313X.2009.03953.x
Acknowledgements
This research work was supported by a start up research grant (FIG 100624 to D. Sircar) from the Indian Institute of Technology Roorkee. SSS is thankful to MHRD-research assistantship (MHRD02-23-200-429) from Indian Institute of Technology Roorkee for perusing his doctoral studies. We thank Dr. Till Beuerle (Technische Universität Braunschweig, Germany) for ESI–MS analyses.
Author information
Authors and Affiliations
Contributions
DS conceived and designed the study. SSS performed experiments, data processing and analyses. MG and DS performed data analyses and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Baochun Li.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Saini, S.S., Gaid, M. & Sircar, D. Benzoate-CoA ligase contributes to the biosynthesis of biphenyl phytoalexins in elicitor-treated pear cell cultures. Plant Cell Rep 39, 207–215 (2020). https://doi.org/10.1007/s00299-019-02484-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02484-0