Abstract
Key message
Transgenic sugarcane expressing CaneCPI-1 exhibits resistance to Sphenophorus levis larvae.
Abstract
Transgenic plants have widely been used to improve resistance against insect attack. Sugarcane is an economically important crop; however, great losses are caused by insect attack. Sphenophorus levis is a sugarcane weevil that digs tunnels in the stem base, leading to the destruction of the crop. This insect is controlled inefficiently by chemical insecticides. Transgenic plants expressing peptidase inhibitors represent an important strategy for impairing insect growth and development. Knowledge of the major peptidase group present in the insect gut is critical when choosing the most effective inhibitor. S. levis larvae use cysteine peptidases as their major digestive enzymes, primarily cathepsin L-like activity. In this study, we developed transgenic sugarcane plants that overexpress sugarcane cysteine peptidase inhibitor 1 (CaneCPI-1) and assessed their potential through feeding bioassays with S. levis larvae. Cystatin overexpression in the transgenic plants was evaluated using semi-quantitative RT-PCR, RT-qPCR, and immunoblot assays. A 50% reduction of the average weight was observed in larvae that fed on transgenic plants in comparison to larvae that fed on non-transgenic plants. In addition, transgenic sugarcane exhibited less damage caused by larval attack than the controls. Our results suggest that the overexpression of CaneCPI-1 in sugarcane is a promising strategy for improving resistance against this insect.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The sugarcane weevil Sphenophorus levis (Coleoptera: Curculionidae) was identified in 1977 (Vaurie 1978) and is one of the insects responsible for the most sugarcane production losses in Brazil (OECD 2016). The yield losses can reach 30 t or more/ha and are caused by the larvae, which feed on the sugarcane clumps, generally above soil level, making irregular galleries and leading to the death of the plants (Fonseca et al. 2012; Santos et al. 2015). No efficient control method is available for S. levis (OECD 2016). Hence, new strategies are necessary to combat this insect.
Knowledge of the digestive enzymes present in the gut of insects has been exploited for pest control strategies using these enzymes as targets of inhibition (Haq et al. 2004). Peptidase inhibitors, as example the cystatins, play a significant role in the natural defense of plants (Ryan 1990; Koiwa et al. 1997; Kessler and Baldwin 2002; Gatehouse 2011; Zhu-Salzman and Zeng 2015) by inhibiting digestive peptidases of insects. The ingestion of these inhibitors by pests/insects interferes with the protein degradation process, reducing the availability of amino acids and thereby hindering the synthesis of proteins necessary for growth, development, and reproduction (Broadway and Duffey 1986; Gatehouse 2011).
The previous studies demonstrated the presence of cysteine peptidases in the gut of some Coleopterans (Murdock et al. 1987; Campos et al. 1989; Terra and Cristofoletti 1996; Cristofoletti et al. 2005; Tribolium Genome Sequencing Consortium 2008; Vorster et al. 2015). According to Soares-Costa et al. (2011), the major digestive peptidases present in S. levis larvae are cathepsins L-like cysteine peptidases that initiate digestion in the anterior and middle midgut. This activity is followed by two minor trypsins in the middle and posterior midgut, and is finished by a negligible membrane-bound aminopeptidase (chymotrypsin) on the surface of middle and posterior midgut cells. Fonseca et al. (2012) reported the recombinant expression and biochemical characterization of a digestive cathepsin L-like enzyme (Sl-CathL) from the midgut of S. levis larvae. The EST for this gene represented the most abundant analyzed EST (10.49%) in the library of the S. levis transcriptome project, which substantiates that cysteine peptidases are the major enzymes of the midgut. Recently, pectinases were identified as accessory digestive enzymes in S. levis (Evangelista et al. 2015). Thus, a strategy based on the overexpression of cysteine peptidase inhibitors (cystatins) in sugarcane plants may be effective against S. levis larval attack.
Transgenic plants expressing cystatins impair the growth and development of insects and can thereby enhance crop production (Leplé et al. 1995; Jouanin et al. 2000; Lawrence and Koundal 2002; Haq et al. 2004; Outchkourov et al. 2004; Ribeiro et al. 2006, Nogueira et al. 2012; Chen et al. 2014). Bioassays with insects that have fed on transgenic plants overexpressing cystatins are an efficient method for evaluating the effect of cysteine peptidase inhibitors on insect growth and development. However, similar studies have not yet been conducted with S. levis.
We reported previously that CaneCPI-1 purified from transgenic sugarcane was able to inhibit the catalytic activity of midgut cysteine peptidase enzymes of S. levis (Ribeiro et al. 2008). In this work, we report for the first time that the overexpression of the CaneCPI-1 gene in transgenic sugarcane retards the growth and development of S. levis larvae that feed on the transgenic plants and that the transgenic plants show less damage caused by larval attack.
Materials and methods
Construction of expression vector
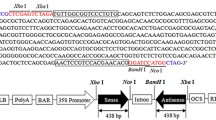
The sugarcane cysteine peptidase inhibitor 1 (CaneCPI-1) gene used in this study was amplified from the pET28canecys plasmid (Soares-Costa et al. 2002), which contained CaneCPI-1 from the sugarcane SP80-3280 cultivar. This cultivar is a commercial hybrid derived from a cross between Saccharum spontaneum and Saccharum officinarum (Vettore et al. 2003). The primers were designed using the Primer 3 program, version 4.0 (Rozen and Skaletsky 1999). The anchored primers CaneF_Bam and CaneR_Bam (Table 1) with BamHI restriction enzyme sites were used. Briefly, 10 ng of the pET28canecys vector, 0.2 mM of dNTPs (Invitrogen), 1 × PCR buffer (20 mM Tris–HCl pH 8.4), 1.5 mM MgCl2, 20 pmol of each primer, and 1 U of Taq DNA Polymerase (Invitrogen) were used in a 100 μL final reaction volume. The following PCR reaction conditions were used: [1×] 94 °C for 1 min, [35×] 94 °C for 30 s, 48 °C for 30 s, 72 °C for 1 min, and [1×] 72 °C for 5 min. The amplification product was recovered from agarose gel slices using the Wizard Gel and PCR Clean-up System (Promega) and digested with the enzyme BamHI (Fermentas). The pAHC17 vector was digested with the same enzyme and dephosphorylated with 2 U of the shrimp alkaline phosphatase (SAP) enzyme (Amersham Biosciences) prior to the ligation. The newly constructed pAHCaneCPI-1 plasmid was confirmed by sequencing, using the DYEnamic ET Terminator Kit (Amersham Biosciences) in an ABI Prism 377 DNA Sequencer (Applied Biosystems).
Transformation, regeneration, and selection of sugarcane plants
The expression vector used for the genetic transformation of sugarcane was pAHC17, which contains the constitutive maize ubiquitin promoter (Ubi-1) for high-level transgene expression in monocots (Christensen and Quail 1996). All procedures related to the transformation and regeneration of sugarcane plants, as well as RT-qPCR and bioassays, were performed at the Sugarcane Technology Center (CTC) (Piracicaba, SP, Brazil). Specimens of S. levis were kindly provided by the Entomology Laboratory of the CTC.
Three-month-old embryogenic calli from the sugarcane cultivar SP80-185 were used for biolistic transformation. Like SP80-3280, the SP80-185 sugarcane cultivar is also a commercial hybrid derived from a cross between S. spontaneum and S. officinarum. Calli were co-bombarded with both pAHCaneCPI-1 and pHA9, which carries geneticin as a selectable marker. The plasmids were co-precipitated with the particles following the method described by Klein et al. (1988). After bombardment, the calli were maintained for 7 days on a medium without antibiotics for recovery. Transgenic calli were selected on a medium containing geneticin for 40 days. Later, the calli were transferred to regeneration medium. Rooted plants were obtained after approximately 90 days. Non-transformed control plants were also obtained through regeneration for comparison. The culture room was maintained at 24 ± 2 °C with a photoperiod of 12 h.
The regenerated plants were tested for CaneCPI-1 integration into the sugarcane genome using the primers Ubi_forward and CaneR_bam (Table 1). Briefly, 20 ng of DNA, 0.2 mM of dNTPs (Invitrogen), 1× PCR buffer (20 mM Tris–HCl pH 8.4), 1.5 mM MgCl2, 8 pmol of each primer, and 1 U of Taq DNA Polymerase (Invitrogen) were used in a 25 µL final reaction volume. The following PCR reaction conditions were used: [1×] 94 °C for 1 min, [40×] 94 °C for 30 s, 48 °C for 30 s, 72 °C for 1 min, and [1×] 72 °C for 5 min. The amplification products were analyzed in a 1% agarose gel stained with ethidium bromide.
Immunoblotting
Total protein was isolated from approximately 200 mg of transgenic and non-transgenic sugarcane leaves using the Trizol Reagent (Invitrogen). The protein concentration was measured using the Bio-Rad Protein Assay Reagent (Bio-Rad). Equal amounts of protein samples from each clone were separated by 15% SDS-PAGE and transferred to an Invitrolon polyvinylidene fluoride (PVDF) membrane (Invitrogen). The membrane was blocked in TBS buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl) with 5% defatted milk and washed with TBS Buffer. Later, the membrane was incubated with the specific anti-CaneCPI-1 antibody (1:3000 dilution) and washed again with TBS. Finally, the membrane was incubated with AP-labeled anti-mouse IgG (Sigma) (1:10,000 dilution) and detected using the AP Conjugate Substrate Kit (Bio-Rad). The employed mouse anti-CaneCPI-1 polyclonal antibody was previously generated by Soares-Costa et al. (2002).
RNA isolation and cDNA synthesis
Total RNA was isolated from 100 mg of fresh sugarcane leaves using the Trizol Reagent (Invitrogen). RNA was quantified in a spectrophotometer, and the integrity was assessed in a 1% agarose gel. Each RNA sample was treated with Amplification Grade DNase I (Invitrogen). The RT reaction was performed with two micrograms of treated RNA using the Improm II Reverse Transcription System Kit (Promega).
Semi-quantitative RT-PCR analysis
The CaneF_bam and CaneR_bam oligonucleotides were used for the CaneCPI-1 gene, and the ST2001F and ST2001R oligonucleotides were used for the polyubiquitin reference gene (Genbank Accession Number: CA179923.1) (Table 1). Six dilutions of cDNA were used in amplification reactions (1:2, 1:4, 1:8, 1:16, 1:32, and 1:64). The PCR was performed under the same conditions used for the CaneCPI-1 gene amplification. The amplification products were analyzed on a 1% agarose gel and visualized using the ChemiDoc™ MP System. The plants that showed high levels of expression were selected for further immunoblot and RT-qPCR analysis.
RT-qPCR analysis
Reverse transcription quantitative PCR (RT-qPCR) was performed according to Bustin et al. (2009). The reaction contained 3 μL of cDNA, 1× Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen), and 0.4 mM of each primer (CaneCPI-1_F and CaneCPI-1_R for the CaneCPI-1 gene and ST2001F_RT-qPCR and ST2001R_RT-qPCR for the polyubiquitin reference gene) in a final volume of 10 µL. The reactions were conducted in triplicate in the ABI 7500 Real Time PCR System (Applied Biosystems). The reaction conditions were 2 min at 50 °C, 10 min at 95 °C, [40×] 95 °C for 15 s, 50 °C for 30 s, 72 °C for 40 s, and [1×] 7 min at 72 °C. The melting curve was analyzed for each reaction to confirm the specificity of the amplification products.
The reactions were optimized for an amplification efficiency between 90 and 110%, a slope between −3.0 and −3.6 and R 2 > 98%. The data were analyzed using the 2−∆∆Ct method (Livak and Schmittgen 2001). The Relative Expression Software Tool (REST) 2005, version 2.0.13, was used to determine the significance of differences in gene expression (Pfaffl et al. 2002). Observed differences in CaneCPI-1 gene expression between transgenic and non-transgenic sugarcane were considered to be significant when p values were less than 0.05 with a 95% confidence interval.
Feeding bioassay
The transgenic sugarcane clones that exhibited higher levels of CaneCPI-1 expression and non-transformed sugarcane (Fig. 1a) were multiplied in vitro to obtain 12 plants for the feeding bioassay. Three independent experiments were performed. Later, the plants were transferred to a greenhouse and maintained until they reached a height of approximately two meters. The S. levis eggs were placed on an artificial diet developed by Degaspari et al. (1987) and were cultured until the 10-day-old larvae stage. Then, a larva was introduced into the stem base of each transgenic and non-transgenic plant. For the inoculation, a manual perforation was made, and a larva was inoculated into each clump (Fig. 1b, c). The hole was sealed with an adhesive tape to prevent the entry of other insects and predators (Fig. 1d). After 10 days, the plants were analyzed for the damage caused by larval feeding, which was represented in terms of centimeters, and the weight of the S. levis larvae was determined.
Feeding bioassay procedures for S. levis larvae introduction into the stem base of sugarcane. a Plants selected for the bioassay; b opening a perforation in the stem base of the plants; c larvae entering the plant with the help of a channel; and d hole closed with adhesive tape. An arrow indicates the location at which the larvae were introduced
Results
Generation of transgenic sugarcane plants and PCR analysis
After selection with geneticin, 35 plants were regenerated and used for PCR analysis. Of these plants, 30 were positive for the CaneCPI-1 gene, which was verified by the presence of an expected amplified fragment of 520 bp, of which 320 bp correspond to CaneCPI-1 and 200 bp correspond to the partial maize ubiquitin promoter (Fig. 2). No amplification was observed for the non-transformed control plant.
Immunoblot assay
The 30 PCR-positive transgenic clones were evaluated using immunoblot assays. The anti-CaneCPI-1 antibody was specific (1:3000 dilution), enabling specific recognition of the cystatin in protein crude extracts of sugarcane leaves. The results showed that transgenic clones 1 and 17 had higher CaneCPI-1 protein expression than the other clones and the non-transformed sugarcane (Fig. 3).
Immunodetection of CaneCPI-1 in crude protein extracts from transgenic and non-transgenic sugarcane leaves. a Coomassie Blue-stained proteins after SDS-PAGE (15%) separation of equal amounts of protein; b the same proteins transferred to a PVDF membrane incubated with polyclonal anti-CaneCPI-1. M bench mark protein ladder, R CaneCPI-1 recombinant protein produced in E. coli and purified, WT crude protein extracts from untransformed plants, 1 and 17 crude protein extracts from the transgenic clones
Semi-quantitative RT-PCR
CaneCPI-1 gene expression levels in the transgenic sugarcane clones selected by immunoblot assays and non-transgenic sugarcane were evaluated by semi-quantitative RT-PCR. An amplification product of 320 bp corresponding to the transgene and 250 bp corresponding to the polyubiquitin reference gene was observed. CaneCPI-1 gene expression was higher in transgenic sugarcane (clones 1 and 17) than in wild-type plants (Fig. 4a). Polyubiquitin gene expression was constant among the transgenic and non-transgenic sugarcane plants (Fig. 4b).
Semi-quantitative RT-PCR. One percent agarose gels showing the PCR amplification products in cDNA from untransformed sugarcane and sugarcane transformed with the pAHCaneCPI-1 construct. a CaneCPI-1 gene expression. b Polyubiquitin reference gene expression. 1 and 17 transgenic sugarcane clones 1 and 17, WT non-transformed sugarcane, M 1 kb DNA ladder and 1, 2, 4, 8, 16, 32, 64 RT-PCR dilutions (1:1, 1:2, 1:4, 1:8, 1:16, 1:32, and 1:64)
RT-qPCR
RT-qPCR analysis was performed to evaluate the CaneCPI-1 expression of transgenic clones 1 and 17 and wild-type sugarcane in independent experiments. An increase in CaneCPI-1 gene expression was observed in transgenic clones 1 (2.75-fold) and 17 (8-fold) in comparison to non-transformed plants (Fig. 5).
Relative expression of CaneCPI-1 gene in transgenic sugarcane clones 1, 17 and non-transformed (WT) sugarcane relative to polyubiquitin in two independent experiments. Error bars were calculated according to Livak and Schmittgen 2001. The differences between transgenic and non-transgenic sugarcane were considered to be statistically significant when p < 0.05
Feeding bioassay
The aim of this experiment was to evaluate the damage caused by the attack of S. levis larvae in transgenic and non-transgenic sugarcane plants. In addition, growth and development of S. levis larvae that fed on transgenic and non-transgenic sugarcane were also analyzed. The damage was evaluated in terms of centimeters for sugarcane and reached an average of 3.9 and 4.3 cm in transgenic clones 1 and 17, respectively, and 12.7 cm in non-transformed plants (Fig. 6a, b). Larvae that fed on the transgenic sugarcane clones 1 and 17 weighed an average of 38.2 and 35.6 mg, respectively, whereas the larvae that fed on the non-transformed sugarcane weighed 74 mg (Fig. 6c, d).
Evaluation of damage caused by larval attack in transgenic and non-transgenic sugarcanes and effect of CaneCPI-1 on S. levis larval growth. a Average damage. b Representative photograph of sugarcane damage caused by larvae. c Average larval weight. d Representative photographs of larvae. S. levis larvae fed on independent transgenic sugarcane clones (1 and 17) expressing CaneCPI-1 and non-transgenic sugarcane (WT). Error bars indicate the standard deviation. The differences between transgenic and non-transgenic sugarcanes were considered to be statistically significant when p < 0.05, as determined by the Student’s t test
Discussion
Brazil is the world’s top producer of sugarcane, with a total production of 665.6 million tons in 2015–16, and a large acreage of 8.65 million hectares is devoted to sugarcane cultivation (Companhia Nacional de Abastecimento—CONAB 2016). Due to the tropical climate and the large land area, the crop is vulnerable to infestation by several insect pests, of which S. levis is known to cause severe damage. The larvae bore into the clumps of sugarcane above soil level, digging irregular galleries. This process reduces sugarcane biomass accumulation and longevity, causing losses that can reach 30 t or more/ha. Infested fields must be destroyed completely to prevent further attack. Because the larvae are present inside the sugarcane stem base and the adults thrive below the soil level, chemical insecticides cannot penetrate efficiently. Very few control methods are currently available for S. levis, and the available methods are not very effective. Sugarcane baits treated with 85% Carbaril can be used as a control measure and to detect the presence of adult weevil populations in the field. Other chemical insecticides, such as Fipronil + Alfa-cypermethrin or Bifenthrin, Imidacloprid, and Fipronil + Carbofuran, can be used in recently harvested areas or over sugarcane ratoons to prevent further infestation. Biological control using baits treated with Beauveria bassiana and Metarhizium anisopliae is also possible but requires optimal conditions for fungal growth (Gallo et al. 2002; Santos et al. 2015). Hence, an alternative strategy that uses transgenic sugarcane resistant to S. levis would be a promising approach.
Several attempts have been made by overexpressing peptidase inhibitors in transgenic plants to impair insect growth and development (Lawrence and Koundal 2002; Habib and Fazili 2007). Transgenic Brassica napus overexpressing oryzacystatin-1 (OC-1) was used to feed aphids of Myzus persicae and reduced the weight and fecundity of the insect. This effect may be due to the presence of cathepsin L/H-type cysteine peptidases which are the predominant digestive enzymes in M. persicae (Rahbé et al. 2003). Moreover, this insect exhibited delays in the time required to reach the adult stage and in the reproductive rate when fed on Arabidopsis thaliana plants overexpressing the Barley cystatin HvCPI-6 (Carrillo et al. 2011). Álvarez-Alfageme et al. (2007) reported that Colorado potato beetle (Leptinotarsa decemlineata) larvae that fed on transgenic potato plants overexpressing the variant HvCPI-1 C68 → G exhibited reduced larval growth. Feeding bioassays with the second and third instars of Phytodecta fornicata revealed significantly increased mortality, which reached approximately 80% of the insects in these instars (Ninkovic et al. 2007).
The CaneCPI-1 used in this study exerts strong inhibitory activity against cysteine peptidases from plants and mammals. Oliva et al. (2004) observed that a HisCaneCPI-1 expressed in Escherichia coli was able to inhibit the mammalian cysteine peptidases cathepsin L (K i = 0.6 nM), cathepsin V (K i = 1.0 nM), and cathepsin K (K i = 0.76 nM) and the plant cysteine peptidases papain (K i = 3.3 nM) and baupain (K i = 21 nM). The HisCaneCPI-1 purified from a transgenic sugarcane showed similar inhibition against human cathepsin L (K i value of 0.9 nM) (Ribeiro et al. 2008). Cysteine peptidases in beetles are biochemically similar to cathepsins L and B in mammals and are usually the main digestive enzymes present in the midgut of larvae, acting at pH 5.0 (Terra and Ferreira 1994). Ribeiro et al. (2008) previously reported the expression and purification of HisCaneCPI-1 in transgenic sugarcane to validate sugarcane as a platform for recombinant protein production. However, those authors did not evaluate the in vivo effect of CaneCPI-1 overexpression on S. levis larvae, which was performed in this study. Moreover, pAHCaneCPI-1 differs from the previous construct of Ribeiro et al. (2008) due to the histidine tag and does not permit purification by affinity chromatography.
Our results showed that CaneCPI-1 protein was detected by immunoblotting, and gene expression was confirmed by semi-quantitative PCR and RT-qPCR in both transgenic and non-transgenic sugarcane cultivar SP80-185. The detection of HisCaneCPI-1 in transgenic and non-transgenic sugarcane was also reported by Ribeiro et al. (2008), which indicates that CaneCPI-1 exhibits basal expression in non-transgenic sugarcane. Finally, our results suggest that the overexpression of CaneCPI-1 in transgenic sugarcane impairs the growth and development of S. levis larvae that feed on the plants. A weight reduction of approximately 50% was observed in larvae that fed on transgenic plants in comparison to non-transgenic sugarcane. Moreover, the transgenic sugarcane inoculated with S. levis larvae exhibited less damage, which is likely due to the role of cystatin in the inhibition of cysteine peptidases. CaneCPI-1 inhibits 85% of cysteine peptidase activity of S. levis larvae midgut in vitro (Ribeiro et al. 2008). CaneCPI-1 also inhibits a cathepsin L-like cysteine peptidase from the midgut of S. levis with a K i value of 2.38 nM. Thus, these evidences and the predominance of cysteine peptidases as midgut enzymes in S. levis explain the observed delay in the growth of larvae that fed on transgenic sugarcane overexpressing CaneCPI-1. No significant developmental alterations in morphology or growth were observed among the transgenic and non-transgenic sugarcane plants. Hence, these results suggest that the overexpression of CaneCPI-1 in transgenic sugarcane is a promising strategy for improving resistance to this insect.
Author contribution statement
FHS and ASCF conceived and designed the study. ASCF, SMC, MCF, and CWR performed the experiments. ASCF, VKS, and MC analyzed the results. VKS, ASCF, MC, and FHS prepared the manuscript. All authors have read and approved the manuscript.
References
Álvarez-Alfageme F, Martínez M, Pascual-Ruiz S, Castañera P, Diaz I, Ortego F (2007) Effects of potato plants expressing a barley cystatin on the predatory bug Podisus maculiventris via herbivorous prey feeding on the plant. Transgenic Res 16:1–13
Broadway RM, Duffey SS (1986) Plant proteinase inhibitor: mechanism of action and effect on the growth and digestive physiology of larval Heliothis zea and Spodoptera exigua. J Insect Physiol 32:827–833
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Campos FA, Xavier-Filho J, Silva CP, Ary MB (1989) Resolution and partial characterization of proteinases and α-amylases from midgets of larvae of the bruchid beetle Callosobruchus maculates (F.). Comp Biochem Physiol 92:51–57
Carrillo L, Martinez M, Álvarez-Alfageme F, Castañera P, Smagghe G, Diaz I, Ortego F (2011) A barley cysteine-proteinase inhibitor reduces the performance of two aphid species in artificial diets and transgenic Arabidopsis plants. Transgenic Res 20:305–319
Chen P, Senthilkumar R, Jane W, He Y, Tian Z, Yeh K (2014) Transplastomic Nicotiana benthamiana plants expressing multiple defence genes encoding protease inhibitors and chitinase display broad-spectrum resistance against insects, pathogens and abiotic stresses. Plant Biotechnol J 12(4):503–515
Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5:213–218
Companhia Nacional de Abastecimento—CONAB (2016) Acompanhamento da Safra Brasileira de Cana-de-açúcar—Safra 2015/16—Quarto Levantamento. http://www.conab.gov.br/OlalaCMS/uploads/arquivos/16_04_14_09_06_31_boletim_cana_portugues_-_4o_lev_-_15-16.pdf. Accessed on: 20/09/2016
Cristofoletti PT, Ribeiro AF, Terra WR (2005) The cathepsin L-like proteinases from the midgut of Tenebrio molitor larvae: sequence, properties, immunocytochemical localization and function. Insect Biochem Mol Biol 35(8):883–901
Degaspari N, Botelho PSM, Dealmeida LC, Castilho HJ (1987) Biology of Sphenophorus levis, Vaurie, 1978 (Col, Curculionidae), with artificial diet and in the field. Pesquisa Agropecuaria Brasil 22:553–558
Evangelista DE, Fonseca FPP, Rodrigues A, Henrique-Silva F (2015) Pectinases from Sphenophorus levis Vaurie, 1978 (Coleoptera: Curculionidae): putative accessory digestive enzymes. J Insect Sci 15:1–8
Fonseca FPP, Soares-Costa A, Ribeiro AF, Rosa JC, Terra WR, Henrique-Silva F (2012) Recombinant expression, localization and in vitro inhibition of midgut cysteine peptidase (Sl-CathL) from sugarcane weevil, Sphenophorus levis. Insect Biochem Mol Biol 42:58–69
Gallo DN, Neto OS, Carvalho S, Baptista RPL (2002) Entomologia agrícola. Fundação de Estudos Agrários Luiz de Queiroz
Gatehouse JA (2011) Prospects for using proteinase inhibitors to protect transgenic plants against attack by herbivorous insects. Curr Protein Pept Sci 12:409–416
Habib H, Fazili KM (2007) Plant protease inhibitors: a defense strategy in plants. Biotechnol Mol Biol Rev 2:68–85
Haq SK, Atif SM, Khan RH (2004) Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: natural and engineered phytoprotection. Arch Biochem Biophys 431:145–159
Jouanin L, Bonadé-bottino M, Girard C, Lerin J, Pham-delegue MH (2000) Expression of protease inhibitors in rapeseed. In: Michaud D (ed) Recombinant protease inhibitors in plants, pp 179–190
Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53:299–328
Klein TM, Harper EC, Svab Z, Sanford JC, Fromm ME, Maliga P (1988) Stable genetic transformation of intact Nicotiana cells by the particle bombardment process. Proc Natl Acad Sci USA 85:8502–8505
Koiwa H, Bressan RA, Hasegawa PM (1997) Regulation of protease inhibitors and plant defense. Trends Plant Sci 2:379–384
Lawrence PK, Koundal KR (2002) Plant protease inhibitors in control of phytophagous insects. Electron J Biotechnol 5:93–109
Leplé JC, Bonadé-Bottino M, Augustin S, Pilate G, Lê Tân VD, Delplanque A, Cornu D, Jouanin L (1995) Toxicity tochrysomela tremulae (Coleoptera: Chrysomelidae) of transgenic poplars expressing a cysteine proteinase inhibitor. Mol Breed 1(4):319–328
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 25:402–408
Murdock LL, Brookhart G, Dunn PE, Foard DE, Kelley S (1987) Cysteine digestive proteinase in Coleoptera. Comp Biochem Physiol 87:783–787
Ninković S, Miljuš-Ðukić J, Radović S, Maksimović V, Lazarević J, Vinterhalter B, Nešković M, Smigocki A (2007) Phytodecta fornicata Brüggemann resistance mediated by oryzacystatin II proteinase inhibitor transgene. Plant Cell Tissue Organ Cult 91:289–294
Nogueira F, Silva CP, Alexandre D, Samuels RI, Soares EL, Aragão FJ, Palmisano G, Domont GB, Roepstorff P, Campos FA (2012) Global proteome changes in larvae of Callosobruchus maculatus Coleoptera: Chrysomelidae: Bruchinae) following ingestion of a cysteine proteinase inhibitor. Proteomics 12(17):2704–2715
OECD (2016) Safety assessment of transgenic organisms in the environment, volume 6: OECD consensus documents, harmonisation of regulatory oversight in biotechnology. OECD Publishing, Paris
Oliva MLV, Carmona AK, Andrade SS, Cotrin SS, Soares-Costa A, Henrique-Silva F (2004) Inhibitory selectivity of canecystatin: a recombinant cysteine peptidase inhibitor from sugarcane. Biochem Biophys Res Commun 320:1082–1086
Outchkourov NS, de Kogel WJ, Wiegers GL, Abrahamson M, Jongsma MA (2004) Engineered multidomain cysteine protease inhibitors yield resistance against western flower thrips (Franklinielia occidentalis) in greenhouse trials. Plant Biotechnol J 2:449–458
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30(9):e36
Rahbé Y, Deraison C, Bonadé-Bottino M, Girard C, Nardon C, Lise Jouanin L (2003) Effects of the cysteine protease inhibitor oryzacystatin (OC-I) on different aphids and reduced performance of Myzus persicae on OC-I expressing transgenic oilseed rape. Plant Sci 164:441–450
Ribeiro AO, Pereira EJ, Galvan TL, Picanco MC, Picoli ET, Silva D, Fari MG, Otoni WC (2006) Effect of eggplant transformed with oryzacystatin gene on Myzus persicae and Macrosiphum euphorbiae. J Appl Entomol 130(2):84–90
Ribeiro CW, Soares-Costa A, Falco MC, Chabregas SM, Ulian EC, Cotrin SS, Carmona AK, Santana LA, Oliva ML, Henrique-Silva F (2008) Production of a His-tagged canecystatin in transgenic sugarcane and subsequent purification. Biotechnol Prog 24(5):1060–1066
Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. In: Stephen M, Stephen AK (eds) Bioinformatics methods and protocols, pp 365–386
Ryan CA (1990) Proteinase inhibitors in plants: genes for improving defenses against insects and pathogens. Ann. Rev. Phytopathol 28:425–449
Santos F, Borém A, Caldas C (2015) Sugarcane: agricultural production, bioenergy and ethanol. Academic Press, London
Soares-Costa A, Beltramini LM, Thiemann OH, Henrique-Silva FA (2002) sugarcane cystatin: recombinant expression, purification, and antifungal activity. Biochem Biophys Res Commun 296:1194–1199
Soares-Costa A, Dias AB, Dellamano M, Fonseca FPP, Carmona AK, Terra WR, Henrique-Silva F (2011) Digestive physiology and characterization of digestive cathepsin L-like proteinase from the sugarcane weevil Sphenophorus levis. J Insect Physiol 57:462–468
Terra WR, Cristofoletti PT (1996) Midgut proteinases in three divergent species of Coleoptera. Comp Biochem Physiol B Biochem Mol Biol 113(4):725–730
Terra WR, Ferreira C (1994) Insect digestive enzymes: properties, compartmentalization and function. Comp Biochem Physiol Part B Comp Biochem 109:1–62
Tribolium Genome Sequencing Consortium (2008) The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955
Vaurie P (1978) Revision of the genus Sphenophorus in South America. American Museum of Natural History, New York
Vettore AL et al (2003) Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res 13(12):2725–2735
Vorster J, Rasoolizadeh A, Goulet MC, Cloutier C, Sainsbury F, Michaud D (2015) Positive selection of digestive Cys proteases in herbivorous Coleoptera. Insect Biochem Mol Biol 65:10–19
Zhu-Salzman K, Zeng R (2015) Insect response to plant defensive protease inhibitors. Annu Rev Entomol 60:13.1–13.20
Acknowledgements
The research was supported by the São Paulo Research Foundation (FAPESP, CBME, CEPID Proc. 98/14138-2). The authors thank Centro de Tecnologia Canavieira, Piracicaba, São Paulo, Brazil, for essential help in sugarcane transformation. FHS is recipient of a productivity scholarship from National Council for Scientific and Technological Development (CNPq) (##311745/2013-0). V.K.S. and A.S.C.F. received a Grant from FAPESP (2013/05370-0 and 2005/59833-5, respectively), and C.W.R. received a Grant from National Council for Scientific and Technological Development (CNPq).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by P. Lakshmanan.
V. K. Schneider and A. Soares-Costa contributed equally to this work.
Rights and permissions
About this article
Cite this article
Schneider, V.K., Soares-Costa, A., Chakravarthi, M. et al. Transgenic sugarcane overexpressing CaneCPI-1 negatively affects the growth and development of the sugarcane weevil Sphenophorus levis . Plant Cell Rep 36, 193–201 (2017). https://doi.org/10.1007/s00299-016-2071-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2071-2