Abstract
Key message
Two Populus bHLH genes ( PtFIT and PtIRO ) were cloned and characterized. The iron deficiency tolerance may be regulated by the PtFIT -dependent response pathway in Populus.
Abstract
Five orthologs of eight Arabidopsis basic helix-loop-helix (bHLH) genes responding to iron deficiency in Populus were analyzed. Open reading frame (ORF) regions of two bHLH genes (PtFIT and PtIRO) were isolated from the iron deficiency tolerant (PtG) and susceptible (PtY) genotypes of Populus tremula ‘Erecta’. Gene sequence analyses showed that each of the two genes was identical in PtG and PtY. Phylogenetic analysis revealed that PtFIT was clustered with the bHLH genes regulating iron deficiency responses, while PtIRO was clustered with another group of the bHLH genes regulating iron deficiency responses in a FIT-independent pathway. Tissue-specific expression analysis indicated that PtFIT was only detected in the root among all tested tissues, while PtIRO was rarely detected in all tested tissues. Real-time PCR showed that PtFIT was up-regulated in roots under the iron-deficient condition. A higher level of PtFIT transcripts was detected in PtG than in PtY. Pearson Correlation Coefficient calculations indicated a strong positive correlation (r = 0.94) between PtFIT and PtIRT1 in PtG. It suggests that the iron deficiency tolerance of PtG may be regulated by the PtFIT-dependent response pathway. The PtFIT-transgenic poplar plants had an increased expression level of PtFIT and PtIRT1 responding to iron deficiency. One PtFIT-transgenic line (TL2) showed enhanced iron deficiency tolerance with higher chlorophyll content and Chl a/b ratio under iron deficiency than the control plants, indicating that PtFIT is involved in iron deficiency response in Populus. The results would provide useful information to understand iron deficiency response mechanisms in woody species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron deficiency-induced chlorosis causes yield loss and poor quality of crops, particularly in calcareous and high pH soils. Iron deficiency is not caused by the lack of iron in the soil, but rather the low availability for plants to absorb (Römheld and Marschner 1986; Guerinot and Yi 1994). Higher plants acquire iron from the soil through two strategies, reduction-based and chelation-based strategies (Römheld and Marschner 1986; Kobayashi and Nishizawa 2012). Non-graminaceous plants (known as Strategy I species) acquire iron through the reduction-based strategy, which is accomplished by reducing Fe(III) to absorbable Fe(II) and then the absorbable Fe(II) is transported into the root by the IRT1 transporter (Eide et al. 1996; Robinson, et al. 1999). Graminaceous plants (known as Strategy II species) produce Fe(III) chelators, mugineic acid family phytosiderophores (MAs), to form a complex of Fe(III)-MAs that is transported into the root by YS1 (Yellow stripe 1) or YSL (Yellow stripe 1-like) transporter (Takagi 1976; Curie et al. 2001; Murata et al. 2006; Inoue et al. 2009). Research has revealed that two groups of genes, Ferric reductase oxidase (FRO) and Iron-regulated transporter (IRT), are involved in iron reduction and transport in Strategy I species (Eide et al. 1996; Robinson, et al. 1999). When plants are exposed to the iron deficiency condition, the FRO and IRT genes are induced to reduce Fe(III) to Fe(II) and to regulate the transport of Fe(II) in plants (Eide et al. 1996; Vert et al. 2001; Jeong and Connolly 2009).
Plants can survive under various stressful conditions by regulating the expression of genes that are involved in many biological changes. Gene regulation could be achieved by many ways, such as DNA methylation, transcriptional and post-transcriptional regulation, or regulation of translation (Latchman 2007). Transcriptional regulation refers to the change at gene expression level via altering the transcription rate. Transcription factors are proteins that bind to a specific region of a promoter to control the transcription rate. As a large family of transcription factors, the basic helix-loop-helix (bHLH) proteins play an important role in regulating genes involved in iron deficiency responses in plants. A bHLH transcription factor (LeFER) isolated from tomato offered the first clue to know how iron deficiency responses are regulated by plants (Ling et al. 2002). The fer mutant in tomato failed to induce LeIRT1 expression under the iron-deficient condition, indicating the direct role of LeFER in regulating the LeIRT1 gene. In addition, LeFER expression was suppressed by iron sufficiency at the post-transcriptional level (Brumbarova and Bauer 2005). An ortholog of LeFER named AtFIT1 (FER-like iron deficiency-induced transcription factor 1, also known as AtbHLH29 or AtFRU) is required for inducing the iron mobilization genes in Arabidopsis (Jakoby et al. 2004; Yuan et al. 2005). The AtFIT1 gene regulates several iron deficiency inducible genes with known or putative functions in iron homeostasis, including AtFRO2 and AtIRT1 (Colangelo and Guerinot 2004; Ivanov et al. 2012). Colangelo and Guerinot (2004) reported that AtFIT1 regulated AtFRO2 at the level of mRNA accumulation and AtIRT1 at the level of protein accumulation. Further study demonstrated that with the co-expression of another two AtbHLH transcription factors, forming AtFIT1/AtbHLH38 and AtFIT1/AtbHLH39 complexes, AtFRO2 and AtIRT1 could be constitutively expressed even under the iron-sufficient condition (Yuan et al. 2008). Additionally, another two AtbHLH transcription factors (AtbHLH100 and AtbHLH101) within the same subgroup of AtbHLH38 and AtbHLH39 were strongly induced by iron deficiency in the root and leaf of Arabidopsis (Wang et al. 2007). Sivitz et al. (2012) proposed that rather than the AtFIT1 target genes, AtbHLH100 and AtbHLH101 likely regulate genes involved in the distribution of iron within the plant, suggesting that AtbHLH100 and AtbHLH101 play an AtFIT1-independent regulation role in iron deficiency responses. Maurer et al. (2014) reported that the triple defective mutant of bHLH39/100/101 showed an up-regulated expression of AtIRT1, AtFRO2, and AtFIT as the wild type under iron deficiency. With a different opinion, Wang et al. (2013) proposed that both AtbHLH100 and AtbHLH101 could interact with AtFIT1 according to the yeast two-hybrid analysis and bimolecular fluorescence complementation assay. Furthermore, Long et al. (2010) reported that in addition to AtFIT1 acting as a master regulator in the iron deficiency response, another AtbHLH transcription factor, POPEYE (AtPYE, also known as AtbHLH047) expressed in the pericycle, was also responding to iron deficiency. AtPYE helps maintain iron homeostasis by regulating the expression of known iron homeostasis genes and other genes involved in transcription, development, and stress response according to microarray data. Additionally, AtPYE interacts with AtPYE homologs, including IAA–Leu Resistant3 (AtILR3, also named as bHLH105) or AtbHLH115 that is involved in metal ion homeostasis to regulate the downstream target genes. In other species, Legay et al. (2012) proposed that in potato, the expression of the FER-like transcription factor that shares 90 % identities with the LeFER gene was also influenced by iron status and a strong positive correlation between the expressions of the FER-like transcription factor and IRT1 was observed. In woody plants, three bHLH genes (MxbHLH01, MxIRO2, and MxFIT) were isolated and characterized in Malus xiaojinesis. The MxbHLH01 expression was restricted to the root and up-regulated under the iron-deficient condition and MxbHLH01 might interact with other proteins to regulate genes in response to iron deficiency (Xu et al. 2011). The MxIRO2 gene (iron-related transcription factor) was induced in the root and leaf of Malus xiaojinesis under iron deficiency. It might form a heterodimer or multimer with other transcription factors to control the expression of genes related to iron absorption (Yin et al. 2013). The MxFIT gene was up-regulated in roots under iron deficiency at both mRNA and protein levels, while almost no expression was detected in leaves irrespective of iron supply. The transgenic Arabidopsis plants with MxFIT had increased AtIRT1 and AtFRO2 transcripts in roots under the iron-deficient condition, showing a stronger resistance to iron deficiency (Yin et al. 2014).
Plant hormones were considered as the upstream signaling molecules that involve in the iron deficiency response by regulating the FIT gene (Brumbarova et al. 2014). The expression of AtIRT1 and AtFRO2 was positively regulated by auxin (Giehl et al. 2012) and ethylene (Lingam et al. 2011). In addition, cytokinin and jasmonic acid showed the suppression of the iron deficiency responses (Seguela et al. 2008; Maurer et al. 2011).
In a previous study (Huang and Dai, personal communication), the PtIRT1 gene from Populus tremula was cloned and its expression was strongly induced by iron deficiency; particularly, the increment of PtIRT1 transcripts was much greater in the iron deficiency tolerant clone (PtG) than in the iron deficiency susceptible clone (PtY) of Populus tremula. However, overexpression of the PtIRT1 gene in other transgenic poplar species did not enhance Fe accumulation compared to the wild type regardless of iron status. It indicated that some transcriptional control mechanisms might be involved in regulating PtIRT1 in iron uptake and transport in poplar. Therefore, we cloned and characterized the bHLH genes from both iron deficiency tolerant and susceptible clones of Populus tremula and overexpressed them in other poplar species. The results would offer a view of how transcription factors regulate genes in response to iron deficiency and further strengthen the understanding of iron deficiency response mechanisms in woody species.
Materials and methods
Discovery of the bHLH candidate genes in response to iron deficiency in Populus
To identify bHLH proteins that may be in response to iron deficiency in Populus, the expression profile of the Arabidopsis bHLH genes in the root of Arabidopsis under iron deficiency at various time points was analyzed based on the published microarray data in NCBI. A total of 167 bHLH genes in Arabidopsis predicted by Carretero-Paulet et al. (2010) were used in this study. Microarray data were downloaded from the Gene Expression Omnibus under the series entry GSE10502 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10502). The expression data of the AtbHLH genes were extracted from GSE10502 using BRB-Array Tools software (Simon et al. 2007). All extracted data were normalized based on the mean expression value of each gene and analyzed and graphed using the mean of Multi-Experiment Viewer software (Saeed et al. 2006). Genes were hierarchically clustered based on ‘Pearson’s correlation’ distance matric and ‘average linkage’ method. Analysis of variance (ANOVA) was performed to identify the AtbHLH genes that expressed at a significantly different level in each treatment at the level of P ≤ 0.01. The output genes were used as queries to BlastP their orthologs in Populus trichocarpa in NCBI. The putative orthologous genes showing high similarity to the AtbHLH genes in response to iron deficiency were considered as the candidates that may also respond to iron deficiency in Populus.
Plant materials and growth conditions
Iron deficiency tolerant (PtG) and susceptible (PtY) clones of Populus tremula ‘Erecta’ were used in this study. Plants were maintained and grown in a hydroponic system comprised a 30-hole PVC plate, black plastic container (42 × 34 × 13 cm), Hoagland’s solution, and an air pump. The PVC plate was floated on the solution that was prepared according to Hoagland and Arnon (1939). Each container contained seven liters of Hoagland’s solution that was aerated with an air pump (TOPFIN Aquarium Air Pump, Model: AIR-8000) and refreshed every week. Each hole on the plate held one plant. The container was then covered by plastic film to maintain the moisture. For acclimation, film was gradually removed from the container after 1 week. For the iron-sufficient treatment, plants were grown in full strength Hoagland’s solution containing 30 µM Fe(II)-ethylenediaminetetraacetic acid (EDTA). For the iron-deficient treatment, Fe(II)-EDTA was removed from the Hoagland’s solution and 200 µM ferrozine was added.
RNA extraction and cDNA preparation
Total RNA was isolated using the QIAGEN RNeasy Plant Mini Kit (QIAGEN Inc, Valencia, CA, USA) according to the manufacturer’s instructions. RNA was isolated from three biological replicates of each treatment. Prior to cDNA synthesis, the RNA was quantified by a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) and agarose gel electrophoresis. A total of 1 µg RNA was treated with gDNA wipeout buffer to eliminate possible contaminating genomic DNA and then subjected to reverse transcription with RT primer mix (oligo-dT and random primers) and unique QIAGEN Omniscript and Sensiscript reverse transcriptases according to the manufacturer’s instructions of the QuantiTect Reverse Transcription Kit (QIAGEN Inc, Valencia, CA, USA).
Gene cloning and sequence analysis
The open reading frame (ORF) region of each candidate gene was cloned from Populus tremula. Primers were designed using the PrimerSelect module of the DNASTAR Lasergene® software package (DNASTAR, Inc., Madison, WI, USA). All primers used for gene cloning and RT-PCR are listed in Table 1. The PCR was performed according to the instructions of the Elongase® Enzyme Mix (Invitrogen™, Carlsbad, CA, USA). Target PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN Inc, Valencia, CA, USA) and then ligated into the pGEM-T easy vector (Promega, Madison, WI, USA). Plasmid DNA was extracted from the white colonies grown on indicator plates containing X-gal and IPTG, using a PerfectPrep™ Spin Mini Kit (5 PRIME Inc., Gaithersburg, MD, USA) and sent for sequencing at the Iowa State University DNA Facility (Ames, IA, USA). The domains of putative proteins were analyzed using SMART (http://smart.embl-heidelberg.de/). A phylogenetic tree was constructed using the predicted amino acid sequences of the bHLH genes from Populus and other species by the MegAlign module of the DNASTAR Lasergene® software package.
Evaluation of the expression profile of the bHLH genes
The expression profile of the bHLH genes in various poplar tissues including the root tip, root, phloem, xylem, mature leaf, young leaf, and shoot tip was analyzed using semi-quantitative RT-PCR. Samples were collected from PtG and PtY grown under the iron-sufficient condition. Primers corresponding to the PtbHLH genes and the Populus actin gene (NCBI accession no: XM_002298674.1) were designed using the PrimerSelect module of the DNASTAR Lasergene® software package and are listed in Table 1. Prior to RT-PCR, the quality of cDNA was assessed by PCR using actin-specific primers designed to span introns to detect genomic DNA contamination. PCR amplification was carried out in a 16 µl reaction that consisted of 5 ng cDNA template, 0.375 µM of each primer, 0.2 mM dNTP, 1.5 mM MgCl2, 1×Green GoTaq® Flexi buffer, and 5 U Taq DNA Polymerase. The amplification conditions were denaturing for 30 s at 94 °C (3 min before the first cycle), annealing for 40 s at 56 °C, and extension for 50 s at 72 °C (5 min after the final cycle) for 30 cycles. PCR products were separated in a 2 % agarose gel at 110 volts (V) for 30 min. The gel was visualized under UV light and images were captured using an AlphaImager® Gel Documentation System (Proteinsimple Inc., Santa Clara, California, USA).
Expression quantification of the bHLH genes responding to iron deficiency
The expression of the bHLH genes in the root of the poplar plant responding to iron deficiency was evaluated by real-time quantitative PCR that was performed on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using a PerfCTa® SYBR Green SuperMix (ROX) kit (Quanta BioSciences, Inc, Gaithersburg, MD, USA). The gene expression was detected in both PtG and PtY plants after they were transferred to the iron-deficient solution for 0, 0.5, 1, 3, and 6 days. Each treatment had three biological replicates with 10 individual plants per replicate. Primers of the PtbHLH genes and internal control (PtTIF5α, NCBI accession no: CV251327.1) used for real-time quantitative PCR are listed in Table 1. The absolute quantity of the target gene was determined based on the standard curves constructed for each gene according to the method of Larionov et al. (2005). Values of expression level were presented as a ratio of the copy number of PtbHLHs to that of PtTIF5α. To evaluate amplification specificity, melting curve analysis was performed at the end of each PCR run according to the manufacturer’s recommendation. Two technical replicates were used for each PCR.
Expression vector construction and plant transformation
The Suppro::PtbHLH expression cassette was constructed using the pCAMBIA S1300 expression vector system (Supplementary Fig. 1) and introduced into P. canescens × P. grandidentata ‘Cl6’ using an Agrobacterium-mediated method according to Dai et al. (2003). Positive transgenic plants were screened on rooting medium (1/2 MS medium with 0.5 µM NAA) supplemented with 5 µM hygromycin. Verified transgenic lines were proliferated in vitro (Dai et al. 2003) and grown in the hydroponic culture system.
Molecular characterization of transgenic events
Transgenic lines were confirmed by PCR with the specific primers located in the flanking regions of the insertions according to the method of Dai et al. (2003) (Table 1). The expression level of PtFIT and PtIRT1 was determined in the root of transgenic plants grown under either iron-sufficient or iron-deficient conditions by real-time quantitative PCR as described above.
Physiological analysis of transgenic plants grown under the iron-deficient condition
The content of chlorophyll and carotenoids, Chl a/b ratio, and the content of Mn, Zn, Fe, and Cu in the control and transgenic plants grown in either iron-sufficient or iron-deficient condition were determined. Leaves were collected at day 9 after the treatment. Pigments were extracted from liquid nitrogen grinded leaf tissues by 80 % acetone and the content of chlorophyll a, chlorophyll b, and carotenoids was determined following the method of Lichtenthaler (1987). The chlorophyll and carotenoid concentration was expressed as mg/kg FW (fresh weight). For determining the content of mineral elements, leaf samples were oven-dried at 65 °C for 2 days and subjected to leaf tests in the North Dakota State University Cereal Science lab.
Statistical analysis
Data were expressed as mean ± SE. An unpaired two-tailed t test was used to analyze significant differences between the treatment and the control.
Results
Discovery of the bHLH candidate genes in response to iron deficiency in Populus
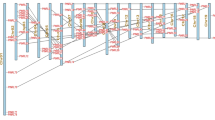
A total of 117 AtbHLH genes had the corresponding probes in the array of GSE10502 containing 22,810 genes based on the microarray gene annotation database. The expression pattern of individual AtbHLH genes is illustrated in Fig. 1. The seven time points of the iron deficiency treatment (0, 3, 6, 12, 24, 48, and 72 h) were clustered into three groups: Group 1 included 0 and 3 h treatment; group 2 included 6, 12, and 24 h treatment; and Group 3 contained 48 and 72 h treatment. A significant difference in gene expression was observed between group 1 and 3, indicating that the bHLH genes might respond to iron deficiency at 3 h after the treatment and such a response could be clearly observed at 48 h after the treatment. According to the ANOVA analysis, 25 AtbHLH genes showed significant differences in the level of expression at seven time points (Fig. 1). Of these genes, nine (At007, At039ORG3, At047, At059UN12, At093, At101, At105ILR3, At115, and At143) were up-regulated and the rest of genes were down-regulated by iron deficiency. Previous researches confirmed that At105ILR3, At101, At115, At039ORG3, and At047 were transcription factors that positively regulate iron deficiency responses in plants (Wang et al. 2007, 2013; Yuan et al. 2008; Long et al. 2010). In other research, the At093 gene was found to be involved in stomatal development (Ohashi-Ito and Bergmann 2006) and At059UN12 may be related to fertilization (Pagnussat et al. 2005). No research was reported regarding the function of At007and At143. Therefore, the orthologs of At105ILR3, At101, At115, At039ORG3, and At047 in Populus trichocarpa were considered as the candidate genes that may regulate responses to iron deficiency in Populus. In addition, At029FRU, At038ORG2, and At100 likely play a role in regulation of iron deficiency (Jakoby et al. 2004; Yuan et al. 2005, 2008; Wang et al. 2007, 2013; Sivitz et al. 2012). However, no corresponding probes of At038ORG2 and At100 in the microarray were identified and the P value of At029FRU was 0.024 (>0.01) according to the ANOVA analysis. All eight orthologs in Populus are listed in Table 2.
Expression profiles of the AtbHLH genes in response to iron deficiency at different time points after the iron deficiency treatment. a Microarray analyses of the 117 AtbHLH genes. Genes and samples are ordered based on a hierarchical clustering analysis; b Expression patterns of 25 AtbHLH genes showing significant differences at different time points
Cloning and sequence analysis of PtFIT and PtIRO
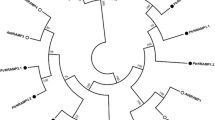
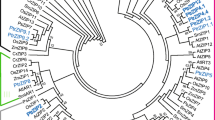
The open reading frame (ORF) regions of two candidate genes were amplified from Populus tremula (PtG and PtY). The PCR primers were designed according to the corresponding nucleotide sequence information of XP_002313541.2 and XP_002323250.2. Nucleotide sequence alignment revealed that either of two genes was identical in both PtG and PtY. Based on their similarity to the bHLH genes, two genes were named PtFIT and PtIRO, respectively (Supplementary Fig. 2). The 939 bp ORF of PtFIT encoded a deduced protein of 313 amino acid residues with a molecular weight of 34.9 kDa and an isoelectric point of 4.74. The 795 bp ORF of PtIRO encoded a deduced protein of 265 amino acid residues with a molecular weight of 30.0 kDa and an isoelectric point of 6.55. Deduced amino acid sequence analysis revealed that PtFIT showed 14.0–57.1 % identity to other bHLH genes (57.15 % to AtbHLH29 and 55.7 % to MxFIT). The PtIRO also showed 14.3–54.7 % identity to other bHLH genes (54.7 % to MxIRO2). Alignment analysis of amino acid sequences showed that PtFIT and PtIRO contained the conserved bHLH domain (Pfam accession: PF00010) (Fig. 2). Phylogenetic analysis suggested that the bHLH proteins could be divided into two subgroups (Fig. 3). PtFIT was clustered with AtbHLH29, LeFER, MxbHLH01, and MxFIT, while PtIRO was clustered with the rest of bHLHs. The distinct structures between these two subgroups might lead to the divergence in their functions. A similar result was reported by Carretero-Paulet et al. (2010) where AtbHLH29 and AtbHLH38/AtbHLH39 belong to two different subfamilies and play different roles.
Amino acid alignment of PtFIT and PtIRO proteins with other bHLH proteins from Arabidopsis thaliana, Malus xiaojinensis, and Lycopersicon esculentum. The bHLH conserved domain is indicated with lines. Shaded areas represent identical residues (black) or similar residues (gray) found in most of the proteins. ‘ ’ indicates the motifs
’ indicates the motifs
A phylogenetic tree of the PtFIT, PtIRO2, and other bHLH genes constructed by ClustalW. The accession numbers of the studied proteins are following the gene name. PtFIT and PtIRO are framed. The corresponding sources are Lycopersicon esculentum (Le), Malus xiaojinensis (Mx), Arabidopsis thaliana (At), Oryza sativa (Os), and Hordeum vulgare (Hv). The scale bar represents 0.2 amino acid substitutions per site
Expression analysis of PtFIT and PtIRO
The expression of PtFIT and PtIRO in the root tip, root, phloem, xylem, mature leaf, young leaf, and shoot tip of both PtG and PtY was determined using semi-quantitative PCR. As shown in Fig. 4, PtFIT was detected only in the root tip and root, while a weak expression of PtIRO was detected in the shoot tip. A real-time quantitative PCR was conducted to evaluate the expression profile of PtFIT in the root responding to iron deficiency at different times after the iron deficiency treatment (Fig. 5). Results showed that a slight decrease of the PtFIT transcript was detected at day 0.5, then gradually increased and peaked at day 6 in PtG. In PtY, the expression of PtFIT showed the first peak at day 1, and then decreased until day 3. The second expression peak of PtFIT in PtY was detected at day 6. The expression level of PtFIT at day 6 was more than 2-fold higher in PtG than in PtY.
Overexpression of the PtFIT gene in P. canescens × P. grandidentata ‘Cl6’
Overexpression of the PtFIT gene was evaluated in five independent transgenic lines of P. canescens × P. grandidentata ‘Cl6’. One line with the empty vector (no PtFIT) was the control (NC). As shown in Fig. 6, the expression level of PtFIT in transgenic lines decreased under the iron-sufficient condition except in TL5 that was similar to NC. Under the iron-deficient condition, the expression of PtFIT significantly increased except in TL13. Two transgenic lines (TL12 and TL5) showed a significantly higher expression level of PtFIT than the control under iron deficiency. The expression pattern of PtIRT1 in PtFIT-transgenic lines was also evaluated. Similar to that of PtFIT, the expression of PtIRT1 was also inhibited under iron sufficiency and dramatically enhanced under iron deficiency (Fig. 6). Compared to NC, PtIRT1 transcripts in transgenic plants were reduced more than half under the iron-sufficient condition, while PtIRT1 transcripts were significantly increased in TL10 compared to NC under iron deficiency. On the other hand, PtIRT1 transcripts were increased 2.44 times in NC and 3.38–13.43 times in transgenic plants under the iron-deficient condition (Supplementary Table 1).
Quantitative assay of PtFIT and PtIRT expression in the control and transgenic poplar plants. Plants were grown under iron sufficiency (Fe+) or deficiency (Fe−). NC the transgenic line with the empty vector (no target gene). The relative expression is quantified by real-time PCR and normalized to the PtTIF5α gene. Values are means of three replicates and standard errors are indicated as a vertical line on the top of each bar
Physiological analysis of the PtFIT-transgenic lines responding to iron deficiency
The contents of chlorophyll and mineral elements in the PtFIT-transgenic leaves were determined. As shown in Figs. 7 and 8, no significant increase of chlorophyll (Chl a + b), Chl a/b ratio, or carotenoids was observed in transgenic lines under the iron-sufficient condition. Under the iron-deficient condition, the chlorophyll content decreased in TL10 and TL13 as well as in NC, but no significant changes in TL12, TL2, and TL5 were detected. The Chl a/b ratio also decreased under iron deficiency in all transgenic plants as well as in NC. TL2 had the highest Chl a/b ratio in all transgenic lines (Fig. 7). Similarly, iron deficiency significantly decreased the content of carotenoids in NC, while no significant decrease in transgenic plants except TL2 and TL10 was found (Fig. 8). The contents of Mn, Zn, Cu, and Fe in leaves of the transgenic plants under the iron-deficient or iron-sufficient condition were determined (Fig. 9). In transgenic plants, iron deficiency showed no effect on Mn content, but slightly lowered the Zn content. Moreover, iron content was significantly decreased and Cu content was significantly increased under iron deficiency. Under the iron-sufficient condition, transgenic lines accumulated more Zn and Fe with the exception that TL5 had a similar Fe content to NC. Under iron deficiency, TL2 and TL5 accumulated more Zn than other lines and NC, while no significant difference in Fe, Mn, or Cu content was found among all transgenic and NC plants.
Chlorophyll content and Chl a/b ratio in the control and PtFIT-transgenic poplar lines under iron-sufficient (Fe+) and iron-deficient (Fe−) conditions. NC the transgenic line with the empty vector (no target gene). Values are means of three replicates and standard errors are indicated as a vertical line on the top of each bar
The carotenoids content in the control and PtFIT-transgenic poplar lines under iron-sufficient (Fe+) and iron-deficient (Fe−) conditions. NC the transgenic line with the empty vector (no target gene). Values are means of three replicates and standard errors are indicated as a vertical line on the top of each bar
The content of Mn, Zn, Fe, and Cu in the leaves of the control and PtFIT-transgenic poplar lines under iron-sufficient (Fe+) and iron-deficient (Fe−) conditions. NC the transgenic line with the empty vector (no target gene). Values are means of three replicates and standard errors are indicated as a vertical line on the top of each bar
Discussion
The basic helix-loop-helix (bHLH) is an important family of transcription factors (Massari and Murre 2000). In this study, the orthologs of the AtbHLH genes responding to iron deficiency in Populus were discovered using microarray data. Microarray is a useful tool to explore the genes responding to changes of the environmental conditions. In Arabidopsis, the microarray method has been used to analyze the genome-wide responses of genes to iron deficiency, such as the differential expression in different tissues (shoots, roots, or leaves) or under different Fe supply conditions (Dinneny et al. 2008; Buckhout et al. 2009; Long et al. 2010; Schuler et al. 2011; Sivitz et al. 2012; Waters et al. 2012). We analyzed the expression profile of the AtbHLH genes in response to iron deficiency in the root using the microarray data submitted by Dinneny et al. (2008). Twenty-five AtbHLH genes showed the responses to iron deficiency (Fig. 1). Among them, three up-regulated genes (At039ORG3, At047, and At101) were also confirmed by other microarray analyses that involved in iron deficiency responses (Sivitz et al. 2012; Waters et al. 2012). Five orthologs of AtbHLH were identified in Populus trichocarpa according to the BlastP result and two out of five (PtFIT and PtIRO) that are homologs of XP_002313541.2 and XP_002323250.2 were cloned from Populus tremula.
Amino acid sequence analyses revealed that the bHLH domain sequences of PtFIT and PtIRO had the highest similarity with AtbHLH29 and MxIRO2, respectively (Supplementary Fig. 2). AtbHLH29 and MxIRO2 functioned as regulators of the genes responding to iron deficiency (Colangelo and Guerinot 2004; Jakoby et al. 2004; Yuan et al. 2005; Yin et al. 2013). Phylogenetic analysis showed that the two iron deficiency regulators were clustered into two groups (Fig. 3). The PtFIT protein contained the typical threonine–glutamate–arginine (T–E–R) motif in the basic region of the bHLH domain at positions 5-9-13, as other members in the same subgroup. Differently, the feature showed in the PtIRO protein was histidine–glutamate–arginine (H–E–R) (Fig. 2). Research showed that H–E–R could bind to the G-box (5′-CACGTG-3′) of a promoter and T–E–R was bound to a variation of the E-box (5′-CANNTG-3′) hexanucleotide sequence, revealing the different roles of the FIT and IRO genes in regulating iron deficiency response in plants (Heim et al. 2003; Li et al. 2006). Information in this study suggests that PtFIT and PtIRO cloned from Populus tremula belong to the bHLH family and may function differently in iron deficiency response in Populus.
The up-regulated expression of PtFIT was observed and more transcripts were detected in roots under iron deficiency (Figs. 5, 6). Similar responses of AtbHLH29 and MxFIT were also reported (Jakoby et al. 2004; Yin et al. 2014). However, very weak expression of PtIRO was detected in the shoot tip of Populus tremula (Fig. 4), which is different from the MxIRO2 gene that expressed in the leaf and root of Malus xiaojinensis under iron sufficiency (Yin et al. 2013). It indicates that PtIRO may have a different function from MxIRO2 and further research is needed to evaluate the possible function of PtIRO related to iron deficiency responses. This study also showed that the expression of PtFIT was higher in PtG than PtY at day 6 after the iron deficiency treatment. Research in Arabidopsis indicated that AtIRT1 is a major downstream gene regulated by AtFIT1 (Colangelo and Guerinot 2004; Jakoby et al. 2004; and Yuan et al. 2005). In this study, the correlation between PtIRT1 and PtFIT responding to iron deficiency was strongly positive (r = 0.94) between PtIRT1 and PtFIT in PtG (Fig. 10), but the correlation in PtY was weak (r = 0.49). This indicates that FIT in PtG functioned well in regulating IRT1 to respond to iron deficiency as reported in Arabidopsis (Colangelo and Guerinot 2004; Jakoby et al. 2004; Yuan et al. 2005); however, in PtY, the regulation function of FIT might be inhibited or needs to be induced by other factors (Yuan et al. 2008) and lack of those factors or connections between those factors may explain such a weak correlation between PtFIT and PtIRT. In addition, the expression profile of these genes in response to iron deficiency and tissue specificity needs to be further compared in both iron deficiency tolerant and susceptible poplar trees.
To further characterize the gene function, PtFIT driven by a constitutive super promoter (a trimer of the octopine synthase transcriptional activating element affixed to the mannopine synthase 2′ transcriptional activating element plus minimal promoter) was transferred into another poplar species (P. canescens × P. grandidentata ‘Cl6’). No enhanced constitutive expression of the PtFIT gene in transgenic lines was detected in this study. The transgenic plants had a relatively lower expression level of PtFIT under the iron-sufficient condition compared to the control, while the expression of PtFIT was induced by iron deficiency in most transgenic plants (Fig. 6). This indicates that there might be other factors regulating the expression of PtFIT and these factors can be activated by iron deficiency. Research showed that FIT also interacts with other elements (interactors) that regulate iron uptake. To date, many AtFIT interactors showed the positive regulation in response to iron deficiency; however, the negative regulation of AtFIT interactors might play an important role in preventing plants from metal toxicity caused by over-accumulation of iron or other metals (Brumbarova et al. 2014). The suppressed expression of PtFIT in transgenic plants under iron sufficiency might indicate the existence of such negative interactors of PtFIT in Populus. Similar interactors may also play roles in regulating the native promoter of the FIT gene; therefore, functional characterization of the gene promoter should be further investigated.
As expected, the expression of PtIRT1 in PtFIT-transgenic plants was also inhibited by iron sufficiency and induced by iron deficiency (Fig. 6). Additionally, the fold change of the PtIRT1 transcript was much greater in transgenic lines than in the control (Supplementary Table 1). Previous research showed a similar result to which AtIRT1 transcripts were not accumulated under the iron-sufficient condition, but increased under the iron-deficient condition in transgenic Arabidopsis plants overexpressing AtFIT1 or MxFIT. This suggests that PtFIT might have the function to induce the expression of PtIRT1 and the similar function was also found in AtFIT1 and MxFIT (Jakoby et al. 2004; Yuan et al. 2008, 2014). We predict that PtFIT could induce the expression of PtIRT1 and might play an important role in regulation of iron transport under iron deficiency. Under iron sufficiency, the PtFIT-transgenic poplar lines accumulated more Fe than the control plant.
In plants, the iron level significantly affects the chlorophyll formation and its structure by regulating the activities of heme enzymes in leaf tissues and the formation of thylakoid membrane in chloroplast. Decreased chlorophyll content was found as the consequence of iron deficiency (Marsh et al. 1963; Terry and Abadia 1986). Notably, one of the PtFIT-trangenic line (TL2) showed an increased chlorophyll (Chl a+b) content and Chl a/b ratio compared to the control under iron deficiency. This result is consistent with the findings in transgenic Arabidopsis in which overexpression of MxFIT increased chlorophyll content (Yuan et al., 2014). Interestingly, the TL2 line showed no enhanced Fe accumulation under iron deficiency, suggesting that TL2 might use iron more efficiently. These findings suggest genetic regulation of PtFIT has the potential to enhance the tolerance to iron deficiency in poplar trees.
In conclusion, two bHLH transcription factors (PtFIT and PtIRO) were cloned from Populus tremula. Functions of PtFIT in regulating iron deficiency were characterized. Results indicated that PtFIT might regulate PtIRT1 that is directly involved in regulation of iron deficiency response in Populus. This study provides useful information for further understanding of the mechanisms of iron deficiency response in poplar trees and other woody species.
Author contribution statement
DH and WD conceived and designed the study. The experiments were conducted by DH. WD supervised the research and guided data interpretation. DH and WD wrote the paper. All authors read and approved the final manuscript.
References
Brumbarova T, Bauer P (2005) Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol 137:1018–1026
Brumbarova T, Bauer P, Ivanov R (2014) Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. doi:10.1016/j.tplants.2014.11.004
Buckhout TJ, Yang TJ, Schmidt W (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genom 10:147
Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153:1398–1412
Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16:3400–3412
Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe 1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346–349
Dai W, Cheng ZM, Sargent W (2003) Plant regeneration and Agrobacterium-mediated transformation of two elite aspen hybrid clones from in vitro leaf tissues. In Vitro Cell Dev Biol Plant 39:6–11
Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320:942–945
Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93:5624–5628
Giehl RF, Lima JE, von Wiren N (2012) Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell 1:33–49
Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104:815–820
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20:735–747
Hoagland DR, Arnon DI (1939) The water culture method for growing plants without soil. Calif Agr Expt Sta Circ 347
Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK (2009) Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem 284:3470–3479
Ivanov R, Brumbarova T, Bauer P (2012) Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol Plant 5(1):27–42
Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (HLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577:528–534
Jeong J, Connolly EL (2009) Iron uptake mechanisms in plants: functions of the FRO family of ferric reductases. Plant Sci 176:709–714
Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation and regulation in higher plants. Ann Rev Plant Biol 63:131–152
Larionov A, Krause A, Miller W (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinform 6:62
Latchman DS (2007) Gene regulation: a eukaryotic perspective. 5th edition. Taylor & Franics Group e-Library, New York
Legay S, Guignard C, Ziebel J, Evers D (2012) Iron uptake and homeostasis related genes in potato cultivated in vitro under iron deficiency and overload. Plant Physiol Biochem 60:180–189
Li XX, Duan XP, Jiang HX, Sun YJ, Tang YP, Yuan Z, Guo JK, Liang WQ, Chen L, Yin JY, Ma H, Wang J, Zhang DB (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141:1167–1184
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic membranes. Meth Enzymol 148:350–382
Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99:13938–13943
Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23:1815–1829
Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22:2219–2236
Marsh HV Jr, Evans HJ, Matrone G (1963) Investigation of the role of iron in chlorophyll metabolism. I. Effect of iron deficiency on chlorophyll and heme content and on the activities of certain enzymes in leaves. Plant Physiol 38:632–638
Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol Cell Biol 20:429–440
Maurer F, Mueller S, Bauer P (2011) Suppression of Fe deficiency gene expression by jasmonate. Plant Physiol Biochem 49:530–536
Maurer F, Naranjo Arcos MA, Bauer P (2014) Responses of a triple mutant defective in three iron deficiency-induced BASIC HELIX-LOOP-HELIX genes of the subgroup Ib(2) to iron deficiency and salicylic acid. PLoS ONE 9(6):e99234
Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T (2006) A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J 46:563–572
Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18:2493–2505
Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132:603–614
Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397:694–697
Römheld V, Marschner H (1986) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80:175–180
Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J (2006) TM4 microarray software suite. Meth Enzymol 411:134–193
Schuler M, Keller A, Backes C, Philippar K, Lenhof H, Bauer P (2011) Transcriptome analysis by GeneTrail revealed regulation of functional categories in response to alterations of iron homeostasis in Arabidopsis thaliana. BMC Plant Biol 11:87
Seguela M, Briat JF, Vert G, Curie C (2008) Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J 55:289–300
Simon R, Lam A, Li M, Ngan M, Menenzes S, Zhao Y (2007) Analysis of gene expression data using BRB-array tools. Cancer Inform 3:11–17
Sivitz AB, Hermand V, Curie C, Vert G (2012) Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7:e44843
Takagi S (1976) Naturally occurring iron-chelating compounds in oat- and rice-root washing. I. Activity measurement and preliminary characterization. Soil Sci Plant Nutr 22:423–433
Terry N, Abadia J (1986) Function of iron in chloroplasts. J Plant Nutr 9:609–646
Vert G, Briat JF, Curie C (2001) Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J 26:181–189
Wang HY, Klatte M, Jakoby M, Baumlein H, Weisshaar B, Bauer P (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226:897–908
Wang L, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and Functional Redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6:503–513
Waters BM, McInturf SA, Stein RJ (2012) Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J Exp Bot 63:5903–5918
Xu HM, Wang Y, Chen F, Zhang XZ, Han ZH (2011) Isolation and characterization of the iron-regulated MxbHLH01 gene in Malus xiaojinensis. Plant Mol Biol Report 29:936–942
Yin L, Wang Y, Yuan M, Zhang X, Pan H, Xu X, Han Z (2013) Molecular cloning, polyclonal antibody preparation, and characterization of a functional iron-related transcription factor IRO2 from Malus xiaojinensis. Plant Physiol Biochem 67:63–70
Yin L, Wang Y, Yuan M, Zhang X, Xu X, Han Z (2014) Characterization of MxFIT, an iron deficiency induced transcriptional factor in Malus xiaojinensis. Plant Physiol Biochem 75:89–95
Yuan Y, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15:613–621
Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18:385–397
Acknowledgments
We thank Drs. Michael Christoffers, Ed Deckard, Xiwen Cai, and Ann Denton for valuable suggestions and comments when we were conducting the research. We also thank Dr. Michael Christoffers for reviewing this manuscript. This research was supported in part by McIntire-Stennis Project ND06212.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Li.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, D., Dai, W. Molecular characterization of the basic helix-loop-helix (bHLH) genes that are differentially expressed and induced by iron deficiency in Populus . Plant Cell Rep 34, 1211–1224 (2015). https://doi.org/10.1007/s00299-015-1779-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1779-8