Abstract
Key message
Activation of SA-dependent signaling pathway and suppression of JA-dependent signaling pathway seem to play key roles in B. thuringiensis -induced resistance to R. solanacearum in tomato plants.
Abstract
Bacillus thuringiensis, a well-known and effective bio-insecticide, has attracted considerable attention as a potential biological control agent for the suppression of plant diseases. Treatment of tomato roots with a filter-sterilized cell-free filtrate (CF) of B. thuringiensis systemically suppresses bacterial wilt caused by Ralstonia solanacearum through systemic activation of the plant defense system. Comparative analysis of the expression of the Pathogenesis-Related 1(P6) gene, a marker for induced resistance to pathogens, in various tissues of tomato plants treated with CF on their roots suggested that the B. thuringiensis-induced defense system was activated in the leaf, stem, and main root tissues, but not in the lateral root tissue. At the same time, the growth of R. solanacearum was significantly suppressed in the CF-treated main roots but not in the CF-treated lateral roots. This distinct activation of the defense reaction and suppression of R. solanacearum were reflected by the differences in the transcriptional profiles of the main and lateral tissues in response to the CF. In CF-treated main roots, but not CF-treated lateral roots, the expression of several salicylic acid (SA)-responsive defense-related genes was specifically induced, whereas jasmonic acid (JA)-related gene expression was either down-regulated or not induced in response to the CF. On the other hand, genes encoding ethylene (ET)-related proteins were induced equally in both the main and lateral root tissues. Taken together, the co-activation of SA-dependent signaling pathway with ET-dependent signaling pathway and suppression of JA-dependent signaling pathway may play key roles in B. thuringiensis-induced resistance to R. solanacearum in tomato.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacillus thuringiensis is an effective bio-insecticide, which is highly toxic to insects but not to mammals, and is not harmful to the environment (Roh et al. 2007; Schnepf et al. 1998). Recently, B. thuringiensis has attracted considerable attention as a biological control agent against plant diseases (Zhou et al. 2008). B. thuringiensis generally produces several compounds including b-exotoxins, antibiotics, degrading enzymes, bacteriocins, and a signal molecule in the bacterial quorum-sensing system (Zhou et al. 2008). Such antimicrobial substances produced by B. thuringiensis are thought to be major components of the disease-suppressive activity of B. thuringiensis against plant pathogens (Cherif et al. 2003, 2008; Dong et al. 2002; Raddadi et al. 2009; Reyes-Ramirez et al. 2004).
However, there is recent evidence that supports the ability of B. thuringiensis to activate the plant defense system (Hyakumachi et al. 2013). Treatment of tomato roots with the extracellular compounds which were secreted by B. thuringiensis into the filter-sterilized cell-free filtrate (CF) results in suppressing the growth of Ralstonia solanacearum and development of wilt symptoms in tomato plants. In tomato plants treated with CF on their roots, the expression of salicylic acid (SA)-responsive Pathogenesis-Related 1(P6) [PR-1(P6)], a typical marker gene for activation of the plant defense system, is systemically induced. Therefore, the disease suppressive activity of the CF seems to be conferred by activation of the plant defense system, rather than by the activity of antibacterial substances.
Systemic acquired resistance (SAR) and induced systemic resistance (ISR) are two forms of induced resistance against a broad range of pathogens in plants. It has been well demonstrated that SA-dependent signaling pathway is involved in SAR through induction of SA-responsive acidic PR gene expression, whereas jasmonic acid (JA) and ethylene (ET)-dependent signaling pathways are done to ISR accompanying up-regulation of JA and ET-responsive basic PR gene expression (Glazebrook 2001; Pieterse et al. 1998). Colonization of some root-colonizing rhizobacteria (Pseudomonas spp. and Bacillus spp.) in plant roots triggers a systemic plant resistance response known as ISR (Pieterse et al. 2002; van Loon 2007). ISR elicited by Pseudomonas spp. has been well characterized at the molecular level (Haas and Défago 2005; Pieterse et al. 2002; van Loon 2007). In root-colonizing Bacillus spp., various strains of species B. amyloliquefaciens, B. subtilis, B. pasteurii, B. cereus, B. pumilus, B. mycoides, and B. sphaericus induce ISR and exhibit significant reduction in the incidence or severity of various diseases on diverse hosts (Choudhary and Johri 2009; Kloepper et al. 2004; Vallad and Goodman 2004; van Loon 2007). Thus, plants seem to have the ability to acquire enhanced level of resistance to pathogens after exposure to extracellular compounds or biotic stimuli provided by some rhizobacteria (De Vleesschauwer and Hofte 2009).
Bacillus thuringiensis seems to be present in an extremely large range of environments. It can be isolated from both bulk and rhizosphere soil, insects, and stored-product dust (Bernhard et al. 1997; Kaelin et al. 1994; Martin and Travers 1989; Meadows et al. 1992) and is also distributed inside various plant tissues in low abundance (Smith and Couche 1991). B. thuringiensis belongs to the spore-forming B. cereus group, which includes the mammalian pathogens B. cereus and B. anthracis (Rasko et al. 2005). B. thuringiensis can be distinguished from the other species by its ability to produce insecticidal crystal proteins during sporulation. Therefore, B. thuringiensis should probably be differentially classified from plant root-colonizing Bacillus spp. Hence, in contrast to ISR elicited by plant root-colonizing Bacillus spp., signaling pathways regulating the defense system induced by B. thuringiensis remain to be investigated.
Ralstonia solanacearum causes bacterial wilt disease resulting in severe losses of solanaceous crops. Because R. solanacearum exists in a viable but non-culturable condition (VBNC) and survives in the soil for many years, chemicals and crop rotation have a limited effect on the control of this disease (Hayward 1991). The most effective management method is to use bacterial wilt-resistant cultivars onto which susceptible but high-quality cultivars are grafted (Lee et al. 1998). Recently, transcriptome analysis of the bacterial wilt-resistant cultivar inoculated with R. solanacearum revealed that bacteria seem to be localized in the primary xylem tissues due to coordinated activation of several defense-related genes in the xylem and pith tissues surrounding xylem vessels (Ishihara et al. 2012; Nakaho 1997; Nakaho et al. 2004). Some beneficial microorganisms also have potential to control bacterial wilt disease in tomato. The colonization of Pseudomonas fluorescence FPT9601-T5, a commercial plant-promoting rhizobacteria (PGPR) in Japan, suppresses bacterial wilt disease (Aino et al. 1997). Transcript profiles of the leaves of FPT9601-T5-colonized Arabidopsis thaliana indicate that FPT9601-T5 activates plant responses in a similar manner to other known PGPR and rhizobia (Wang et al. 2005). Treatment of tomato roots with non-pathogenic Pythium oligandrum (PO), which is commercially available as Polygardron (Polyversum) in the Slovak Republic (Brozova 2002; Butt and Copping 2000), is also effective for controlling bacterial wilt in tomato through PO-induced resistance to R. solanacearum (Hase et al. 2006, 2008; Takahashi and Takenaka 2011). The systemic activation of the JA- and ET-signaling pathways in this model was demonstrated by global gene expression analysis of PO-treated tomato roots (Takahashi et al. 2006). Hence, transcriptome analysis of tomato plants showing resistance to R. solanacearum improved our understanding of the molecular events underlying plant immune system activated by B. thuringiensis. In the present study, to gain further insight into the possible molecular mechanism underlying B. thuringiensis-induced disease resistance in tomato, the global gene expression profile and growth of R. solanacearum in the CF-treated tomato roots were analyzed.
Materials and methods
Growth conditions of plants and bacteria
Solanum lycopersicum cv. ‘Oogata-fukuju’ plants were grown in 9-cm pots containing quartz sand at 24 °C in a growth chamber under continuous fluorescent light (70 μmol/m2/s) and fertilized with 1,000-fold-diluted Hyponex solution (Hyponex Japan) at 3-day intervals (Takahashi et al. 2005). Bacillus thuringiensis serovar fukuokaensis B88-82 was cultured in Nutrient Broth (NB) medium (Nissui, Tokyo, Japan) without NaCl at 25 °C for 2 days. Ralstonia solanacearum isolate 8242R (race 1, biovar 4), which was used for challenge inoculation, was incubated at 30 °C in liquid CPG medium containing 0.001 % tetrazolium chloride and 50 μg/ml rifampicin for 48 h on a rotary shaker (Hendrick and Sequeira 1984).
Treatment with filter-sterilized cell-free filtrate (CF) of B. thuringiensis
Cultures of B. thuringiensis adjusted to a final density of 1.8 × 108 cfu/ml were briefly centrifuged at 7,000 rpm for 10 min at 25 °C to pellet the bacterial cells. The supernatant was filtered through a nitrocellulose membrane (0.22 μm pore size, Merck Millipore, MA, USA) to produce the CF. The roots of two-week-old tomato plants were carefully removed so as to minimize injury to the root tissue. After rinsing the roots with distilled water (DW) three times, the roots were dipped into 200 ml of CF. The CF-treated plants were incubated in a growth chamber at 25 °C for 48 h under 14 h light (70 μmol/m2/s):10 h dark conditions. The roots of control tomato plants were treated with diluted NB liquid medium and grown under the same conditions. For analyzing the response of tomato leaves directly treated with the CF, the leaves of two-week-old plants were sprayed with 10 ml of the CF or diluted NB liquid medium and kept under the same growth conditions for 48 h. After those treatments, total RNA was isolated from main and lateral roots and stem tissues for analysis of gene expression by northern hybridization, quantitative RT-PCR with a set of defense gene primers (Online Resource Table S1) or microarray method.
To analyze the time course of defense gene expression in response to CF, after treatment with CF or diluted NB liquid medium as a control for 48 h, the treated plant roots were washed with DW three times, placed into 50-ml Falcon tubes containing DW and then further incubated in a growth chamber until 10 days after CF treatment. Total RNA was isolated from main roots at 0, 2, 4, 8, and 10 days after CF treatment.
For assessment of bacterial wilt disease, the plants which were treated with CF or diluted medium at their roots for 48 h were used for inoculation with R. solanacearum as described below.
Inoculation with R. solanacearum
After treatment of tomato roots with the CF of B. thuringiensis for 48 h, or diluted NB medium as the control, the roots were rinsed with DW three times. The plants were cut so that each retained 1 cm of root, and they were then transferred to 50 ml Falcon tubes containing a bacterial suspension of 1 × 107 cfu/ml of R. solanacearum. The inoculated plants were further incubated at 30 °C in a growth chamber under 14 h light (70 μmol/m2/s):10 h dark conditions. At 12 h after inoculation, the inoculated plants were washed with DW three times and then transferred to 50 ml Falcon tubes containing DW. Plants were grown at the same growth chamber and inspected daily for wilting symptoms. Disease severity, based on foliar symptoms of wilting, was monitored daily for 7 days after inoculation with the pathogen.
To measure the growth of bacteria in root and stem tissues of CF-treated tomato and control, after further washing the plants with DW three times, tissue segments of main roots, lateral roots and stems were excised from five inoculated plants, respectively, at 5 or 7 days after inoculation. After measurement of fresh weight of each tissue segment sample, the segments were ground with DW that is tenfold weight of each segment sample. The original homogenate and tenfold serial dilutions of the homogenate were spread onto three plates of CPG agar medium containing 50 μg/ml rifampicin. The colonies were counted after 48 h of incubation at 30 °C. Measurement of bacterial growth was repeated three successive trials, and the representative was shown. Student’s t test was used to determine significant difference in bacterial growth between CF treatment and control (α = 0.05).
Detection of defense-related gene expression by northern hybridization
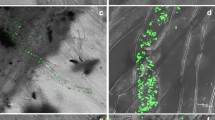
A representative tomato plant used for RNA isolation is shown in Fig. 1a, b. Total RNA was isolated from the leaf tissue (L1), two stem sections (S1: 10 mm in length from near the root base, S2: 10 mm in length located 50–60 mm above the root base), a lateral root section (R1: a 10-mm section including root tip), and a main root section (R2: 10 mm in length from under the soil surface) of three tomato plants treated with the CF, or three control plants, using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Transcripts of tomato PR-1(P6) were detected by northern hybridization as described previously (Hyakumachi et al. 2013).
Differential expression of the Pathogenesis-Related 1 [PR-1(P6)] gene in leaf, stem, and root tissues of tomato plants (Solanum lycopersicum cv. ‘Oogata-fukuju’) treated with filter-sterilized cell-free filtrate (CF) of B. thuringiensis on their leaves or roots. a A tomato plant used for RNA extraction. For northern hybridization analysis, total RNA was isolated from leaf tissue (L1), two stem sections [one 10 mm in length from near the root base (S1) and another 10 mm in length located 50–60 mm above the root base (S2)], and lateral root [10 mm section including root tip (R1)]. The broken line shows an enlargement of b. A scale bar shows the length in 2 cm. b Main root used for RNA isolation for northern hybridization analysis. Total RNA was isolated from main root section [10 mm in length under the soil surface (R2)]. The scale bar shows the length in 0.5 cm. c Transcripts of tomato PR-1(P6) in the leaf (L1), stem, (S1 and S2), and root (R1 and R2) tissues of tomato plants treated with the CF of B. thuringiensis on their leaves or roots were detected by northern hybridization (leaf and root). Tomato plants treated with distilled NB medium on their roots were used as controls (Cont). rRNA was used as an internal control for loading of RNA samples
Microarray analysis of tomato root tissues
To investigate changes in the gene expression pattern of CF-treated main and lateral roots and diluted medium-treated controls, global gene expression analysis using a Tomato Gene Expression Microarray 4 × 44K (Agilent Technologies, Santa Clara, CA, USA) composed of 43,000 non-redundant genes was conducted. Total RNA was isolated from two sets of the main and lateral root samples using an RNeasy Plant Mini Kit (Qiagen). Each sample was combined together the root tissues of three independent tomato plants treated with CF or diluted NB medium as a control. Isolated total RNA was amplified and labeled using a Low Input Quick Amp Labeling (LIQAL) Kit (Agilent Technologies). Cyanine 3 (Cy3)-labeled cRNA was purified using RNeasy Mini Spin Column (Qiagen). After fragmentation of 1,650 ng of Cy3-labeled cRNA, the cRNA was applied to the Tomato Gene Expression Microarray and incubated at 65 °C for 17 h. After the hybridization, the array was washed with Agilent Washing Buffer 1 at 25 °C for 1 min and then with Agilent Washing Buffer 2 at 37 °C for 1 min. The probe array was scanned at 5 μm resolution using an Agilent Technologies Microarray Scanner. One-color images of the scanned array were extracted using Agilent Feature Extraction (FE) Software 10.7.3.1. Raw intensity values from each chip were normalized to the 75th percentile of the measurements using GeneSpring GX 9.0 Software. Relative changes in gene expression levels were compared between the samples treated with CF and diluted medium control. Genes up-regulated greater than five-fold or down-regulated less than 1/5-fold relative to controls in either duplicate samples are listed in Supplemental Table S2 and S3. Then, main-root-specific up-regulated genes induced more than two-fold by CF treatment relative to CF-treated lateral roots and main-root-specific down-regulated genes suppressed less than 1/2-fold by CF treatment relative to CF-treated lateral roots were shown by a heat map in Fig. 4. Annotations for each gene product were obtained from the RefSeq, Unigene, TIGR Plant Transcript Assemblies, and TIGR Gene Indices databases. The results of this analysis were depicted as the heat map using MultiExperiment Viewer (MeV) version 4.8.1 software (Saeed et al. 2003). The data discussed in this publication were deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/).
Measurement of defense-related gene expression levels by quantitative PCR
One microgram total RNA was reverse transcribed into cDNA with random hexamer primers using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara-Bio, Shiga, Japan) according to the instruction manual. Gene-specific primers for quantitative RT-PCR are listed in Supplemental Table S1. Quantitative RT-PCR amplification was performed in triplicate 20 μl reactions containing template cDNA (2 μl), 0.4 μM gene-specific primer set, 1× ROX Reference Dye and 1× SYBR Premix Ex Taq II (Tli RNase H Plus) (Takara-Bio). The reaction was run on a 7300 Fast Real-Time PCR System (Applied Biosystems, Foster, CA, USA) with the cycling program: 30 s at 95.0 °C, followed by 40 cycles of 95.0 °C for 5 s and 60.0 °C for 31 s. Fluorescence was measured at the end of each cycle. As a control, a reaction lacking template cDNA was carried out. The absence of nonspecific products and primer dimers was confirmed by analysis of melting curves of RT-PCR products. The number of cycles at the threshold level (C t) for each gene was converted to values relative to those of the tomato actin gene used as an internal standard for normalization. Data are shown as fold change in defense gene expression in CF-treated plants relative to diluted NB medium-treated control plants. Each experiment of defense gene expression was repeated three times. Data were subjected to analysis of variance between CF treatment and control were compared by Student’s t test (α = 0.01).
Results and discussion
Induced systemic resistance in tomato plants treated with filter-sterilized cell-free filtrate (CF) of B. thuringiensis on their roots
When the main and lateral root tissues of tomato plants shown in Fig. 1a, b were treated with CF of B. thuringiensis, the expression of PR-1(P6) encoding antimicrobial protein was induced in main root tissue, but not in lateral root tissue (Fig. 1c). The induction of PR-1(P6) was also observed in leaf and stem tissues of the tomato plants treated with the CF on their roots. On the other hand, as shown in Fig. 1c, in tomato plants whose leaves were directly treated with the CF, PR-1(P6) expression was locally induced in only leaf tissues, but not other tissues. These results suggested that treatment of roots with CF efficiently activates defense gene expression in leaf, stem, and main root tissues, but not in the lateral root tissue, whereas treatment of leaves with CF does not activate it systemically. Leaves and roots are developmentally distinct, but integrated as a structural element, and have an integral role in plant defense. Recently, the evidence that root-derived plant toxins, e.g., nicotine, furocoumarins, and terpenoid aldehydes are implicated in the leaf defense against pathogens, was presented (Erb et al. 2009). Thus, perception of CF in tomato roots may effectively trigger to transduce systemic defense signals to activate defense system on aboveground part of tomato plants thereby inducing PR-1(P6) in stems and leaves. Furthermore, main and lateral roots reportedly respond to plant hormones, nutrient composition, and environmental stresses in a differential manner (Ivanchenko et al. 2008; Jain et al. 2007; Malamy 2005; Tien et al. 1979). Therefore, this differential induction of PR-1(P6) expression between the main and lateral root tissues suggests that CF-treated main and lateral roots of tomato plants may be differentially responsive to infection with soil-borne pathogens.
To assess the susceptibility of main and lateral root tissues of CF-treated tomato plants to inoculation with R. solanacearum, the number of bacterial cells was compared among stem, main, and lateral root tissues of tomato plants treated with CF on their roots following inoculation with R. solanacearum. At 5 days after inoculation, wilt symptoms developed in control plants, but not in CF-treated plants (Fig. 2a), and the growth of bacteria was significantly suppressed in stem, main root, and lateral root tissues of CF-treated plants (Fig. 2c). Bacterial wilt symptoms did not appear on CF-treated plants even at 7 days after inoculation (Fig. 2b). However, bacteria in CF-treated lateral root tissue grew to similar extent as in lateral root tissue of control plants at 7 days after inoculation, whereas the suppression of bacterial growth continued in both stem and main root tissues of CF-treated plants (Fig. 2d). In the assay for antimicrobial activity against R. solanacearum 8242R, CF of B. thuringiensis did not directly suppress their growth on the medium (Online Resource Fig. S1), suggesting there is less possibility that antimicrobial activity of CF is a major factor for suppressing the growth of R. solanacearum. Thus, CF treatment seems to activate the plant defense system in stem and main root tissues, thereby suppressing the growth of R. solanacearum in those tissues. On the other hand, in the lateral root tissue, CF treatment apparently cannot induce the defense system, or only partially induces it, and allows R. solanacearum to grow.
Bacterial growth in root and stem tissues of tomato plants and wilt symptom development. Tomato plants were treated with the CF (CF) or diluted medium control (Cont) on their roots. a, b Development of wilt symptoms in CF-treated tomato plants and their control at 5 or 7 days after inoculation with R. solanacearum (5 and 7 dpi). The scale bar shows the length in 1 cm. c, d Growth of bacterial cells in stem (S1 in Fig. 1a), main root, and lateral root tissues of CF-treated tomato plants and their control at 5 or 7 days after inoculation with R. solanacearum. The mean and standard deviation (SD) values of bacterial cell counts were calculated in each experiment. An asterisk (*) indicates a statistically significant difference of bacterial growth between CF treatment and control (Student’s t test, α = 0.05, n = 5)
In bacterial wilt-resistant tomato cultivars, R. solanacearum is localized in the primary xylem tissues by the coordinated activation of several defense-related, hormone signaling, and lignin biosynthesis genes in the xylem and pith tissues surrounding xylem vessels (Ishihara et al. 2012). Thus, the restriction of bacterial growth to root tissues through activation of the defense system seems to effectively control bacterial wilt diseases caused by R. solanacearum in tomato. Transcriptome analysis of CF-induced resistance in tomato may be more broadly applicable to address a common molecular mechanism shared by B. thuringiensis-treated tomato and bacterial wilt-resistant tomato cultivars for resistance to R. solanacearum.
This differential activation between main and lateral root tissues of the plant defense system in response to the CF of B. thuringiensis is intriguing. The CF seems to contain the elicitor molecule(s) having the activity to induce defense reaction in tomato as shown in Figs. 1 and 2. In addition, the CF may contain various factors produced by B. thuringiensis, excluding the elicitor molecule(s) inducing defense reactions in tomato. Such compounds may alter plant gene expression which is not directly associated with the activation of the plant defense reaction by the CF. To better understand the molecular events of plant defense reactions in response to the CF, it is necessary to eliminate such unexpected gene expression induced by the concomitant compounds in the CF. Comparative analysis of global gene expression between CF-treated main root tissue resistant to R. solanacearum and CF-treated lateral root tissue susceptible to R. solanacearum is an appropriate strategy to focus on the molecular aspects of resistance specifically induced by the CF of B. thuringiensis.
Specific changes in gene expression in CF-treated main root tissue
All data of microarray analysis of global gene expression of CF-treated main and lateral root tissues compared with that of the corresponding tissues treated with diluted medium as a control have been deposited in NCBIs Gene Expression Omnibus and are accessible through GEO Series accession number GSE50402. 166 genes in CF-treated main root tissue and 661 genes in CF-treated lateral root tissue were up-regulated more than five-fold (Fig. 3a). The expression of 82 genes among these up-regulated genes was coordinately regulated in both CF-treated main and lateral root tissues. On the other hand, 84 genes of the 166 up-regulated genes were uniquely induced in CF-treated main root tissue and might be directly associated with the mechanism of resistance to R. solanacearum (Fig. 3a).
Venn diagram of up- and down-regulated genes in CF-treated tomato roots. Numbers of genes up-regulated more than five-fold (a) and down-regulated less than 1/5-fold (b) at main and lateral root tissues treated with the CF, respectively, were shown. Up-regulated (n = 166) and down-regulated (n = 224) genes in the main root are presented with black circles. Up-regulated (n = 661) and down-regulated (n = 453) genes in the lateral root are indicated by gray circles. Main-root-specific up- or down-regulated genes, lateral-root-specific up- or down-regulated genes and coordinately up- or down-regulated genes in the main and lateral roots are indicated in circles
Compared with diluted medium-treated controls, 224 genes in CF-treated main root tissue and 453 genes in CF-treated lateral root tissue were down-regulated less than 1/5-fold (Fig. 3b). Among these down-regulated genes, 98 genes were coordinately regulated in both CF-treated main and lateral root tissues, and 126 of 224 down-regulated genes were uniquely suppressed in CF-treated main root tissue and could also be directly associated with resistance to R. solanacearum (Fig. 3b).
In CF-treated lateral root tissues, total 661 and 453 genes were up- and down-regulated, respectively, whereas total 166 and 224 genes were done in CF-treated main root tissues (Fig. 3). Increased number of up-and down-regulated genes in CF-treated lateral roots relative to CF-treated main roots indicates that lateral roots seems to be more sensitive to CF than main roots, which may be concerned with differential responsiveness of main and lateral roots to CF.
To further characterize the main-root-specific gene expression profile during CF-induced resistance, the expression levels of 84 and 126 genes, which were specifically up- or down-regulated in CF-treated main roots relative to the diluted medium-treated main roots as a control, were compared with those of the corresponding genes in CF-treated lateral roots. As shown in Fig. 4 and Online Resource Table S2, the expression of 33 of 84 genes up-regulated in CF-treated main roots were induced more than two-fold relative to those in CF-treated lateral roots. These 33 genes included genes encoding pathogenesis-related (PR) proteins PR-2, PR-1b1(p14), PR-1(P6), P4, PR-4, PR-P69E, PR-P69G, and β-1,3-glucanase which have antimicrobial activity (Fig. 4). Specific induction of P4 and PR-2 in CF-treated main roots was also confirmed by analysis of the relative level of their transcripts with quantitative RT-PCR (Fig. 5). Because these genes are SA-inducible (Danhash et al. 1993; van Kan et al. 1992), SA-mediated signaling pathways appear to be activated in CF-treated main roots. Interestingly, the expression of an IAA-responsive SAUR gene and IAA11 which encodes transcriptional repressors of auxin-regulated genes (Audran-Delalande et al. 2012; Chapman and Estelle 2009; Reed 2001), was also induced in CF-treated main roots (Fig. 4). Since recent evidence suggests that auxin signaling and SA signaling are negatively cross-talked during plant defense (Kazan and Manners 2009), up-regulation of the expression of genes encoding transcriptional repressors of auxin-regulated genes in CF-treated main roots seems to be consistent with the mutually antagonistic manner underlying auxin and SA signalings.
Main-root-specific genes up- or down-regulated by CF treatment. Main-root-specific up-regulated genes (n = 33) that were induced more than two-fold by CF treatment relative to CF-treated lateral roots are shown in the upper portion of the heat map. Main-root-specific down-regulated genes (n = 32) suppressed less than 1/2-fold by CF treatment relative to CF-treated lateral roots are shown in the lower portion of the heat map. Gene expression fold ratio is shown as a matrix with rows representing genes and columns representing the two sets of the main and lateral root tissues (Main 1, Main 2, Lateral 1, and Lateral 2) on the heat map [up-regulated (red), down-regulated (green), and no change (black)]. Corresponding gene products are listed (color figure online)
Quantitative analysis of CF-induced or suppressed gene transcripts by quantitative RT-PCR. Relative amounts of P4, PR-2, PI-II, and CEVI57 transcripts in CF-treated main or lateral root tissues were measured by quantitative RT-PCR. The amount of each transcript is shown relative to the transcript amount of the tomato actin gene. The mean and SD values of transcript levels were calculated in each experiment. An asterisk (*) indicates a statistically significant difference of gene expression between CF treatment and control (Student’s t test, α = 0.01, n = 3)
Of the 126 genes whose expression was down-regulated in CF-treated main roots, there were 32 genes whose expression was suppressed less than 1/2-fold relative to the CF-treated lateral roots (Fig. 4 and Online Resource Table S3). These 32 genes included genes encoding proteinase inhibitors II (PI-II) and CEVI57 (PI-CEVI57) (Fig. 4 and Online Resource Table S3). Quantitative analysis of the levels of PI-II and PI-CEVI57 transcripts suggested that their expression was suppressed in CF-treated main roots, but not in CF-treated lateral roots (Fig. 5). The expression of PI genes in tomato is responsive to jasmonic acid (JA) (Gadea et al. 1996); thus the JA signaling pathway seems to be suppressed in main root tissue in response to CF treatment.
Coordinate changes in gene expression in CF-treated main and lateral root tissues
Annotation of 82 genes up-regulated in both main and lateral roots of CF-treated plants suggests that the expression of genes encoding ethylene (ET)-responsive transcription factors and genes related to ethylene synthesis were coordinately induced (Online Resource Table S2). This result indicates that the ET-mediated signaling pathway seems to be activated in main and lateral root tissues by treatment with the CF. Up-regulation of genes encoding auxin-responsive transcription factors, glutathione S-transferases (GSTs), and mitochondrial small heat shock protein was also observed in CF-treated main and lateral root tissues (Online Resource Table S2). Induction of auxin-responsive transcription factors may result from the cross-talk with activation of SA signaling pathway in response to CF. GSTs have been well documented to be involved in diverse aspects of biotic and abiotic stresses, especially detoxification processes (Dixon et al. 2002). Accumulation of small heat shock proteins is induced by oxidative stress and adaptive response (Banzet et al. 1998). Thus, up-regulation of those gene expressions seems to be associated with CF-activated defense reaction.
On the other hand, among 98 genes coordinately down-regulated in CF-treated main and lateral root tissues, the expression of genes encoding enzymes involved in JA biosynthesis, and a gene encoding 2-oxoglutarate (2OG)-Fe(II) oxygenase-like protein decreased reproducibly in both main and lateral root tissues of CF-treated plants (Online Resource Table S3). The decreased level of transcripts related to JA biosynthesis appears to be linked with specific down-regulation of JA-responsive genes in CF-treated main roots. 2OG-Fe(II) oxygenases have also been identified as catalyzing steps in the biosynthesis of plant signaling molecules including ethylene and the gibberellins (Prescott and Lloyd 2000; van Damme et al. 2008). Therefore, down-regulation of those gene expressions may be involved in CF-induced resistance to pathogens.
Time course of P4 and PR-2 gene expression in CF-treated main and lateral root tissues
The expression of various defense genes leads to the production of defensive compounds, such as pathogenesis-related (PR) proteins and enzymes involved in the biosynthesis of protective secondary metabolites. In addition to PR-1(P6), tomato P4 encoding tobacco PR-1a-like protein having antimicrobial activity and PR-2 encoding β-1,3-glucanase which can inhibit pathogen growth are also known to be marker genes for enhanced resistance (van Kan et al. 1992). As shown in Fig. 4 and Online Resource Table S2, the expression of P4 and PR-2 in main roots is clearly up-regulated at 2 days after CF treatment. The level of P4 and PR-2 was monitored at 0, 2, 4, 6, 8, and 10 days after treatment with CF. Induction of PR-2 expression continued until 10 days after CF treatment (Fig. 6a). The expression ratio of P4 also increased at 2, 4, and 6 days after CF treatment (Fig. 6b). These results suggest that the change in gene expression observed at 2 days after CF treatment as shown in Figs. 1c and 4 continues at the point in time when the suppression of bacterial wilt disease was observed, as shown in Fig. 2.
Time course of P4 and PR-2 expression in CF-treated main root by quantitative RT-PCR. a Relative amounts of P4 and PR-2 transcripts in CF-treated main root (CF) and diluted NB medium-treated control (C) were measured at 0, 2, 4, 8, and 10 days after CF treatment by quantitative RT-PCR. The amount of each transcript is shown relative to the amount of tomato actin gene transcript used as an internal standard for normalization. The mean and SD values of transcript level were calculated in each experiment. An asterisk (*) indicates a statistically significant difference of gene expression between CF treatment and control (Student’s t test, α = 0.01, n = 3). b Change fold of P4 and PR-2 gene expression in CF-treated main roots (CF) relative to diluted NB medium-treated control (C) at 2, 4, 8, and 10 days after CF treatment was shown. The mean and SD values of transcript level were calculated in each experiment
Signaling pathways conferring CF-induced resistance to R. solanacearum
In tomato plants challenge-inoculated with R. solanacearum after pretreatment with CF on their roots, the growth of R. solanacearum in the stem and main root tissues was suppressed and the induction of defense-related gene expression in CF-treated roots was observed. The gene expression profile specific to the CF-treated main root tissue indicates that the SA-dependent signaling pathway is activated and that the JA-dependent signaling pathway seems to be suppressed. SA, JA, and ET are involved to different extents in defense responses against a broad range of pathogens (Durrant and Dong 2004; Glazebrook 2001; Pieterse et al. 2002). SA signaling is mainly associated with systemic acquired resistance (SAR) that is induced in plant cultivars carrying particular resistance (R) genes upon infection with necrotizing pathogens carrying the corresponding avirulence (Avr) genes (Durrant and Dong 2004). On the other hand, the JA/ET-dependent signaling pathways primarily mediate ISR elicited by plant growth-promoting rhizobacteria (PGPR) (Pieterse et al. 1998; Thomma et al. 1998; Ton et al. 2002) although recent evidence also indicates partial involvement of SA-dependent signaling during ISR in some cases (Niu et al. 2011). In general, the SA-dependent signaling pathway interacts antagonistically with the JA-dependent signaling pathway (Glazebrook 2001, 2005; Niki et al. 1998; Takahashi et al. 2004) although basal resistance against certain pathogen is controlled by the combined actions of SA, JA, and ET-dependent signaling pathways (Xu et al. 1994). The activation of SA signaling and suppression of JA signaling in CF-treated main roots are consistent with antagonistic interactions between these signaling pathways observed in several other examples of induced resistance.
With regard to the ET-dependent signaling pathway, the expression of ET-related genes is clearly induced in CF-treated main and lateral root tissues (Online Resource Table S2). The activation of the ET-dependent signaling pathway seems insufficient to induce resistance to R. solanacearum, as the bacterial growth was not significantly suppressed in CF-treated lateral roots (Fig. 2). However, the activation of ET signaling by CF treatment in combination with the activation of SA signaling may induce effective resistance to R. solanacearum. Taken together, the co-activation of SA-dependent signaling and ET-dependent signaling, with suppression of JA-dependent signaling, may play key roles for B. thuringiensis-induced resistance to R. solanacearum in tomato plants.
The B. thuringiensis is a well-known insecticide. At the same time, it can be useful as a microbial biocontrol agent to suppress plant diseases. Thus, B. thuringiensis has potential as a bifunctional biopesticide to control a broad range of insects and pathogens for plant protection. The transcriptome analysis of tomato roots in response to the CF of B. thuringiensis presented here provides clues to the molecular events induced by the CF of B. thuringiensis. To further understand the mechanism of B. thuringiensis-induced resistance to R. solanacearum, it will be important to identify the specific substances present in the CF that can induce resistance to R. solanacearum in tomato. Interestingly, volatile compounds and lipopeptides produced by Bacillus spp. have been identified as elicitors in ISR (Ryu et al. 2004). Therefore, the characterization of the substances able to induce disease resistance will provide new insights for further evaluation of the practicality of B. thuringiensis as an effective biocontrol agent.
Abbreviations
- CF:
-
Filter-sterilized cell-free filtrate
- ET:
-
Ethylene
- JA:
-
Jasmonic acid
- SA:
-
Salicylic acid
References
Aino M, Maekawa Y, Mayama S, Kato H (1997) Biocontrol of bacterial wilt of tomato by producing seedlings colonized with endophytic antagonistic pseudomonas. In: Ogoshi A, Kobayashi K, Homma Y, Kodama F, Kondo N, Akino S (eds) Proceedings of the 4th international workshop on PGPR. Nakanishi Printing, Sapporo, pp 120–123
Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53:659–672
Banzet N, Richaud C, Deveaux Y, Kazmaier M, Jean Gagnon J, Triantaphylide C (1998) Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J 13:519–527
Bernhard K, Jarrett P, Meadows M, Butt J, Ellis DJ, Roberts GM, Pauli S, Rodgers P, Burges HD (1997) Natural isolates of Bacillus thuringiensis: worldwide distribution, characterization, and activity against insect pests. J Invertebr Pathol 70:59–68
Brozova J (2002) Exploitation of the mycoparasitic fungus Pythium oligandrum in plant protection. Plant Prot Sci 38:29–35
Butt TM, Copping LG (2000) Fungal biological control agents. Pestic Outlook 11:186–191
Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43:265–285
Cherif A, Chehimi S, Limem F, Hansen BM, Hendriksen NB, Daffonchio D, Boudabous A (2003) Purification and characterization of the novel bacteriocin entomocine 9, and safety evaluation of its producer, Bacillus thuringiensis subsp. entomocidus HD9. J Appl Microbiol 95:990–1000
Cherif A, Rezgui W, Raddadi N, Daffonchio D, Boudabous A (2008) Characterization and partial purification of entomocin 110, a newly identified bacteriocin from Bacillus thuringiensis subsp. entomocidus HD110. Microbiol Res 163:684–692
Choudhary DK, Johri BN (2009) Interactions of Bacillus spp. and plants—with special reference to induced systemic resistance (ISR). Microbiol Res 164:493–513
Danhash N, Wagemakers CA, van Kan JA, de Wit PJ (1993) Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Mol Biol 22:1017–1029
De Vleesschauwer D, Hofte M (2009) Rhizobacteria-induced systemic resistance. Adv Bot Res 51:224–266
Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol 3:reviews3004.1–reviews3004.10
Dong YH, Gusti AR, Zhang Q, Xu JL, Zhang LH (2002) Identification of quorum-quenching N-acylhomoserine lactonases from Bacillus species. Appl Environ Microbiol 68:1754–1759
Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42:185–209
Erb M, Lenk C, Degenhardt J, Turlings TCJ (2009) The underestimated role of roots in defense against leaf attackers. Trends Plant Sci 14:653–659
Gadea J, Mayda ME, Conejero V, Vera P (1996) Characterization of defense-related genes ectopically expressed in viroid-infected tomato plants. Mol Plant Microbe Interact 9:409–415
Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr Opin Plant Biol 4:301–308
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319
Hase S, Shimizu A, Nakaho K, Takenaka S, Takahashi H (2006) Induction of transient ethylene and reduction in severity of tomato bacterial wilt by Pythium oligandrum. Plant Pathol 55:537–543
Hase S, Takahashi S, Takenaka S, Nakaho K, Arie T, Seo S, Ohashi Y, Takahashi H (2008) Involvement of jasmonic acid signalling in bacterial wilt disease resistance induced by biocontrol agent Pythium oligandrum in tomato. Plant Pathol 57:870–876
Hayward AC (1991) Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol 29:65–87
Hendrick C, Sequeira L (1984) Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl Environ Microbiol 48:94–101
Hyakumachi M, Nishimura M, Arakawa T, Asano S, Yoshida S, Tsushima S, Takahashi H (2013) Bacillus thuringiensis suppresses bacterial wilt disease caused by Ralstonia solanacearum with systemic induction of defense-related gene expression in tomato. Microbes Environ 28:128–134
Ishihara T, Mitsuhara I, Takahashi H, Nakaho K (2012) Transcriptome analysis in resistant and susceptible cultivars of tomato infected with Ralstonia solanacearum. PLoS ONE 7:e46763
Ivanchenko MG, Muday GK, Dubrovsky JG (2008) Ethylene–auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J 55:335–347
Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG (2007) Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 144:232–247
Kaelin P, Morel P, Gadani F (1994) Isolation of Bacillus thuringiensis from stored tobacco and Lasioderma serricorne (F.). Appl Environ Microbiol 60:19–25
Kazan K, Manners JM (2009) Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci 14:373–382
Kloepper JW, Ryu CM, Zhang S (2004) Induce systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Lee JM, Bang HJ, Ham HS (1998) Grafting of vegetables. J Japan Soc Hortic Sci 67:1098–1104
Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28:67–77
Martin PAW, Travers RS (1989) Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl Environ Microbiol 55:2437–2442
Meadows MP, Ellis DJ, Butt J, Jarrett P, Burges HD (1992) Distribution, frequency, and diversity of Bacillus thuringiensis in an animal feed mill. Appl Environ Microbiol 58:1344–1350
Nakaho K (1997) Distribution and multiplication of Ralstonia solanacearum in stem-inoculated tomato rootstock cultivar LS-89 resistant to bacterial wilt. Ann Phytopathol Soc Jpn 63:341–344
Nakaho K, Inoue H, Takayama T, Miyagawa H (2004) Distribution and multiplication of Ralstonia solanacearum in tomato plants with resistance derived from different origins. J Gen Plant Pathol 70:115–119
Niki T, Mitsuhara I, Seo S, Ohtsubo N, Ohashi Y (1998) Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol 39:500–507
Niu D-D, Liu H-X, Jiang C-H, Wang Y-P, Wang Q-Y, Jin H-L, Guo J-H (2011) The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol Plant Microbe Interact 24:533–542
Pieterse CMJ, van Wees SCM, Ton J, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580
Pieterse CMJ, van Wees SCM, Ton J, van Pelt JA, van Loon LC (2002) Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol 4:535–544
Prescott AG, Lloyd MD (2000) The iron(II) and 2-oxoacid-dependent dioxygenases and their role in metabolism. Nat Prod Rep 17:367–383
Raddadi N, Belaouis A, Tamagnini I, Hansen BM, Hendriksen NB, Boudabous A, Cherif A, Daffonchio D (2009) Characterization of polyvalent and safe Bacillus thuringiensis strains with potential use for biocontrol. J Basic Microbiol 49:293–303
Rasko DA, Altherr MR, Han CS, Ravel J (2005) Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev 29:303–329
Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6:420–425
Reyes-Ramirez A, Escudero-Abarca BI, Aguilar-Uscanga G, Hayward-Jones PM, Eleazar-Barbozacorona J (2004) Antifungal activity of Bacillus thuringiensis chitinase and its potential for the biocontrol of phytopathogenic fungi in soybean seeds. Food Microbiol Saf 69:M131–M134
Roh JY, Choi J, Li MS, Jin BR, Je YH (2007) Bacillus thuringiensis as a specific, safe, and effective tool for insert pest control. J Microbiol Biotechnol 17:547–559
Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Schnepf E, Crickmore N, van Rie Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH (1998) Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806
Smith RA, Couche GA (1991) The phylloplane as a source of Bacillus thuringiensis variants. Appl Environ Microbiol 57:311–315
Takahashi H, Takenaka S (2011) Defense system induced by elicitin-like proteins of biocontrol agent Pythium oligandrum. In: Walpert T, Shiraishi T, Akimitsu K, Glazebrook J (eds) Genome-enabled integration of research in plant-pathogen systems. APS Press, St Paul, pp 39–46
Takahashi H, Kanayama Y, Zheng MS, Kusano T, Hase S, Ikegami M, Shah J (2004) Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to Cucumber mosaic virus. Plant Cell Physiol 45:803–809
Takahashi H, Shimizu A, Arie T, Rosmalawati S, Fukushima S, Kikuchi M, Hikichi Y, Kanda A, Ohnishi K, Ichinose Y, Yasuda C, Kodama M, Egusa M, Masuta C, Sawada H, Shibata D, Hori K, Watanabe Y (2005) Catalog of Micro-Tom tomato responses to common fungal, bacterial and viral pathogens. J Gen Plant Pathol 71:8–22
Takahashi H, Ishihara T, Hase S, Chiba A, Nakaho K, Arie T, Teraoka T, Iwata M, Tugane T, Shibata D, Takenaka S (2006) Beta-cyanoalanine synthase as a molecular marker for induced resistance by fungal glycoprotein elicitor and commercial plant activators. Phytopathology 96:908–916
Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95:15107–15111
Tien TM, Gaskins MH, Hubbell DH (1979) Plant growth substances produced by Azospirillum brasilense and their effect on the growth of pearl millet (Pennisetum americanum L.). Appl Environ Microbiol 37:1016–1024
Ton J, van Pelt JA, van Loon LC, Pieterse CMJ (2002) Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol Plant Microbe Interact 15:27–34
Vallad GE, Goodman RM (2004) Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci 44:1920–1934
van Damme M, Huibers RP, Elberse J, van den Ackerveken G (2008) Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J 54:785–793
van Kan JAL, Joosten MHAJ, Wagemakers CAM, van den Berg-Velthuis GCM, De Wit PJGM (1992) Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol Biol 20:513–527
van Loon LC (2007) Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol 119:243–254
Wang Y, Ohara Y, Nakayashiki H, Tosa Y, Mayama S (2005) Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol Plant Microbe Interact 18:385–396
Xu Y, Chang PLC, Liu D, Narasimhan ML, Kashchandra GR, Hasegawa PM, Bressan RA (1994) Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6:1077–1085
Zhou Y, Choi YL, Sun M, Yu Z (2008) Novel roles of Bacillus thuringiensis to control plant diseases. Appl Microbiol Biotechnol 80:563–572
Acknowledgments
This research was supported in part by a Grant-in-Aid for development of mitigation and adaptation techniques to global warming in the sectors of agriculture, forestry, and fisheries, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Toriyama.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 3 (XLSX 46 kb)
Genes which were up-regulated in either two sets of the main (Main 1 and Main 2) or lateral root (Lateral 1 and Lateral 2) more than five-fold relative to control, are listed. Change fold ratio of up-regulated genes in four root samples (Main 1, Main 2, Lateral 1 and Lateral 2) are shown respectively. Accession number and description of each gene, which were assigned by public databases (Ref, Unigene, TIGR Plant Transcript Assemblies and TIGR Gene Indices databases), are presented. Deduced function of each gene product is described at right column. The function that should be unknown is left blank. Main-root-specific up-regulated genes listed in Fig. 4 are shown in gray color line. Ethylene-related genes (yellow color line), Glutathione S-transferase (blue color line), Heat shock protein (green color line), and auxin-responsive genes (pick color line), which were coordinately up-regulated in the main and lateral roots, are highlighted.
299_2013_1515_MOESM4_ESM.xlsx
Supplementary material 4 (XLSX 52 kb)
Genes which were down-regulated in either two sets of the main (Main 1 and Main 2) or lateral root (Lateral 1 and Lateral 2) less than 1/5-fold relative to control, are listed. Change fold ratio of down-regulated genes in four root samples (Main 1, Main 2, Lateral 1, and Lateral 2) are shown, respectively. Accession number and description of each gene, which were assigned by public databases, and deduced function of each gene product are described. The function that should be unknown is left blank. Main-root-specific down-regulated genes listed in Fig. 4 are shown by gray line. Genes for JA biosynthesis (purple line) and oxidoreductase, 2OG-Fe(II) oxygenase (light blue purple line), which were coordinately down-regulated in main and lateral roots, are highlighted.
Rights and permissions
About this article
Cite this article
Takahashi, H., Nakaho, K., Ishihara, T. et al. Transcriptional profile of tomato roots exhibiting Bacillus thuringiensis-induced resistance to Ralstonia solanacearum . Plant Cell Rep 33, 99–110 (2014). https://doi.org/10.1007/s00299-013-1515-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1515-1