Abstract
Chalcone isomerase (CHI, EC 5.5.1.6) is one of the key enzymes in the flavonoid biosynthesis pathway catalyzing the stereospecific isomerization of chalcones into their corresponding (2S)-flavanones. In this investigation, both the cDNA sequence and the genomic sequence encoding the chalcone isomerase from Ginkgo biloba L. (designated as GbCHI) were isolated from the leaves. The GbCHI gene contained two introns and three extrons and encoded a peptide of 244 amino acids with a predicted molecular mass of 26.29 kDa and a pI of 7.76. RQPCR showed that GbCHI was expressed in a tissue-specific manner in G. biloba. Expression of GbCHI was also up-regulated by UV-B irradiation or treatment with 5-aminolevulinic acid or three plant growth regulator—ethylene, abscisic acid, and chlormequat—and these effects were consistent with analysis of the GbCHI promoter region. The recombinant protein was successfully expressed in an E.coli strain with the pET-28a vector. In vitro enzyme activity, assayed by HPLC, indicated that recombinant GbCHI protein could catalyze the formation of naringenin from 6′-hydroxychalcone. RQPCR analysis showed that CHI activity correlated with changes in transcription level of the CHI gene, GbCHI activity was also positively correlated with total flavonoid levels in ginkgo leaves, suggesting CHI as a key gene regulating flavonoid accumulation in ginkgo leaves.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flavonoids are ubiquitous plant secondary products that have diverse functions in plant physiology and ecology (Mol et al. 1998; Winkel-Shirley 2002), including protection against UV-B radiation and pathogen attack, attraction of pollinating insects, regulation of male fertility, cell cycle regulation, and auxin transport. Flavonoids also have many pharmaceutical uses in human health, with pharmacological properties that have been especially valuable in the treatment of early-stage Alzheimer’s disease, vascular dementia, and many other diseases (Diamond et al. 2000; Oken et al. 1998; Smith and Luo 2004). Flavonoids are synthesized by the well-studied phenylpropanoid pathway (Koes et al. 2005) and most of the enzymes and genes involved in flavonoid biosynthesis have been characterized (Li et al. 2006).
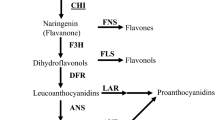
Chalcone isomerase (also called chalcone flavanone isomerase) is one of the key enzymes in the flavonoid biosynthesis pathway and catalyzes conversion of chalcones to (2S)-naringenin. Although this step can occur spontaneously, CHI catalyzes it 107-fold more efficiently (Bednar and Hadcock 1988). CHIs can be classified into two types of isozymes: one is an elicitor-inducible form that acts on both 6′-hydroxychalcone and 6′-deoxychalcone and presumably is involved in the legume-specific 5-deoxyflavonoid pathway. The second form acts on only 6′-hydroxychalcone and is generally found in non-legumes (Kimura et al. 2001; Shimada et al. 2003). At present, CHI genes have been cloned from several plants: Callistephus chinensis, Medicago sativa, Petunia hybrida, Oryza sativa, Hordeum vulgare, and Saussurea medusa (Druka et al. 2003; Kuhn et al. 1978; Li et al. 2006; McKhann and Hirsch 1994; Van Tunen et al. 1988). One previous report has suggested that chalcone isomerase may be a regulatory enzyme in the biosynthesis of flavonoids (Mehdy and Lamb 1987). In Glycyrrhiza uralensis, the enhanced accumulation of flavonoids was correlated with the elevated levels of CHI transcripts and CHI activity (Zhang et al. 2009). Overexpression of petunia CHI gene in tomato results in an increase of up to 78-fold in fruit peel flavonols, mainly due to an accumulation of rutin (Muir et al. 2001). Therefore, efforts to study the induction and developmental or tissue-specific regulation of flavonoids biosynthetic activity have been focused mainly on the mechanisms controlling CHI gene expression.
Ginkgo biloba is often called “a living fossil” of the plant kingdom, and has been well studied owing to the many active ingredients, such as flavonoids, contained in its leaves. At present, about 38 different flavonoids, especially, multiform glycosides of flavonols, have been isolated from G. biloba (van Beek 2002; van Beek and Montoro 2009). However, the concentrations of these economically important compounds are often insufficient to justify commercial exploitation of ginkgo leaves. In recent years, a number of investigations have focused on the biological mechanism of flavonoid formation, the in vitro chemical synthesis of flavonoids, and the use of tissue culture for producing flavonoids, among others aspects (Cheng et al. 2004b; Xu 2008; Xu et al. 2008a, b). Few studies have yet focused on synergistic effects of genetic modification and the production of flavonoids in Ginkgo biloba leaves. In the present study, a gene, designated as GbCHI, and shown previously to encode a putative CHI enzyme, was isolated from the leaves of Ginkgo biloba as a useful tool for studying the regulation of flavonoid biosynthesis. Based on the catalytic function of CHI at the branch point of (2S)-naringenin metabolism, the expression profile, and the correlation between GbCHI expression and flavonoid content in Ginkgo biloba, we suggest that GbCHI might be a specific key gene in the regulation of flavonoid accumulation in ginkgo leaves, as well as being a crucial in vivo factor in responses to UV stress, hormones, or plant growth regulator.
Materials and methods
Plant materials and treatments
Twelve-year old of grafted G. biloba seedlings were growing in a greenhouse in Jingzhou (E, 111°54′–112°19′, N, 30°6′–30°39′, Hubei province, central of China) were sampled as plant materials. For gene cloning and tissue expression, diverse tissues including young leaves, mature leaves, ovules, stamens, albumen, gynoecia, stems, and roots were collected for DNA and RNA extraction as described by Xu et al.(2008a). Tissues were immediately frozen in liquid nitrogen and kept at −80°C prior to total RNA extraction.
One year old cuttings from the same genotypic strain of G. biloba were subjected to treatments with UV-B, chlormequat chloride (CCC), 5-aminolevulinic acid (ALA), abscisic acid (ABA), gibberellin (GA), and ethephon (ETH). For UV-B treatment, seedlings were exposed to 1500 μJ/m2 UV-B irradiation in a closed chamber, and the control cuttings were placed in a dark closed chamber. The CCC (10 mM), ALA (100 μM), ABA (10 mM), GA (15 mM), and ETH (20 mM) were dissolved in 0.01% Tween 20 and sprayed onto young leaves. The control leaves were sprayed with an equivalent volume of 0.01% (v/v) Tween 20.
Cloning of the full-length cDNA and genomic DNA of GbCHI
Given species conservation of the functional domain of the CHI protein, a pair of degenerate primers, CHIDF and CHIDR, was designed according to the conserved peptide sequences, LGGAGYR and PGSSILFT, to amplify a conservative fragment of the GbCHI gene. The purified PCR product was cloned into the pMD18-T vector (TaKaRa, Dalian China), and the positive clone was confirmed by a BLASTN query of the NCBI Database to verify the homology of this cloned fragment with CHI sequences from other species.
Based on the sequence of cloned GbCHI fragments, the specific primer pairs (CHI5R and CHI3R) and the nested primer pairs (CHI5N and CHI3N) were designed to amplify the 5′ and 3′ end of GbCHI using the SMARTTM RACE cDNA Amplification Kit (Clontech, USA). The PCR products were purified and cloned into the pMD18-T vector for sequencing. After comparing and aligning the sequence of 5′RACE, 3′RACE, and the internal fragment, the full-length cDNA sequence of GbCHI was obtained. Two gene-specific primers, CHIGF and CHIGR, designed based on the cDNA sequence, were used to amplify the genomic sequence of GbCHI.
Amplification of the 5′-promoter region of GbCHI
The promoter region was isolated by genomic walk strategy using a Universal GenomewalkerTM kit (Clontech, USA) according to the manufacturer’s instructions. The first turn of PCR was carried out using AP1 and CHI5R as primers, and constructed libraries as a template using the following protocol: 94°C for 4 min; 30cycles (98°C for 10 s, 68°C for 3 min) and 68°C for 10 min. The products were 20-fold-diluted and used for nested PCR, which was performed using AP2 and CHI5N as primers under the following conditions: 94°C for 4 min; 32 cycles (94°C for 30 s, 56°C for 50 s, and 72°C for 3 min); and 72°C for 10 min. The PCR products were cloned into pMD18-T for sequencing (Sangon, China).
Bioinformatics analysis and molecular evolution analyses
The obtained sequences were analyzed using bioinformatics tools at websites (http://www.ncbi.nlm.nih.gov and http://www.xpasy.org). Vector NTI Suite 10 was used for sequence alignment and analysis. The cis acting element in the promoter region was predicted using PlantCARE (Lescot et al. 2002). Phylogenetic tree analysis of GbCHI and known CHIs from other plant species retrieved from GenBank were aligned with Mega 4.0 (Tamura et al. 2007). The phylogenetic tree was constructed by a neighbor-joining (NJ) method and measured by bootstrap analysis with 1000 replicates. SPSS 16 was used for statistical analysis and graphing.
Southern blot analysis
Genomic DNA (20 μg/sample) was digested overnight at 37°C with Sma I, Sac I, and Hind III. The digested DNA was fractionated by 0.9% agarose gel electrophoresis and transferred onto a PVDF membrane (Roche Applied Science, Germany). At total of 50 ng purified GbCHI was used as a template in a total volume of 20 μl for probe labeling. Probe labeling (DIG), hybridization, and signal detection were performed following the manufacturer’s instruction for the DIG High Primer DNA Labeling and Detection Starter Kit II (Roche Applied Science, Germany).
Relative quantification by RQPCR
The transcription levels of GCHI were determined in different G. biloba tissues, as well as in young seedling leaf samples collected at different time points after stress and hormone treatments. RQPCR was carried out using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, American) with SYBR Green PCR Master Mix (Applied Biosystems, American) according to the manufacturer’s protocol. The G. biloba glyceraldehydes-3-phosphate dehydrogenase gene (GbGAPDH, L26924) (Jansson et al. 1994), was used as the reference gene as described by Xu (2008b).
The gene-specific primers (CHIF, CHIR) and reference primers (GAPF, GAPR) for RQPCR were listed in Table 1. The RQPCR conditions were: 10 min at 95°C, and 40 cycles (95°C for 15 s, 60°C for 1 min). Before performing RQPCR, primer efficiency was evaluated using both GbCHI and GbGAPDH at 100, 150, 200, 250, and 300 nM combinations. A 150 nM concentration was chosen as most suitable combination for both genes. For each plant sample, aliquots of 150 ng total RNA was analyzed for each gene and the two genes (GbCHI and GbGAPDH) were always analyzed simultaneously. Each sample was amplified three times and all reactions were performed on ABI PRISM 7500 Sequence Detection System. With a housekeeping gene GbGAPDH, the relative amount of the GbCHI transcript is presented as 2(−dCt) according to the CT method (dCt = Ctsample − Ctcontrol) described in the RQPCR Application Guide (Applied Biosystems). When comparing the expression of GbCHI in different tissues, the relative expression of GbCHI was achieved by calibrating its transcription level to that of the reference gene, GbGAPDH.
Expression of CHI recombinant proteins in Escherichia coli
To clone GbCHI into an expression vector, a pair of primers, CHIZF and CHIZR, were designed and synthesized to amplify the coding region by RT-PCR with the incorporation of a restriction enzyme site and a protective base to simplify later vector construction. After confirmation by sequencing, the resulting recombinant plasmid was introduced into BL21 (DE3) by the heat shock method. A single colony of E.coli BL21 cells harboring the expression plasmid pET28a-GbCHI was inoculated at 37°C in Luria–Bertani medium containing kanamycin (50 mg l−1) and were grown with shaking (150 rpm) at 37°C until the optical density (OD600) reached about 0.6. For induction, IPTG was added at a final concentration of 1 mM and the cells were further cultured at 30°C for 2 h. The cells were lysed by sonication for 10 s at 4°C, and centrifuged at 7000×g for 15 min. Supernatants and pellets were analyzed by SDS-PAGE and Coomassie Brilliant Blue R250 staining. The recombinant GbCHI protein from induced cells was purified using Nickel-CL agarose affinity chromatography (Bangalore Genei) and used for in vitro enzyme assays.
Western blotting was carried out to verify expression of a GbCHI protein having a His-tag in the N-terminus. After electrophoresis, the proteins were electrotransferred onto a PVDF membrane and detected with antimouse RGS-His antibody (Santa Cruz, American), and a secondary antibody (goat anti-mouse IgG), conjugated to alkaline phosphatase (AP). Western Blue Stabilized Color Substrate for AP (Promega, USA) was used for the color reaction.
Assay of GbCHI activity
For assay of GbCHI activity, 6′-hydroxychalcone or 6′-deoxychalcone (10 μg each in 10 μl of ethanol) was incubated at 30°C for 5 min with 0.49 ml of 50 mM potassium phosphate (pH 7.5) containing 10 μl of crude enzyme (total 0.5 ml). 6′-hydroxychalcone or 6′-deoxychalcone was purchased from Advanced Technology and Industrial Corporation (Hong Kong, China). The reaction mixture was then extracted with ethyl acetate and the products were analyzed by HPLC on a Shim-pack CLC-O DS column (6.0 × 150 mm; Shimadzu, Kyoto, Japan). The chromatography conditions were a mobile phase of 40% (v/v) methanol and 4% (v/v) acetic acid in water at a flow rate of 0.8 ml min−1 at 40°C, and the eluate was monitored at 304 nm. To analyze the stereochemistry of CHI reaction products, 6′-hydroxychalcone and a crude extract from E. coli expressing CHI isozymes were incubated at 30°C for 3 h. An extract of E. coli transformed by the vector without insert was used for the control under the same reaction conditions. The reaction product was developed on a silica-gel thin-layer chromatography plate (Merck, Darmstadt, Germany) with the solvent toluene:ethyl acetate:methanol:light petroleum (6:3:1:3 [v/v]), and the naringenin spot (R F = 0.44) was recovered. The sample was then analyzed by HPLC on a Chiracel OD-RH column (4.6 × 150 mm; Daicel, Tokyo) with 35% (v/v) aqueous acetonitrile at a flow rate of 0.6 ml min−1 at 30°C, and the eluate was monitored at 290 nm.
CHI activity in G. biloba leaves was measured by the methods of Jez et al. (2000) and Fouché and Dubery (1994), with minor modifications. The standard CHI assay reaction mixture contained the following components in a final volume of 1 ml: 50 mM Tris–HCl buffer (pH 7.6) containing 1% ethanol, 100 μM narigenin chalcone in 2-ethoxy-ethanol (18.5 μM), and an appropriate amount of protein extract. The rate of spontaneous cyclization of the substrate was determined beforehand and was subtracted from the rate in the presence of enzyme. Naringenin chalcone was synthesized from naringenin according to the procedure of Moustafa and Wong (1967) and authentic naringenin was purchased from Herbfine (Nanchang, China).
Extraction and determination of flavonoid content
Flavonoids were extracted identified and quantified as previously described by Xu et al. (2008a) with some modifications. Briefly, 100 mg of dried leaf powder was dissolved in 40 ml of acidified methanol. The samples solutions were then suitably diluted with methanol and filtered through 0.45 μm filter membranes (Millipore, Nylon) for high performance liquid chromatography analysis. Selected samples were analyzed by HPLC (model LC-10, Shimadzu, Kyoto, Japan) with a SPD-10AVP UV–Vis detector. Flavonoids were analyzed on a Shim-pack VP-ODS column (250 ml × 4.6 mm), using a mobile phase of methanol: 0.4% (v/v) phosphoric acid (60:40) for elution. The flow rate was 1.0 ml min−1 and injection volume was 20 μl. The UV detector was set at λ = 370 nm. Three extraction samples were prepared for HPLC analysis and each sample was injected three times. A standard mixture of quercetin, kaempferol, and isorhamnetin was selected because the many different flavonol glycosides that occur in ginkgo leaves are predominantly derivatives of these three flavonol aglycones (van Beek 2002). Based on the methods used to determine flavonoids concentrations described by van Beek (2002), flavonoid contents were calculated by multiplication of the total content of quercetin, kaempferol, and isorhamnetin by a factor 2.51 and were expressed as percentages.
Results
Characterization of the full-length cDNA, genomic DNA, and promoter region of the GbCHI gene
A full-length cDNA of the CHI gene was obtained from G. biloba leaf tissue by degenerate PCR and RACE methods. The cDNA sequence was 926 bp with a poly A tail, and contained a 732 bp open reading frame(ORF) encoding a 244 amino acid protein. A 20 bp 5′ untranslated region was upstream of the start codon, and the coding region was followed by a 160 bp 3′ untranslated region downstream from the stop codon (Fig. 1). A phylogenetic tree generated by the neighbor-joining method based on the putative amino acid sequences of CHIs showed that all members of the CHI family could be sorted into three monophyletic group. The G. biloba CHI was included in a polyphyletic group with other CHIs from various non-leguminous plants (Fig. 2).
The full-length GbCHI sequence, promoter region sequence, intron sequence and deduced amino acid sequence of GbCHI gene. The exons are indicated in capital letter and the introns are indicated in italic letter. The start condon (ATG), the stop (TAG), and putative polyadenylation signals are underlined. TBFs and cis-elements of the GbCHI promoter region are shown in black box
A phylogenetic tree based on deduced amino acid sequences of various CHIs. Amino acid sequences were analyzed using the Mega 4.0 program. The numbers at each node represented the bootstrap value, with 1000 replicates. Monocotyledons and most other species are classified into the type I CHI group. Known typeII CHIs form a subgroup, and legumes CHIs belong to this group. GbCHI does not belong to any group. The CHI-like protein of Pasteurella multocida is introduced as an external frame of reference. The relative evolutionary time is displayed under the tree. GenBank accession number are as follows: Hordeum vulgare, AF474923; Oryza sativa, AF474922; Zea mays, Q08704; Vitis vinifera, P51117; Citrus sinensis, BAA36552; Gossypium hirsutum, ABM64798; Camellia sinensis, AAZ17563; Saussurea medusa, AF509335; Pyrus communis, ABQ08639; Lotus japonicus chi2, CAD69022; Ipomoea purpurea, ABW69677; Lotus japonicus chi4, BAC53984; Medicago sativa, P28012; Phaseolus vulgaris, CAA78763; Glycine max, ABA86740; Glycyrrhiza uralensis, ABM66533; Pasteurella multocida, NP245761
The genomic GbCHI gene was isolated from a pair of specific primers derived from the start and stop condon regions of the cDNA. The full-length genomic DNA of GbCHI gene was 1290 bp long and had 100% identity with the coding region of the full-length cDNA sequence. Exon 1 (351 bp), exon 2 (225 bp), and exon 3 (156 bp) were separated by intron 1 (97 bp) and intron 2 (59 bp). The putative splicing site also obeyed the GU/AG rule (Fig. 1). By genome walking, we isolated a 1011 bp fragment up stream of the ATG codon which corresponded to the putative promoter region. The 5′ upstream region of GbCHI possessed the typically high A + T content (65.6%), commonly found in other plant promoters. Computational analysis revealed six potential TATA box sequences within the upstream region of GbCHI, at positions −140, −206, −224, −346, −639, and −963. Several conserved eukaryotic promoter elements, including a CAAT box, were also observed at positions −58, −143, −299, −424, −570, −956, and −968. A number of phenylpropanoid regulation sequences were also noted. Two putative E-boxes were identified: the first one located at positions −950 to −956, and one near the transcription initiation site at position −418 to −424 (Fig. 1). Two MYB-binding sites (EEC) with a consensus sequence GANTTNC were identified on the GbCHI 5′-upstream sequence (Solano et al. 1995; Yoshioka et al. 2004). Sequence analysis of the 5′-upstream region of CHI with PLACE revealed two pollen-expression-related sequence motifs (POLRE) (Bate and Twell 1998) at positions −739 and −859. Two putative SEBF-like elements with the central YTGTCWC core (Boyle and Brisson 2001; Yamagata et al. 2002) were present in the promoter region. Four GATA box elements with the core sequence GATA were found at positions −23, −157, −181, and −911 these showed a tissue specific expression-related sequence. Several LHC-related cis-elements (CAANNNNATC) (Piechulla et al. 1998) known to play important roles in the tomato circadian rhythms, were also identified in the 5′ upstream region of GbCHI (Fig. 1).
A core motif (GARE), described as a dehydration and GA-response expression element (Sutoh and Yamauchi 2003), was identified at position −237 to −230. The GbCHI contained a conserved 5′UTR sequence ACGTGCGC similar to the well known ABRE or G-Box elements found in ABA responsive genes such as the wheat Em (Guiltinan et al. 1990), rice rab16 (Mundy et al. 1990), and barley Hva1 genes (Straub et al. 1994). A DPBF binding core element (ACACNNG) was also found at position −554, which functions as an ABA-responsive cis-element in Arabidopsis (Lopez-Molina and Chua 2000). Four GT-1 motifs (GRWAAW) were observed at positions −157, −181, −699, and −769 and one SA-IN transcription factor binding site (GGAAAA) was identified at position −699. Both of these are involved in induction of genes by salicylic acid (SA) (Park et al. 2004; Zhou 1999), an important component of the signal transduction cascades that activate plants defense responses against pathogen attack. Four GATA boxes with tissue specific expression-related sequences and an ethylene-responsive element (ERE) motif (Itzhaki et al. 1994; Rawat et al. 2005) were found at positions −23, −157, −181, −911, and −710.
To examine if the GbCHI gene belongs to a multigene family, aliquots of 20 μg genomic DNA were digested with Sma I, Hind III, and Sac I, which did not cut within the coding region. These fragments were then hybridized with the coding sequence of GbCHI under high stringency conditions. In total, 5–7 specific hybridization bands ranging from 0.5 to 10 kb were recovered in each of the restriction enzyme-digested lanes (Supplementary Fig. 1S), this indicated that GbCHI belonged to a multigene family.
The expression of the GbCHI gene and accumulation of flavonoids in different tissues
The transcription level of GbCHI and accumulation of flavonoids varies according to tissues, growth conditions, and tissue developmental stage. The expression pattern of the GbCHI gene was examined in different tissues of grafted plants by extracting total RNA from young leaves (buds), mature leaves (130 days after budding), ovules, fruit albumen, gynoecia, stamens, stems, and roots. The results of RQPCR analysis are shown in Fig. 3a. The transcription of GbCHI was clearly tissue specific and development-dependent. The highest level of GbCHI mRNA was observed in mature leaves, with moderate levels in stamens, gynoecia, and stems, but GbCHI was only weakly expressed in ovules and young leaves, and expression was lowest in albumen. No GbCHI transcripts were detected in the roots. These results agreed with the pattern of flavonoids accumulation in G. biloba (Fig. 3b). As seen in Fig. 3b, flavonoids were detected abundantly in mature leaves, and to a lesser extent in the stamens and gynoecia, but was not detected in roots. Thus, expression of the GbCHI gene appears to be one of the critical factors determining the flavanone accumulation pattern in G. biloba.
Comparison among different tissues in G. biloba regarding relative amount of GbCHI mRNA and accumulation of flavonoids. a Expression pattern of GbCHI gene in different tissues with GAPDH gene as control. b Accumulation pattern of flavonoids in different tissues. Data are mean values of triplicate tests ± SD
Effect of UV-B, GA, CCC, ABA, ETH, and ALA on GbCHI expression
RQPCR of ginkgo seeding following diverse chemical and stress treatments showed that GbCHI was inducibly expressed. Following a UV-B treatment, the amount of GbCHI mRNA was quickly increased during the 0–12 h of treatment and then declined thereafter, returning to a similar level to that of control at 96 h (Fig. 4a). Exogenous application of CCC caused a rapid and durable induction of the GbCHI gene at 12 h, and that level was maintained until 24 h, when the transcript level began to increase slowly, reaching a maximal level at 96 h that was 9.6-fold higher than the control (Fig. 4b).
Relative quantities of GbCHI mRNA at various time points post-treatment with UV-B (a), CCC (b), ALA (c), ABA (d), GA (e), and ETH (f). Each sample was individually assayed in triplicate. Values shown represent the mean reading from three treated plants and the error bars indicated the standard errors of the mean
As shown in Fig. 4c, d, and f, the expression of GbCHI was apparently up-regulated by ALA, ABA, and ETH. Transcription levels of GbCHI increased along with the treatment time and reached the highest level at the end of 24 or 48 h, at 9.2-fold, 8.2-fold, and 2.8-fold, respectively, compared with the controls. The GbCHI transcription was down-regulated by GA and the mRNA transcript showed a quick decrease within the first 24 h, followed by a slow rise (Fig. 4e). Exogenous GA, therefore, appeared to decrease the transcription level of GbCHI. Overall, all five tested abiotic stresses appeared to cause significant accumulation of GbCHI mRNA.
Characterization of the deduced GbCHI protein
The deduced GbCHI protein sequence of GbCHI consisted of 244 amino acids residues, with an isoelectric point of 7.76. The amino acid sequence of the GbCHI shared 62.7% identity with that of Camellia sinensis, 64.4% with Gossypium hirsutum, 65.0% with Pyrus communis, 52.5% with Phaseolus vulgaris, 50.3% with Medicago sativa, 52.5% with Glycyrrhiza uralensis, and 61.0% with Lotus japonicus. Similar to other non-legume plants, the four conserved amino acid residues essential for GbCHI active sites were Thr80, Tyr138, Gly145, and Ser221. This was unlike the case in Saussurea medusa, where the corresponding residues are Thr50, Tyr108, Met115, and Ser192, or in Medicago sativa (legume), where the corresponding residues are Thr48, Tyr106, Asn113, and Thr190 (Gensheimer and Mushegian 2004; Jez et al. 2000; Li et al. 2006). A GenBank CDD search and function analysis revealed that GbCHI belongs to the chalcone superfamily (Supplementary Fig. 2S).
Expression and purification of GbCHI
In order to express GbCHI in E.coli, we cloned the coding sequence of GbCHI into pET-28a, an expression vector with the T7 promoter and a His-tag, yielding pET28a-GbCHI. The expression construct was checked for in-frame fusion by DNA sequencing (Sangon, China). Upon induction by IPTG, GbCHI was expressed as a major soluble protein product (Supplementary Fig. 3S, lanes 2–5). Supplementary Fig. 3S Lane 6 shows the insoluble proteins after IPTG induction for 2.0 h. The molecular weight of the expressed recombinant protein was estimated as a 27 kDa band with a His-tag: this size was in agreement with that predicted by bioinformatics methods. The recombinant GbCHI protein was purified using Nickel-NTA agarose affinity chromatography and the purified protein showed the expected size (Supplementary Fig. 3S, lane 7). Western blotting of purified recombinant GbCHI protein confirmed its specific immune reactivity to anti-His antibodies (Supplementary Fig. 3S, lane western blot).
The activity of GbCHI in vitro
GbCHI activities were assayed using extracts of the recombinant expressing CHI cDNA. The reaction product of recombinant GbCHI was detected by HPLC monitored at 304 nm and its identity confirmed as naringenin and naringenin chalcone by comparison to known standards (Supplementary Fig. 4S, a). The elution profiles of ethylacetate-extracted reaction products showed that CHI yielded 5-hydroxyflavanone (naringenin) from the incubation with 6′-hydroxychalcone (naringenin chalcone) as substrates (Supplementary Fig. 4S, b). On the other hand, the control added with crude enzyme extract from E.coli + pET28a vector had more residual substrate and less spontaneous product (Supplementary Fig. 4S, c). To confirm the enzymatic cyclization by GbCHI proteins, the stereochemistry of naringenin at the C-2 position was estimated by HPLC on a chiral column. As shown in Supplementary Fig. 5S a and b, (2S)-naringenin was the predominant product compared with (2R)-naringenin in the presence of CHI protein, whereas only a racemic mixture of naringenin was found in the control reaction (Supplementary Fig. 5S, c). Compared with a standard sample (Supplementary Fig. 6S, a), the reaction with 6′-deoxychalcone (isoliquiritigenin) as substrate showed no product resulting from GbCHI activity (Supplementary Fig. 6S, b) and control (Supplementary Fig. 6S, c). Thus, our data suggest that GbCHI is a type I CHI, which only accepts 6′-hydroxychalcone as a substrate.
The relationship between GbCHI expression, GbCHI activity, and flavonoid accumulation during ginkgo leaf growth
To determine the temporal expression pattern of the CHI gene in ginkgo leaves, we examined CHI gene transcription, flavonoid content, and CHI activities at different leaf developmental stages. RQPCR of leaves at 15 developmental stages (Fig. 5) showed curves for changes in CHI expression that paralleled the curves for relative GbCHI activities at different developmental stages. The CHI gene mRNA was detected at all stages, with a gradual increase occurring from stage 1 to stage 4 and a peak at stage 4. After a brief decline in July 18, the transcript level continued to rise to an annual peak by August 30; the highest transcript level was 45.1107. A third peak (40.8804) appeared on September 29, followed by a subsequent slow decline.
The GbCHI activity showed a similar pattern during annual leaf growth, again indicating three peaks. CHI activity began to increase upon emergence of the leaf bud. The first peak of activity occurred around June 13 (14.6167 U/min mg protein) and CHI activity declined thereafter to a minimum by July 4 (12.0816 U/min mg protein). A second peak and the highest yearly activity occurred on September 11 (29.7331 U/min mg protein). A decline followed with minimal activity on September 29. A third peak (28.7272 U/min mg protein) was followed by a rapid decline. The peaks of annual activity showed slight lags behind the peak transcript levels.
Flavonoid content correlated with the seasonal patterns of GbCHI activity. An obvious peak was noted on July 4 (0.8039), on August 13 (0.7763), on September 29(0.9383), and on November 1(1.0994), with the annual peak appearing on November 1. The first peak appearing on July 4 also correlated with a peak in GbCHI transcript level. The annual peaks of flavonoid content also lagged behind the peaks of GbCHI activity. From a commercial standpoint, the best harvest stage for flavonoids appeared to be from the end of September to early November. Linear regression indicated that flavonoid content (Y1) and CHI activity (X1) were linearly correlated (Table 2), with an F value of 80.622, a correlation coefficient of 0.373, and a linear equation represented by Y 1 = (0.014 ± 0.002)X 1 + (0.459 ± 0.032). Linear regression of CHI transcription levels (X 2) and CHI activity (Y 2) showed a high correlation coefficient R 2 = 0.421; the F value was 96.755, and the linear equation was represented by Y 2 = (0.467 ± 0.047)X 2 + (5.352 ± 1.463) (Table 3). From this, we deduced that the CHI activity and accumulation of flavonoids in Ginkgo biloba leaves would be affected by CHI gene transcription levels.
Discussion
Characterization and function analysis of GbCHI
In this study, a G. biloba gene encoding a type I chalcone isomerase was isolated and characterized. Many genes encoding CHI from angiosperm plant species have been isolated and characterized at the genetic, chemical, and enzymological levels. However, cloning of a CHI gene from a gymnosperm plant such as G. biloba has not yet been reported. This work verifies for the first time the presence of a CHI in a gymnosperm. According to the analysis and comparison, GbCHI could be classified as a member of the type I CHI family.
Classification of GbCHI as to type I was not possibly based only on the phylogenetic tree. Studies of the three-dimensional structure of the Medicago sativa CHI (Jez et al. 2000) (a type I CHI) indicated the presence of four conserved amino acid residues (Supplementary Fig. 2S) that were essential for its activity. Amino acid analysis and three-dimensional structural configurations of GbCHI indicated that this enzyme was similar to a type I CHI. Previous observations (Dixon et al. 1988; Shimada et al. 2003) have shown that CHI converts both 6′-deoxychalcone and 6′-hydroxynchalcone to isoflavonoids and flavonoids (referred to as type II CHI), while GbCHI is capable of converting only 6′-hydroxynchalcone to 5-hydroxyflavanone (as a type I CHI). In addition, GbCHI controls the preferred formation of the biologically active (2S)-flavanones. Thus, GbCHI was clearly similar to non-leguminous types, based on amino acid sequence and type I substrate specificity. The non-enzymatic conversion of chalcones yields racemic (2R/S)-flavanones. Since only (2S)-flavanones are intermediates of the subsequent flavonoid pathway, CHI specificity guarantees the efficient formation of biologically active (2S)-flavanones. Only trace amounts of flavonoids were detected in the Arabidopsis transparent testa 5 mutants lacking CHI activity (Shirley et al. 1995). These results indicate that CHI activity is essential for the in vivo production of flavonoids.
Effects of UV-B and plant growth regulator on GbCHI expression
The ultraviolet absorbing characteristics of flavonoids have been considered as evidence for a role in UV protection. The induction of GbCHI by UV-B should not be surprising, since a light-responsive element and homologous MYB/MYC recognition sites, present in most of the phenylpropanoid biosynthesis gene promoters studied (Solano et al. 1995; Yoshioka et al. 2004; Xu et al. 2008b), were identified in the 5′-upstream region of GbCHI. UV exposure could induce the expression of flavonoid synthesis-related genes allowing flavonoids to play a role in resistance to UV damage. The first direct evidence in support of a role for flavonoids in UV protection came from experiments with Arabidopsis mutants, which showed that lesions in chalcone synthase or chalcone isomerase resulted in UV-hypersensitive phenotypes (Li et al. 1993). A role for flavonoids in UV protection is further supported by Bieza and Lois’ (2001) isolation of an Arabidopsis mutant that is tolerant of extremely high UV-B levels. This mutant shows constitutively high levels of a number of phenolics, including flavonoids. Interestingly, the CHI mutants were the most sensitive to UV light and showed a corresponding decrease in flavonoids and sinapate esters (Li et al. 1993). Indeed, flavonoids are often present in the epidermal cell layers of leaves and in tissues that are susceptible to UV light, such as pollen and the apical meristem. This may be the reason that the outer leaves of a ginkgo tree have a higher flavonoid content (Cheng 2001). In response to UV irradiation, the leaves of Ginkgo biloba showed a rapid increase in the transcription of GbCHI, this result indicated that the expression level of GbCHI could improve the activity of GbCHI, further increasing the accumulation of flavonoids in the leaves.
Changes in flavonoid levels in response to environmental stimuli and at specific developmental stages were mediated by regulated CHI expression. The synthesis of flavonoids compounds in plants is controlled by various factors, with enzymes and internal phytohormones as the major regulators (Cheng et al. 2000; Cheng 2001). However, there have been few studies of exogenous hormone responses with respect to the regulation of CHI expression.
Plant hormones play roles in flavonoid formation. For example, ethylene (ETH) has been shown to regulate the expression of several genes in flavonoid biosynthesis. Kush et al. (1990) reported that ETH treatment induced transcript accumulation of CHI, chalcone synthase (CHS), and phenylalanine ammonia lyase (PAL) in the rubber tree (Hevea brasiliensis). Ardi et al. (1998) observed that exogenous ETH led to the activation of transcript accumulation of CHS and flavanone-3-hydroxylase (F3H) in Persea americana. El-Kereamy et al. (2003) discovered that exogenous ETH could stimulate the transcription of flavonoid biosynthesis gene in grape (Vitis vinifera), including CHS, F3H, and anthocyanidin synthase (ANS). The in vitro spraying of ETP (ethephon) promoted ABA and ETH synthesis to various degrees while it hindered GA synthesis, but increased flavonoid content when supplied at various levels (Cheng et al. 2004a). In the present work, transcription of GbCHI was induced by ETH, consistent with the observation of ethylene-responsive elements in promoter region of GbCHI, These features suggested that GbCHI is likely to respond to ethylene-responsive signal transduction.
One common element between the ABA-mediated pathways and the flavonoid pathway is the role of the MYB and MYC classes of transcription activators on target gene expression (Abe et al. 1997). Some ABA-response elements are also present in the 5′-upstream region of the CHI genes. Cheng et al. (2004a) reported that treatment with ABA had significantly increased flavonoid content. In our present study, the exogenous application of ABA triggered a significant induction of GbCHI, together with the identification of homologous MYB/MYC recognition sites. These results suggested that the increased flavonoid content in ginkgo leaves might be related to the triggering of GbCHI transcription by ABA. These correlative data indicate that ABA may play an important role in the accumulation of flavonoids.
5-aminolevulinic acid (ALA) is a key precursor in the biosynthesis of porphyrins such as chlorophyll and heme, and its formation may be the rate limiting step (Von Wettstein et al. 1995). Leaf chlorophyll and flavonoid content are closely related (Awad et al. 2001; Cheng 2001). Treatment with ALA can promote the biosynthesis of flavonoids and anthocyanins (Saure 1990; Xu 2008). The present study showed that the transcription of GbCHI could be induced by ALA. Therefore, the expression of CHI played an important role in the promotion of flavonoid content.
Gibberellin (GA) speeds growth and reduces CHS activity, lowering the accumulation of flavonoids (Carrier et al. 1990). GA also reduced the content of flavonoids in the root of Scutellaria baicalensis (Cai et al. 2008). Reduced GA levels promote flavonoid accumulation (Cheng et al. 2004b). In this experiment, exogenous gibberellins restrain the gene expression level of GbCHI. Meanwhile, a primary study reveals that the exogenous GA process can adjust and control the reduction of accumulated content of ginkgo flavones, which reveals that the reduction of ginkgo flavones by gibberellic has something to do with the adjustment and control of the reduction of GbCHI gene transcription level. Therefore, GA treatment was not suitable for the cultivation of ginkgo and other medicinal plants whose leaves are to be used as the medicinal materials.
Cycocel (CCC) is known to inhibit gibberellins responses (Saure 1990) and thus might stimulate formation of flavonoids (Cheng et al. 2004a). Cheng et al. (2004a) reported that CCC greatly increased flavonoid contents of G. biloba leaves. In contrast, in Scutellaria baicalensis, CCC reduced the content of flavonoids in the root while elevating the content of lignin in the aerial parts (Cai et al. 2008). In our study, CCC greatly induced the transcription of GbCHI consistent with accumulation of flavonoids (Cheng et al. 2004a).
The expression patterns of the GbCHI gene and accumulation of flavonoids in different tissues and leaf developmental stages
The pattern of GbCHI transcript expression matched the accumulation pattern of flavonoids (Figs. 3, 5), indicating that the GbCHI is one of the key enzymes in the flavonoid biosynthetic pathway in G. biloba, and is responsible for the formation of flavonoids in different tissues. Two POLRE cis-elements were found on the 5′ upstream region, which act as cis factors to direct pollen-specific expression in dicots (Bate and Twell 1998). Some SEBF-like elements were also found in the upstream region, some of these consensus sequences are present in promoters of fruit-specific expression genes (Boyle and Brisson 2001; Yamagata et al. 2002). Therefore, it can be speculated that high levels of expression of GbCHI and enzyme activity serve to allow accumulation of flavonoids in ovules and stamens. Thus, control of the expression level of CHI can be viewed as a potential approach to increase flavonoid production in ginkgo.
Little is known regarding the expression of CHI genes at different developmental stages in ginkgo leaves. Previous work on temporal expression profiles reported that the transcription of flavonoid synthesis genes was associated with the accumulation of flavonoids (Xu et al. 2007, 2008a). In the present study, we showed that the qualitative differences in competence for CHI activity at different developmental stages are primarily the result of the differences in the transcription of CHI in developing leaves. Significant correlation was found between expression level of CHI genes and CHI activity at all of the annual cycle stages of ginkgo leaves. Transcriptional controls play an important role in regulating the overall activity of flavonoid biosynthesis (Jaakola et al. 2002). The same correlation was seen between CHI activity and flavonoid content (Tables 2, 3). In early July, plants have more leaves than they do in November, thus July would be the best time to harvest ginkgo leaves for highest flavonoid recoveries. Our study showed that the mRNA levels of the CHI gene and the accumulation of flavonoids both were correlated with CHI activity. The CHI activity paralleled the GbCHI transcription level but not the flavonoid accumulation, which might be explained by (1) The enzyme activities in the various branch pathways are highly regulated, the accumulation of flavonoids is a complex processed or (2) Flavonoid concentrations in ginkgo leaves are not only regulated by the involvement of genes, but also by other factors, such as photosynthesis, tree age, season, quantity of fertilizer application, etc. (Cheng et al. 2009).
Ginkgo biloba L. is a medicinal plant in China, and its flavonoids can be found in standardized ginkgo leaf extract preparations (24% flavonoids). Thus, the flavonoids are considered to be important for the beneficial pharmacological effects of this plant (Smith and Luo 2004; van Beek 2002). Ecological factors influence flavonoid accumulation primarily during the young stages of ginkgo leaf development. Among all ecological factors, light, seasonal variations, and hormones are the most important (Cheng et al. 2009). This study used G. biloba leaves as experimental material to explore the effects of regulating factors on the synthesis of flavonoids compounds and on the modulation of GbCHI expression and activity. The overall aim was to provide a theoretical and practical basis for increasing the flavonoid content of G. biloba leaves used in commercial production. The best time for application of flavonoid-promoting regulators and the optimum application concentrations still need further study. However, with the development of molecular biology techniques that now allow the successful cloning of genes encoding some key enzymes, the prospect for improved synthesis of flavonoids is optimistic.
Abbreviations
- bp:
-
Base pair
- HPLC:
-
High performance liquid chromatography
- IPTG:
-
Isopropyl β-d-thiogalactoside, C9H18O5S
- PCR:
-
Polymerase chain reaction
- RACE:
-
Rapid amplification of cDNA ends
- RQPCR:
-
Real-time quantitative PCR
- RT-PCR:
-
Reverse transcriptase polymerase chain reaction
- S.D.:
-
Standard deviation
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- UV:
-
Ultraviolet
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868
Ardi R, Kobiler I, Jacoby B, Keen NT, Prusky D (1998) Involvement of epicatechin biosynthesis in the activation of the mechanism of resistance of avocado fruits to Colletotrichum gloeosporioides. Physiol Mol Plant Pathol 53:269–285
Awad MA, Wagenmakers PS, de Jager A (2001) Effects of light on flavonoid and chlorogenic acid levels in the skin of ‘Jonagold’ apples. Sci Hortic 88:289–298
Bate N, Twell D (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37:859–869
Bednar RA, Hadcock JR (1988) Purification and characterization of chalcone isomerase from soybeans. J Biol Chem 263:9582–9588
Bieza K, Lois R (2001) An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol 126:1105–1115
Boyle B, Brisson N (2001) Repression of the defense gene PR-10a by the single-stranded DNA binding protein SEBF. Plant Cell 13:2525–2537
Cai G, Guo Y, Yao H, Chen S, Zhou T (2008) Impacts of cycocel and gibberellin on the biomass and flavonoid production in Scutellaria baicalensis Georgi. Chin Agric Sci Bull 24:213–217
Carrier D, Cosentino G, Neufeld R, Rho D, Weber M, Archambault J (1990) Nutritional and hormonal requirements of Ginkgo biloba embryo-derived callus and suspension cell culture. Plant Cell Rep 8:635–638
Cheng SY (2001) Studies on the main facrtors influencing flavonoid formation and its regulation in Ginkgo biloba leaf. Dissertation, Shandong Agricultural University
Cheng SY, Gu MR, Shu HR (2000) Advances in research on flavonoids in Ginkgo biloba leaf. Scientia Silvae Sinicae 36:110–115
Cheng SY, Wang Y, Fei YJ, Zhu GC (2004a) Studies on the effects of different treatments on flavonoids contents in Ginkgo biloba leaves and their regulating mechanism. J Fruit Sci 21:116–119
Cheng SY, Wang Y, Li JK, Fei YJ, Zhu GC (2004b) Study on the relationship between the endogenous hormones and flavonoids. Scientia Silvae Sinicae 40:45–49
Cheng SY, Xu F, Wang Y (2009) Advances in the study of flavonoids in Ginkgo biloba leaves. J Med Plants Res 3:1248–1252
Diamond BJ, Shiflett SC, Feiwel N, Matheis RJ, Noskin O, Richards JA, Schoenberger NE (2000) Ginkgo biloba extract: mechanisms and clinical indications. Arch Phys Med Rehabil 81:668–678
Dixon RA, Blyden ER, Robbins MP, Van Tunen AJ, Mol JN (1988) Comparative biochemistry of chalcone isomerases. Phytochemistry 27:2801–2808
Druka A, Kudrna D, Rostoks N, Brueggeman R, von Wettstein D, Kleinhofs A (2003) Chalcone isomerase gene from rice (Oryza sativa) and barley (Hordeum vulgare): physical, genetic and mutation mapping. Gene 302:171–178
El-Kereamy A, Chervin C, Roustan JP, Cheynier V, Souquet JM, Moutounet M, Raynal J, Ford C, Latché A, Pech JC (2003) Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol Plant 119:175–182
Fouché S, Dubery I (1994) Chalcone isomerase from Citrus sinensis: purification and characterization. Phytochemistry 37:127–132
Gensheimer M, Mushegian A (2004) Chalcone isomerase family and fold: no longer unique to plants. Protein Sci 13:540–544
Guiltinan M, Marcotte W Jr, Quatrano R (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250:267–271
Itzhaki H, Maxson JM, Woodson WR (1994) An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. PNAS 91:8925–8929
Jaakola L, Maatta K, Pirttila A, Torronen R, Karenlampi S, Hohtola A (2002) Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol 130:729–739
Jansson S, Meyer-Gauen G, Cerff R, Martin W (1994) Nucleotide distribution in gymnosperm nuclear sequences suggests a model for GC-content change in land-plant nuclear genomes. J Mol Evol 39:34–46
Jez JM, Bowman ME, Dixon RA, Noel JP (2000) Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat Struct Biol 7:786–791
Kimura Y, Aoki T, Si A (2001) Chalcone isomerase isozymes with different substrate specificities towards 6-hydroxy- and 6-deoxychalcones in cultured cells of Glycyrrhiza echinata, a leguminous plant producing 5-deoxyflavonoids. Plant Cell Physiol 42:1169–1173
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Kuhn B, Forkmann G, Seyffert W (1978) Genetic control of chalcone-flavanone isomerase activity in Callistephus chinensis. Planta 138:199–203
Kush A, Goyvaerts E, Chye ML, Chua NH (1990) Laticifer-specific gene expression in Hevea brasiliensis (rubber tree). PNAS 87:1787–1790
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Vande Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acids Res 30:325–327
Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5:171–179
Li F, Jin Z, Qu W, Zhao D, Ma F (2006) Cloning of a cDNA encoding the Saussurea medusa chalcone isomerase and its expression in transgenic tobacco. Plant Physiol Biochem 44:455–461
Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41:541–547
McKhann HI, Hirsch AM (1994) Isolation of chalcone synthase and chalcone isomerase cDNAs from alfalfa (Medicago sativa L.): highest transcript levels occur in young roots and root tips. Plant Mol Biol 24:767–777
Mehdy M, Lamb C (1987) Chalcone isomerase cDNA cloning and mRNA induction by fungal elicitor, wounding and infection. EMBO J 6:1527–1533
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3:212–217
Moustafa E, Wong E (1967) Purification and properties of chalcone-flavanone isomerase from soya bean seed. Phytochemistry 6:625–632
Muir S, Collins G, Robinson S, Hughes S, Bovy A, De Vos C, van Tunen A, Verhoeyen M (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19:470–474
Mundy J, Yamaguchi-Shinozaki K, Chua N (1990) Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. PNAS 87:1406–1410
Oken BS, Storzbach DM, Kaye JA (1998) The efficacy of Ginkgo biloba on cognitive function in Alzheimer disease. Arch Neurol 55:1409–1415
Park H, Kim M, Kang Y, Jeon J, Yoo J, Kim M, Park C, Jeong J, Moon B, Lee J (2004) Pathogen-and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol 135:2150–2161
Piechulla B, Merforth N, Rudolph B (1998) Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol Biol 38:655–662
Rawat R, Xu ZF, Yao KM, Chye ML (2005) Identification of cis-elements for ethylene and circadian regulation of the Solanum melongena gene encoding cysteine proteinase. Plant Mol Biol 57:629–643
Saure MC (1990) External control of anthocyanin formation in apple. Sci Hortic 42:181–218
Shimada N, Aoki T, Sato S, Nakamura Y, Tabata S, Ayabe S (2003) A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy (iso) flavonoids in Lotus japonicus. Plant Physiol 131:941–951
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Smith JV, Luo Y (2004) Studies on molecular mechanisms of Ginkgo biloba extract. Appl Microbiol Biotechnol 64:465–472
Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J (1995) Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB. Ph3) from Petunia hybrida. EMBO J 14:1773–1784
Straub P, Shen Q, Ho T (1994) Structure and promoter analysis of an ABA-and stress-regulated barley gene, HVA1. Plant Mol Biol 26:617–630
Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J 34:635–645
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
van Beek TA (2002) Chemical analysis of Ginkgo biloba leaves and extracts. J Chromatogr A 967:21–55
van Beek T, Montoro P (2009) Chemical analysis and quality control of Ginkgo biloba leaves, extracts, and phytopharmaceuticals. J Chromatogr A 1216:2002–2032
Van Tunen AJ, Koes RE, Spelt CE, Van der Krol AR, Stuitje AR, Mol JN (1988) Cloning of the two chalcone flavanone isomerase genes from Petunia hybrida: coordinate, light-regulated and differential expression of flavonoid genes. EMBO J 7:1257–1263
Von Wettstein D, Gough S, Kannangara CG (1995) Chlorophyll biosynthesis. Plant Cell 7:1039–1057
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223
Xu F (2008) Cloning and expression of GbPAL and GbANS genes and effect of ALA on the content of flavonoids in Ginkgo biloba. Dissertation, Shandong Agriculture University
Xu F, Cheng SY, Cheng SH, Wang Y, Du HW (2007) Time course of expression of chalcone synthase gene in Ginkgo biloba. J Plant Physiol Mol Biol 33:309–317
Xu F, Cai R, Cheng SY, Du HW, Wang Y, Cheng SH (2008a) Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. Afr J Biotechnol 7:721–729
Xu F, Cheng H, Cai R, Li LL, Chang J, Zhu J, Zhang FX, Chen LJ, Wang Y, Cheng SH (2008b) Molecular cloning and function analysis of an anthocyanidin synthase gene from Ginkgo biloba, and its expression in abiotic stress responses. Mol Cells 26:536–547
Yamagata H, Yonesu K, Hirata A, Aizono Y (2002) TGTCACA motif is a novel cis-regulatory enhancer element involved in fruit-specific expression of thecucumisin gene. J Biol Chem 277:11582–11590
Yoshioka S, Taniguchi F, Miura K, Inoue T, Yamano T, Fukuzawa H (2004) The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell 16:1466–1477
Zhang H, Liu J, Lu H, Gao S (2009) Enhanced flavonoid production in hairy root cultures of Glycyrrhiza uralensis Fisch by combining the over-expression of chalcone isomerase gene with the elicitation treatment. Plant Cell Rep 28:1205–1213
Zhou D (1999) Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4:210–214
Acknowledgments
This work was supported by the Natural Science Foundation of China (30971974), the Program for New Century Excellent Talents in University (NCET-04-0746), and University–industry Cooperation Fund of Hubei Educational Office (CXY2009B009).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Quiros.
First two authors contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cheng, H., Li, L., Cheng, S. et al. Molecular cloning and function assay of a chalcone isomerase gene (GbCHI) from Ginkgo biloba . Plant Cell Rep 30, 49–62 (2011). https://doi.org/10.1007/s00299-010-0943-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0943-4