Abstract
Citrus, rich in carotenoids, is the most important fruit crop based on the total annual production. In the carotenoid biosynthesis pathway, phytoene synthase (PSY, EC 2.5.1.32) catalyzes the dimerization of two molecules of geranylgeranyl pyrophosphate (GGPP) to phytoene and has been shown to be a rate-limiting enzyme for the synthesis of carotenoids. In this study, we investigated catalytic activity of CsPSY from Cara Cara navel orange (Citrus sinensis Osbeck) by heterologous expression in Escherichia coli containing a GGPP-producing plasmid. Moreover, the effects of CsPSY overexpression on carotenoid accumulation were also functionally analyzed in transgenic Hongkong kumquat (Fortunella hindsii Swingle). The resulting transgenic plants produced orange fruits, and extracts from the fruits of four overexpressing plants had a 2.5-fold average increase of phytoene with the content approximately 71.38 μg/g fresh weight. Lycopene, β-carotene, and β-cryptoxanthin in transgenic fruits were also markedly increased, whereas the levels of lutein and violaxanthin kept nearly unchanged with 1.1–1.3 folds variation. Transcript levels of carotenoid biosynthetic genes in the CsPSY overexpressed plants remained unaltered except that PDS and ZDS showed a minor increase. This study suggests that CsPSY plays a crucial role in citrus carotenoid biosynthesis and could be used as a means of engineering fruit crop for the production of carotenoids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids are an important family of isoprenoid pigments synthesized by plants and certain algae, bacteria, and fungi. In photosynthetic organisms, carotenoids play essential functions as components of the light-harvesting system, and protecting plant cells from photo-oxidative damage (Demmig-Adams and Adams 2002). Carotenoids provide the yellow, orange or red coloration characteristic of many flowers and fruits, to attract animals for pollination or for the dispersal of seeds (Bartley and Scolnik 1995). Carotenoids also serve as precursors for the biosynthesis of the plant hormone abscisic acid (Schwartz et al. 1997) and for the production of volatile compounds for fruit and flower flavor and aroma (Bouvier et al. 2003; Simkin et al. 2004). In addition, plant carotenoids are the primary dietary source of provitamin A and are generally believed to offer numerous benefits to human health (Botella-Pavía and Rodríguez-Concepión 2006).
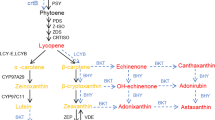
Carotenoids are produced in the plastids from isopentenyl diphosphate precursors originating from the glyceraldehyde phosphate pyruvate pathway (Cunningham and Gantt 1998). The first committing step of carotenoid biosynthesis is a head-to-head coupling of two molecules of geranylgeranyl pyrophosphate (GGPP) by phytoene synthase (PSY) to yield phytoene, which is desaturated by phytoene desaturase (PDS) and zeta-carotene desaturase (ZDS) and isomerized by carotenoid isomerase (CRTISO) to form the linear all-trans-lycopene (Fig. 1). Lycopene is cyclized twice by two individual cyclases, producing α- and β-carotene, which are then subject to oxygenation reactions to produce different xanthophylls, such as lutein, zeaxanthin, violaxanthin, and neoxanthin.
Schematic diagram of the biosynthesis pathway of carotenoids in plants. Summarized based on Cunningham and Gantt (1998), Kato et al. (2004) and Alquezar et al. (2008, 2009). IPP isopentenyl diphosphate, DMAPP dimethylallyl diphosphate, PSY phytoene synthase, PDS phytoene desaturase, ZDS ζ-carotene desaturase, CRTISO carotene isomerase, LCYb lycopene β-cyclase, LCYe lycopene ε-cyclase, HYb β-carotene hydroxylase, ZEP zeaxanthin epoxidase
The high economic value of carotenoids as nutritional sources of vitamin A and health-promoting compounds has prompted some attempts to increase carotenoids biosynthesis in plants. Seed-specific overexpression of bacterial PSY targeted to the plastid led to a 50-fold increases in carotenoid levels in the mature seeds of canola (Brassica napus) (Shewmaker et al. 1999). Similarly, the expression of a bacterial PSY in tomato in a fruit-specific manner resulted in elevated levels of phytoene, lycopene, lutein, and β-carotene (Fraser et al. 2002). Manipulation of the PSY activity has also resulted in increased levels of carotenoids in tomato (Fray et al. 1995), tobacco (Busch et al. 2002), Arabidopsis (Lindgren et al. 2003), rice (Paine et al. 2005), potato (Ducreux et al. 2005) and flaxseed (Fujisawa et al. 2008). However, little is known about PSY genes’ function and their potential of carotenoid improvement in fruit tree.
Cara Cara, an interestingly pink-fleshed mutant originated from Washington navel orange (Citrus sinensis Osbeck), has been characterized and identified as a unique navel orange accumulating high amounts of lycopene and β-carotene in the pulp (Lee 2001). Cara Cara navel orange had drawn extensive attentions and investigations for its attractive red pulp color, high antioxidant activity, and numerous benefits to human health (Xu et al. 2006; Tao et al. 2007; Alquezar et al. 2008). Previously, we isolated cDNA encoding PSY from Cara Cara navel orange, but the function of the gene product had not been demonstrated (Tao et al. 2007). In the carotenoid biosynthetic pathway, PSY is thought to be a key regulatory enzyme in the production of carotenoids; therefore, it has been the potential target of genetic manipulation in plants for enhancing carotenoid levels (Shewmaker et al. 1999; Ye et al. 2000; Fraser et al. 2002). In the present study, we characterized the CsPSY gene from Cara Cara navel orange and demonstrated its catalytic activities by heterologous expression in E. coli cells. Moreover, the CsPSY gene has also been introduced into Hongkong kumquat (Fortunella hindsii Swingle), a citrus relative, and the transgenic fruits have been produced with enhanced levels of carotenoids accumulation, a level about 2.0-fold higher than the non-transformed controls. These results could help us gain more knowledge on the regulatory role of CsPSY in carotenoid biosynthesis in citrus fruits, as will also benefit the improvements of the carotenoid content and composition in citrus fruits in the future.
Materials and methods
Sequence analysis of cDNA encoding PSY
Total RNA was extracted from the pulp of Cara Cara navel orange according to the method described by Liu et al. (2006) and mRNA was purified by the PolyATtract® mRNA Isolation SystemsIII Kit (Promega, USA). The first strand cDNA was synthesized with the RevertAidTM-MuLV Kit (MBI, Lithuania). Gene-specific primers (5′-CAGGGGCCTCAAAATTTTCTT-3′ and 5′-GGCCTTGGATTGTGAATTGG-3′) were designed based on the sequences published previously (Tao et al. 2007). The amplified cDNA was cloned into pMD18-T Easy vector (Takara, Japan) and the sequence was determined on the ABI PRISM TM 310 genetic analyzer (Applied Biosystems, Foster City, CA, USA).
Expression of CsPSY in E. coli
For bacterial expression analysis, we amplified coding sequences of CsPSY by PCR using the forward primer 5′-CGGGATCCATGTCTGTTACA-3′ with a BamHI site (underlined) and the reverse primer 5′-CCCTCGAGTTAAGCCTTACT-3′ with an XhoI site (underlined). The resulting PCR product was cloned into the BamHI and XhoI sites in pET-28a (+) (Novagen, Madison, WI). Plasmid pET-CsPSY and pET-28a (+) (empty vector) as a negative control were transformed into E. coli BL21 (DE3) containing expression plasmid pACCRT-E for GGPP biosynthesis (Misawa et al. 1990). Another plasmid pACCRT-EB (Misawa et al. 1990) that allows bacteria to produce phytoene was transformed into E.coli BL21 (DE3) and used as the positive control. The transformed E. coli were grown overnight at 37°C on Luria–Bertani (LB) solid medium containing 50 μg/mL of kanamycin and 35 μg/mL chloramphenicol. Selected colonies were inoculated in LB liquid medium with appropriate antibiotics, and shaken at 25°C for 4 days in darkness for expression. The cells were harvested by centrifugation (3,000g for 15 min at 4°C), washed in sterile distilled water, lyophilized and used for carotenoid extraction.
Plasmid construction and plant transformation
The complete coding region of CsPSY was amplified by PCR with primers: 5′-CGGGATCCATGTCTGTTACA-3′ (BamHI site underlined) and 5′-GGGGTACCTTAAGCCTTACT-3′ (KpnI site underlined). The amplified products were digested with BamHI and KpnI, inserted into the corresponding sites of the binary plant expression vector PMV (a pBI121 derivative which has been modified by deleting the β-GUS fragment and introducing a new multiple cloning sites: 5′-BamHI, XhoI, SalI, …, XbaI, BamHI, KpnI-3′), resulting in final constructs pBI-CsPSY for transformation. The construct was electroporated into Agrobacterium tumefaciens strain EHA105. Epicotyl segments from 4-week-old etiolated seedlings of Hongkong kumquat (F. hindsii Swingle) were infected with Agrobacterium according to the method of Duan et al. (2007). Briefly, explants after 3 days of co-cultivation were put on an MT medium of supplemented with 3% sucrose, 1.0 mg/L BA, 0.5 mg/L KT, 0.1 mg/L NAA, 50 mg/L kanamycin (Km), 400 mg/L cefotaxime (Cef) for shoot regeneration and first selection. Explants were subcultured on an MT medium supplemented with 3% sucrose, 0.5 mg/L BA, 0.5 mg/L KT, 0.1 mg/L NAA, 100 mg/L Km, 400 mg/L Cef for shoot elongation and second selection. The adventitious shoots that emerged on the segments were detached and cultured on an 1/2 MT medium supplemented with 3% sucrose, 0.5 mg/L NAA, 0.1 mg/L IBA, 0.5 g/L activated charcoal, 50 mg/L Km, 200 mg/L Cef for rooting. Rooted plantlets were transplanted into pots containing a commercial substrate with organic matter and vermiculite under greenhouse facilities.
PCR and Southern blot for transgenic analysis
Standard PCR techniques were used to detect the selective marker gene sequences in leaf samples from regenerated transgenic plants. The NPTII gene primers were: 5′-AGGACCATGTGGTCTCTCTT-3′ and 5′-TGGCCAACACTTGTCACTAC-3′, which produced a 500-bp fragment. The thermal condition was 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C.
Total DNA was isolated from leaves of five individual PCR-positive lines and non-transformed control plant in the greenhouse according to Cheng et al. (2003). For Southern blot analysis, 15 μg DNA sample was completely digested with BamHI, separated on 0.8% (w/v) agarose gel, blotted onto a nylon membranes (Hybond-NX, Amersham-Pharmacia Biotech, Little Chalfont, UK). Probe labeling by Digoxigenin (DIG), hybridization, and detection were conducted according to manufacturer’s instructions (Roche Diagnostics GmbH, Mannheim, Germany).
Carotenoid extraction and HPLC analysis
Carotenoids from bacteria or citrus full-colored fruits and leaves were extracted and analyzed essentially as described by Liu et al. (2007) with slight modifications. Briefly, 100 mg of bacterial cells or 300 mg of citrus fruits and leaves was ground to fine powder and extracted three times with 5 mL of hexane:acetone (1:1, v/v) by vortexing for 30 min until the samples were colorless. The extracts were combined, partitioned by adding 5 mL of NaCl-saturated solution, and centrifuged at high speed for 5 min. The carotenoid containing upper phase was transferred into a new tube and dried under a gentle stream of nitrogen gas.
The dried sample was resuspended in 400 μL of MeCN:MeOH:CH2Cl2 (45:5:50, v/v/v). Separation and identification of carotenoids were carried out on a reverse phase Analytical YMC Carotenoid Column C30 (150 × 4.6 mm i.d., 3 μm, Wilmington, NC, USA) using a Waters HPLC system with a photodiode array detector (Waters, Milford, MA). Identification of carotenoids was achieved by comparison of the individual characteristic absorption spectrum and the retention time with known standards. Quantification was performed using a calibration curve generated with commercially available phytoene, lycopene, β-carotene, α-carotene, β-cryptoxanthin, lutein and violaxanthin standards (Sigma-Aldrich).
Expression analysis of the carotenoid pathway genes in transgenic Hongkong kumquat fruits
Total RNA from Hongkong kumquat fruits was extracted as previously described (Liu et al. 2006). The RNA samples were treated with DNase I and reverse-transcribed with RevertAidTM-MuLV Kit (MBI, Lithuania) to generate the first strand cDNA. The cDNA samples were used as templates in real-time PCR assay in the presence of a SYBR Green PCR Master Mix (Applied Biosystems) and carotenogenic gene-specific primers (Table 1). The reactions were performed in an ABI 7500 Real Time System (Applied Biosystems). Thermal cycling conditions were an initial incubation at 50°C for 2 min and 95°C for 1 min, followed by 40 cycles of denaturation for 15 s at 95°C and annealing/extension for 1 min at 60°C. Relative expression levels were calculated using the ΔΔCT method as described by Lyi et al. (2007). All data were normalized first with the level of the β-actin internal transcript control and then with the expression of the non-transformed controls.
Statistical analyses
Statistical differences between WT and transgenic lines were assessed based on the analysis of variance ANOVA using SPSS (Chicago, IL, USA). Differences were considered significant at a probability level of P ≤ 0.05.
Results
Sequence analysis of CsPSY cDNA
A 1,520-bp CsPSY cDNA from Cara Cara navel orange was isolated using the RT-PCR technique (Tao et al. 2007). It contained an open reading frame (ORF) of 1,311 bp, and encoded a polypeptide of 436 amino acids with an estimated molecular mass of 49.5 kDa. A putative chloroplastic transit sequence was predicted to be the N-terminal residues 1–44 through the Chlorop 1.1 program based on the website: http://www.cbs.dtu.dk/services/ChloroP/ (Fig. 2a). The transmembrane domain of CsPSY was analyzed with DNAman program, which located in 260–282 amino acids (Fig. 2b).
a Deduced amino acid sequence of phytoene synthase cDNA (CsPSY) from Cara Cara navel orange. The putative signal of chloroplastic transit peptide is underlined. b Transmembrane domain of CsPSY located in 260–282 amino acids. c The phylogenetic tree of CsPSY and other related sequences. The tree was constructed by MEGA 4.0 program (Tamura et al. 2007) using the neighbor-joining method (Saitou and Nei 1987). The GenBank accession numbers for the PSY amino acids sequences are as follows: Citrus sinensis (CsPSY), Citrus unshiu (CuPSY, BAB18514), Citrus paradisi (CpPSY, AAD38051), Cucumis melo (CmPSY, CAA85775), Capsicum annuum (CaPSY, CAA48155), Lycopersicon esculentum (LePSY, AAA34153), Helianthus annuus (HaPSY, CAC27383), Arabidopsis thaliana (AtPSY, AAN17427), Zea mays (ZmPSY, AAR08445), Oryza sativa (OsPSY, AAS18307), Narcissus pseudonarcissus (NpPSY, CAA55391), Daucus carota (DcPSY, BAA84763), Erwinia uredovora (EuPSY, BAA14128)
The deduced amino acid sequence of CsPSY was compared with those of other known PSY proteins. The CsPSY sequence shared 99% identity with PSY clone from Citrus unshiu (Kim et al. 2001; Ikoma et al. 2001). CsPSY also had a high degree of homology with other higher plant species, such as Cucumis melo (82%), Lycopersicon esculentum (80%), Capsicum annuum (79%), Helianthus annuus (78%), Arabidopsis thaliana (77%), Narcissus pseudonarcissus (76%), Dunaliella salina (75%), Zea mays (73%) and Oryza sativa (72%). However, a low level of homology (30%) was observed with Erwinia uredovora. The phylogenetic analysis demonstrated similar patterns (Fig. 2c), which indicated that PSY isolated from fruits such as citrus, tomato, capsicum, and melon can be categorized into one group. Thus, these results suggested that an evolutionary link did exist among the fruit-producing plants.
Expression of CsPSY in Escherichia coli
To investigate the catalytical activity of CsPSY, the full-length ORF of CsPSY was cloned into pET-28a (+) and transformed into an E. coli strain engineered to accumulate GGPP (Misawa et al. 1990). HPLC analysis revealed that the GGPP-accumulating E. coli cells transformed with the CsPSY construct showed a peak (Fig. 3b, peak 1) that was not detected in the empty vector control, pACCRT-E/pET-28a (+) (Fig. 3a). The retention time and absorption spectrum of the peak 1 matched well with those of the peak 2 presented in the positive control carrying pACCRT-EB for phytoene biosynthesis (Fig. 3c, peak 2). These results confirmed the functionality of CsPSY gene product, which could catalyze a head-to-head condensation of two molecules of GGPP produced by the E. coli cells to form phytoene.
Pigments produced in E. coli GGPP-accumulating strains. Pigments extracted from E. coli cells harboring plasmids pACCRT-E, the engineered plasmid producing GGPP, and pET-28a, the empty vector (a); plasmids pACCRT-E and pET-CsPSY, which encodes CsPSY (b); and plasmid pACCRT-EB, the engineered plasmid producing phytoene as a positive control (c). The absorption spectra of specific peaks are presented to the boxes with retention times of 23.5 min
Production of transgenic Hongkong kumquat plants expressing CsPSY
To further understand the functional roles of CsPSY gene and the metabolic processes affected in plants, the CsPSY cDNA under the control of the 35S CaMV promoter was transformed into Hongkong kumquat (Fortunella sp.), a closest relative genus of Citrus and had a short juvenile period of about 2 years. Following selection on the basis of kanamycin resistance, approximately 50 putatively independent transgenic Hongkong kumquat plantlets were obtained. Out of them, only 5 lines were positive for the kanamycin resistance gene (NPTII) (Fig. 4a), and the rest 45 were negative for NPTII gene, indicating they were probably escapes. Southern analysis with the NPTII gene probe revealed the integration of 1–2 copies of the transgene in these lines L-11, L-17, L-21 and L-28; however, hybridization signal was not detected in transgenic line L-15 (Fig. 4b).
a Identification of transgenic Hongkong kumquat by PCR reaction. b Southern analysis of wile type (WT) and transgenic lines L-11, L-15, L-17, L-21 and L-28. c The plant growth of WT and transgenic line L-11. d The fruit phenotype of WT and transgenic lines L-11, L-17, L-21 and L-28. Scale bar 1.0 cm
In these transgenic lines, there were no apparently differences in the growth habits or the shape and size of leaf, flower and fruit compared to wild-type plants (Fig. 4c). However, a change of color was observed in the fruits of transgenic plants. The wild-type Hongkong kumquat plants usually formed yellow fruits, while transgenic plant lines showed fruit colors varying from deep yellow to orange (Fig. 4d), suggesting that overexpression of CsPSY in Hongkong kumquat conferred carotenoid accumulation to the transgenic plants to different extent.
Carotenoid analyses in transgenic Hongkong kumquat fruits
To examine the effect of the CsPSY transgene on carotenoid accumulation, four independent transgenic lines with varied copy numbers as confirmed by Southern blot were selected for further investigation. There was no obvious correlation between copy number and carotenoid content (Table 2). The fruits from CsPSY transgenic lines were found to contain up to 171.9 μg/g FW of total carotenoids, a level about 2.0-fold higher than the non-transformed controls. The phytoene content of the CsPSY transgenic lines was estimated to be 58.60–102.18 μg/g FW, representing the greatest increase (1.7–3.0-fold) above non-transformed controls. A similar increase was also observed in lycopene accumulation (2.2–2.9-fold). The fruits of the transgenic lines contained 6.45–7.40 μg/g FW of lutein and 40.32–48.06 μg/g FW of violaxanthin, corresponding to remaining almost unchange (1.1–1.3-fold), compared with those in non-transformed controls. Less abundant Hongkong kumquat carotenoids such as β-carotene and β-cryptoxanthin content were also elevated 2.0- and 2.3-fold, respectively, in most of the CsPSY transgenics. Moreover, there were slight changes in the proportion of carotenoid composition in these lines compared with the wild-type plants. No significant differences in carotenoid amount in the leaves were found between transgenic and non-transformed plants (data not shown).
Perturbations in carotenoid biosynthetic gene expression
Alteration of expression of some carotenoid biosynthetic genes has been shown to affect the transcript levels of other carotenoid genes in plants. To examine the effect of the CsPSY transgene on the expression of carotenogenic genes in the transgenic plants, the transcript levels of a panel of genes associated with carotenoid biosynthesis were examined by quantitative real-time RT-PCR. The genes examined include PSY, PDS, ZDS, LCYb, LCYe, HYb, and ZEP (Fig. 1).
For a set of four plants, a detailed analysis was made to show possible reactions of these genes in response to the overexpression of CsPSY (Fig. 5). A slight increase in the mRNA levels of PDS and ZDS was observed in the CsPSY transgenic lines; however, CRTISO was nearly unaltered. Moreover, transcript levels of other downstream genes (LCYe, LCYb, HYb and ZEP) showed no significant difference in comparison with levels in non-transformed control plants, although there was an increase in PSY transcript by a factor ranging from 4 to 6 (Fig. 5).
Quantitative RT-PCR analysis of the expression of carotenoid biosynthetic genes in transgenic Hongkong kumquat fruits. The levels of expression were normalized to β-actin relative to the non-transformed controls which was set to 1. Data are presented as mean ± SE from three experimental replicates. Biosynthetic gene symbols are as described in the legend of Fig. 1
Discussion
An increased emphasis on dietary sources of health-promoting foods (functional foods) that contain carotenoids is leading to improvement in plant species using traditional breeding methods and biotechnology (Fraser and Bramley 2004; Botella-Pavía and Rodríguez-Concepión 2006). Citrus fruits contain a considerable amount of carotenoids and form part of the daily nutrition of human being. Recently, lycopene-accumulating citrus mutants, such as Star Ruby grapefruit, Cara Cara navel orange, and Hong Anliu sweet orange, have drawn extensive attentions and investigations for their attractive red pulp color, significant antioxidant activity, and numerous benefits to human health (Lee 2001; Fraser and Bramley 2004; Xu et al. 2006; Liu et al. 2007; Alquezar et al. 2008, 2009). Whether citrus fruit could accumulate more specific carotenoids for human health applications or the composition of citrus fruit carotenoids could be purposely controlled has become an interesting breeding objective. For this purpose, a PSY gene that encodes an enzyme in the early part of the carotenoid biosynthetic pathway was considered to be an important target.
PSY is a rate-limiting enzyme in the carotenoid biosynthetic pathway (Cunningham and Gantt 1998). The contribution of transcript abundance of PSY to the carotenoid accumulation was studied in many plant species, including tomato fruit (Giuliano et al. 1993), pepper fruit (Bouvier et al. 1994) and C. unshiu fruits (Ikoma et al. 2001; Kato et al. 2004). Similarly, CsPSY was also observed to be up-regulated with the accumulation of carotenoids in Cara Cara navel orange (Tao et al. 2007). In this research, we further characterize the function of CsPSY to investigate its possibilities in carotenoid improvement in citrus. After cloning of the gene, sequence analysis of the CsPSY indicated that there is one transmembrane domain located in 260–282 amino acids (Fig. 2b), which demonstrate that CsPSY is a membrane-bound protein. Moreover, a typical cleavable transit sequence for plastid targeting in CsPSY (Fig. 2a) was predicted to be the N-terminal residues 1–44; this result is consistent with previous results in Arabidopsis (Scolnik and Bartley 1994), tomato (Ray et al. 1992), and sunflower (Salvini et al. 2005) which are 1–70, 1–62, and 1–67 residues, respectively.
The catalytic activity of carotenoid biosynthetic enzymes could be investigated by heterologous expression in E. coli, which has proven to be a powerful tool for characterizing carotenogenic genes (Chamovitz et al. 1992; Cunningham et al. 1993). Many carotenoid biosynthetic enzymes from plants, such as tomato, Arabidopsis, and pepper, are functionally expressed in E. coli systems (Misawa et al. 1994; Pecker et al. 1996; Sun et al. 1996; Bouvier et al. 1998; Tian and DellaPenna 2001; Cunningham and Gantt 2001), suggesting that enzymes from phylogenetically distant species can be associated into a functional multi-enzyme complex. In the present paper, the expression of CsPSY in E. coli cells confirmed that it encodes a functional PSY, which converted two molecular GGPP to phytoene. Kim et al. (2003) have reported a citrus PSY from C. unshiu, which truncated 89 amino acids in its N-terminus when compared with CsPSY herein; and its catalytic activity is similar to CsPSY in this study, suggesting that the presence of transit peptide did not severely inhibit its catalytic activities. The amount of phytoene produced by CsPSY in E. coli cells was very much lower (15–16%) than that produced by E. uredovora crtB (Fig. 3). This might have been caused by the different solubility between CsPSY and CrtB proteins or enzymatic activities.
PSY is a key enzyme in carotenoid biosynthesis and it is the branching enzyme that determines the flux of carbon source toward carotenoids (Bramley et al. 1992; Shewmaker et al. 1999); therefore, it has been the target of genetic manipulation in many plants. Increasing the activity of PSY leads to substantial increases in carotenoid accumulation and/or results in carotenoid accumulation in plant tissues that do not normally produce carotenoid pigments in tobacco, tomato, rice, Arabidopsis, and rapeseed (Fraser et al. 2002; Busch et al. 2002; Ye et al. 2000; Lindgren et al. 2003; Shewmaker et al. 1999). In the present study, the overexpression of the CsPSY gene in Hongkong kumquat resulted in a significant accumulation of phytoene in transgenic fruits. Increases of lycopene, β-carotene, and β-cryptoxanthin were also observed in the transgenic fruits. These results suggest that the flux of phytoene synthesis from GGPP was first promoted by expressed CsPSY gene product (CsPSY) in the fruits, and phytoene was consecutively decomposed to the downstream metabolites lycopene, β-carotene, β-cryptoxanthin, violaxanthin and lutein, as catalyzed by endogenous carotenoid biosynthetic enzymes such as PDS, ZDS, CRTISO, LCYb, LCYe, HYb, and ZEP in fruits (Fig. 1), which was also suggested in other plant species (Shewmaker et al. 1999; Kato et al. 2004). Increasing carbon flux into the carotenoid pathway can produce other phenotypic effects on plant development because the GGPP is an intermediate common to many pathways (Fig. 1). For example, tomato plants with constitutive overexpression of the fruit-related PSY1 showed dwarfism, spindly vegetative tissue, smaller fruit and reduced gibberellin levels because of redirecting GGPP from the respective pathway (Fray et al. 1995). Also, in tobacco, the constitutive expression of a PSY gene resulted in severe phenotypic effects including dwarfism, altered leaf morphology, and pigmentation (Busch et al. 2002). However, besides the fruits color, no significant phenotypic differences were observed in the CsPSY transgenic lines showing elevated carotenoids in the ripe fruit. It is possible that Hongkong kumquat plants, as a wild fruit tree, may possess some strong ability to mask the detrimental effects caused by expression of a foreign gene during their growth and development.
In conclusion, we functionally characterized a PSY cDNA from Cara Cara navel orange. The high sequence similarities, but more importantly the accumulation of phytoene in CsPSY-expressing transgenic lines and in E. coli cell transformed with the CsPSY construct, indicate that the CsPSY gene encodes a functional PSY and could be an important regulatory step in carotenoid accumulation in citrus fruit. Therefore, CsPSY will have potential for metabolite engineering toward altering pigmentation and enhancing nutritional value of citrus, and producing valuable carotenoids in microorganism.
Abbreviations
- PSY:
-
Phytoene synthase
- cDNA:
-
Complementary DNA
- GGPP:
-
Geranylgeranyl pyrophosphate
- HPLC:
-
High-performance liquid chromatography
- BA:
-
6-Benzyladenine
- IBA:
-
Indole butyric acid
- KT:
-
Kinetin
- NAA:
-
α-Napthaleneacetic acid
- NPT II:
-
Neomycin phosphotransferase II
References
Alquezar B, Rodrigo MJ, Zacarías L (2008) Regulation of carotenoid biosynthesis during fruit maturation in the red-fleshed orange mutant Cara Cara. Phytochemistry 69:1997–2007
Alquezar B, Zacarias L, Rodrigo MJ (2009) Molecular and functional characterization of a novel chromoplast-specific lycopene β-cyclase from Citrus and its relation to lycopene accumulation. J Exp Bot 60:1783–1797
Bartley GE, Scolnik PA (1995) Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7:1027–1038
Botella-Pavía P, Rodríguez-Concepión M (2006) Carotenoid biotechnology in plants for nutritionally improved foods. Physiol Plant 126:369–381
Bouvier F, Hugueney P, d’Harlingue A, Kuntz M, Camara B (1994) Xanthophyll biosynthesis in chromoplast: isolation and molecular cloning of an enzyme catalyzing the conversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant J 6:45–54
Bouvier F, Keller Y, d’Harlingue A, Camara B (1998) Xanthophyll biosynthesis: molecular and functional characterization of carotenoid hydroxylase from pepper fruits (Capsicum annuum L.). Biochim Biophys Acta 1391:320–328
Bouvier F, Suire C, Mutterer J, Camara B (2003) Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 15:47–62
Bramley P, Teulieres C, Blain I, Bird C, Schuch W (1992) Biochemical characterization of transgenic tomato plants in which carotenoid synthesis has been inhibited through the expression of antisense RNA to pTOM5. Plant J 2:343–349
Busch M, Seuter A, Hain R (2002) Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol 128:439–453
Chamovitz D, Misawa N, Sandmann G, Hirschberg J (1992) Molecular cloning and expression in Escherichia coli of a cyanobacterial gene coding for phytoene synthase, a carotenoid biosynthesis enzyme. FEBS Lett 296:305–310
Cheng YJ, Guo WW, Yi HL, Pang XM, Deng XX (2003) An efficient protocol for genomic DNA extraction from Citrus species. Plant Mol Biol Rep 21:177a–177g
Cunningham FX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Phys 49:557–583
Cunningham FX, Gantt E (2001) One ring or two? Determination of ring number in carotenoids by lycopene ε-lycopene cyclase. Proc Natl Acad Sci USA 98:2905–2910
Cunningham FX Jr, Chamovitz D, Misawa N, Gantt E, Hirschberg J (1993) Cloning and functional expression in Escherichia coli of a cyanobacterial gene for lycopene cyclase, the enzyme that catalyzes the biosynthesis of β-carotene. FEBS Lett 328:130–138
Demmig-Adams B, Adams WW III (2002) Antioxidants in photosynthesis and human nutrition. Science 298:2149–2153
Duan YX, Liu X, Fan J, Li DL, Wu RC, Guo WW (2007) Multiple shoot induction from seedling epicotyls and transgenic citrus plant regeneration containing the green fluorescent protein gene. Bot Stud 48:165–171
Ducreux LJM, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA (2005) Metabolic engineering of high carotenoid potato tubers containing enhanced levels of β-carotene and lutein. J Exp Bot 409:81–89
Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43:228–265
Fraser PD, Rőmer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM (2002) Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit specific manner. Proc Natl Acad Sci USA 99:1092–1097
Fray RG, Wallace A, Fraser PD, Valero D, Hedden P, Bramley PM, Grierson D (1995) Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J 8:693–701
Fujisawa M, Watanabe M, Choi SK, Teramoto M, Ohyama K, Misawa N (2008) Enrichment of carotenoids in flaxseed (Linum usitatissimum) by metabolic engineering with introduction of bacterial phytoene synthase gene crtB. J Biosci Bioeng 105:636–641
Giuliano G, Bartley GE, Scolnik PA (1993) Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5:379–387
Ikoma Y, Komatsu A, Kita M, Ogawa K, Omura M, Yano M, Moriguchi T (2001) Expression of a phytoene synthase gene and characteristic carotenoid accumulation during citrus fruit development. Physiol Plant 111:232–238
Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in Citrus fruit. Plant Physiol 134:824–837
Kim IJ, Ko KC, Kim CS, Chung WL (2001) Isolation and expression patterns of a cDNA encoding phytoene synthase in Citrus. J Plant Physiol 158:795–800
Kim IJ, Ko KC, Nam TS, Kim YW, Chung WII, Kim CS (2003) Expression and activity of citrus phytoene synthase and β-carotene hydroxylase in Escherichia coli. J Microbiol 41:212–218
Lee HS (2001) Characterization of carotenoids in juice of red navel orange (Cara Cara). J Agric Food Chem 49:2563–2568
Lindgren LO, Stålberg KG, Höglund AS (2003) Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol 132:779–785
Liu YZ, Liu Q, Tao NG, Deng XX (2006) Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck). J Huazhong Agric Univ 25:300–304
Liu Q, Xu J, Liu YZ, Zhao XL, Deng XX, Guo LL, Gu JQ (2007) A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis Osbeck). J Exp Bot 58:4161–4171
Lyi SM, Zhou X, Kochian LV, Li L (2007) Biochemical and molecular characterization of the homocysteine S-methyltransferase from broccoli (Brassica oleracea var. italica). Phytochemistry 68:1112–1119
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expression in Escherichia coli. J Bacteriol 172:6704–6712
Misawa N, Truesdale MR, Sandmann G, Fraser PD, Bird C, Schuch W, Bramley PM (1994) Expression of a tomato cDNA coding for phytoene synthase in Escherichia coli, phytoene formation in vivo and in vitro, and functional analysis of the various truncated gene products. J Biochem 116:980–985
Paine JA, Shipton CA, Chaggar S (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23:482–487
Pecker I, Gabbay R, Cunningham FX, Hirschberg J (1996) Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol Biol 30:807–819
Ray J, Moureau P, Bird C, Bird A, Grierson D, Maunders M, Truesdale M, Bramley M, Schuch W (1992) Cloning and characterization of gene involved in phytoene synthesis from tomato. Plant Mol Biol 19:401–404
Saitou N, Nei N (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salvini M, Bernini A, Fambrini M, Pugliesi C (2005) cDNA cloning and expression of the phytoene synthase gene in sunflower. J Plant Physiol 162:479–484
Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276:1872–1874
Scolnik PA, Bartley GE (1994) Nucleotide sequence of an Arabidopsis cDNA for phytoene synthase. Plant Physiol 104:1471–1472
Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20:401–412
Simkin AJ, Schwartz SH, Auldridge M, Taylor MG, Klee HJ (2004) The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J 40:882–892
Sun Z, Gantt E, Cunningham FX (1996) Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271:24349–24352
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tao NG, Hu ZY, Liu Q, Xu J, Cheng YJ, Guo LL, Guo WW, Deng XX (2007) Expression of phytoene synthase gene (Psy) is enhanced during fruit ripening of Cara Cara navel orange (Citrus sinensis Osbeck). Plant Cell Rep 26:837–843
Tian L, DellaPenna D (2001) Characterization of a second carotenoid beta-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol Biol 47:379–388
Xu CJ, Fraser PD, Wang WJ, Bramley PM (2006) Differences in the carotenoid content of ordinary citrus and lycopene-accumulating mutants. J Agric Food Chem 54:5474–5481
Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Acknowledgments
We are grateful to Dr. Norihiko Misawa (Marine Biotechnology Institute, Iwate, Japan) for the kindly providing us the plasmids. This research was supported by NSFC (30830078).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Lu.
Rights and permissions
About this article
Cite this article
Zhang, J., Tao, N., Xu, Q. et al. Functional characterization of Citrus PSY gene in Hongkong kumquat (Fortunella hindsii Swingle). Plant Cell Rep 28, 1737–1746 (2009). https://doi.org/10.1007/s00299-009-0774-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-009-0774-3