Abstract
Citrus psorosis is a serious viral disease affecting citrus trees in many countries. Its causal agent is Citrus psorosis virus (CPsV), the type member of genus Ophiovirus. CPsV infects most important citrus varieties, including oranges, mandarins and grapefruits, as well as hybrids and citrus relatives used as rootstocks. Certification programs have not been sufficient to control the disease and no sources of natural resistance have been found. Pathogen-derived resistance (PDR) can provide an efficient alternative to control viral diseases in their hosts. For this purpose, we have produced 21 independent lines of sweet orange expressing the coat protein gene of CPsV and five of them were challenged with the homologous CPV 4 isolate. Two different viral loads were evaluated to challenge the transgenic plants, but so far, no resistance or tolerance has been found in any line after 1 year of observations. In contrast, after inoculation all lines showed characteristic symptoms of psorosis in the greenhouse. The transgenic lines expressed low and variable amounts of the cp gene and no correlation was found between copy number and transgene expression. One line contained three copies of the cp gene, expressed low amounts of the mRNA and no coat protein. The ORF was cytosine methylated suggesting a PTGS mechanism, although the transformant failed to protect against the viral load used. Possible causes for the failed protection against the CPsV are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Citrus psorosis, first described by Swingle and Webber (1896), is a serious viral disease affecting citrus trees in many countries (Roistacher 1993). The distribution of the disease is well documented and its presence has been confirmed in the Americas, Africa and the Mediterranean Basin. In Argentina, a certification program has not been sufficient to control the disease that is causing important losses (Danós 1990). It appears that the causal agent is spread probably by an unknown vector (Beñatena and Portillo 1984). New reports have been published describing the presence of psorosis in Texas, USA (Palle et al. 2004), another region of the world where psorosis appears to spread naturally. Usually, the symptoms appear on 10–15-year-old trees, when they are at the stage of maximum productivity. In the field, the characteristic symptoms on sweet orange, mandarin and grapefruit are bark scaling on the trunk and chlorotic flecks and spots on young leaves (Roistacher 1991, 1993) These foliar symptoms are also observed in experimentally graft-inoculated sweet orange seedlings that are commonly used as biological indicators for the indexing of psorosis (Roistacher 1991). In this test, a shock reaction with shoot necrosis can be observed in the first flush, followed by chlorotic leaf flecking and spotting in subsequent flushes.
The causal agent of the disease is Citrus psorosis virus (CPsV), the type member of genus Ophiovirus (García et al. 1994; Milne et al. 2003). The virus is tripartite and its genome consists of three ssRNAs of negative polarity. CPsV RNA 1 codifies for two ORFs, a 24 kDa polypeptide of unknown function (24 K), and separated by an intergenic region, the putative RNA polymerase (280 kDa) (Naum-Onganía et al. 2003). RNA 2 contains one ORF, encoding a polypeptide of 54.7 kDa of unknown function (54 K), and RNA 3 codes for the coat protein (CP) of 48.6 kDa (48 K) (Sánchez de la Torre et al. 1998, 2002).
At present, CPsV is identified using a triple sandwich immunoassay (TAS-ELISA) detecting the CP (Zanek et al. 2006) or by RT-PCR for viral RNA detection. CPsV infects most Citrus varieties and has also been found in Poncirus trifoliata L. Raf. and in Troyer and Carrizo citrange hybrids (Poncirus trifoliata L. Raf. x Citrus sinensis L. Osbeck), that are used as rootstocks in many countries, including Argentina and Uruguay. There are no known sources of natural resistance or tolerance to the disease.
Pathogen-derived resistance (PDR) has been used as a means to control viral diseases through the incorporation of viral-derived sequences into transgenic plants (Sanford and Johnston 1985; Powell-Abel et al. 1986; Beachy et al. 1990; Baulcombe 1996; Goldbach et al. 2003). In this paper, we describe the production of transgenic plants of sweet orange expressing the cp gene of CPsV, their challenge with the virus, and their evaluation for resistance.
Materials and methods
Virus isolates, plasmid constructs and bacterial strain
In this work, isolate CPV-4 was used. This isolate is considered from Florida (USA) where it was isolated first (Garnsey and Timmer 1980), although it probably originated in Texas (Garnsey et al. 1976).
The full-length ORF of cp gene from isolate CPV-4 was amplified by RT-PCR using the CPV3 (5′ CCTAACGCTAGCATGTCGATTCCTATTAAAG 3′) and the CPV4 (5′ GACGAATTCTTAAAAGCATAACATGCAAGC 3′) primers (Barthe et al. 1998) and it was cloned subsequently in the pGEM-T vector. The resulting plasmid was digested with NheI and EcoRI to release the cp gene. The protruding ends thus formed were filled and inserted between the Cauliflower mosaic virus (CaMV) 35S promoter, with double enhancer plus Alfalfa mosaic virus (AMV) RNA 4 leader sequence and the nopaline synthase gene (nos) terminator sequence of plasmid pMOG180 (Mogen International). Correct cloning and insert orientation were confirmed by sequencing. The resulting expression cassette was then subcloned into the binary vector p35SGUSINT (Vancanneyt et al. 1990) at the EcoRI site, generating p35SGUSINT/CP (see Fig. 1a). This plasmid was transformed into Agrobacterium tumefaciens strain EHA 105 by electroporation.
a Schematic representation of the T-DNA from the binary plasmid p35SGUSINT/CP engineered to express the cp gene (2,440 bp) from the CPsV strain CPV-4 in transgenic sweet orange plants. The gene is controlled by the CaMV 35S promoter (35SP) containing a duplicated enhancer region, the Alfalfa mosaic virus RNA4 leader sequence (AlMV-RNA4-leader) and the nos terminator (nosT). The cp coding region (ORF-cp) is flanked by the nptII gene between the nos promoter (nosP) and the nosT, and by the uidA gene (GUS) between the 35SP and the nosT. Transcription orientation of each gene is indicated by the arrows, and EcoRI and HindIII restriction sites are designated by vertical lines. The relative size of the CP probe and the expected hybridization products are indicated. RB right border, LB left border. b Southern blot analysis of total DNA extracted from the transgenic lines (1, 18, 20, 21, 23, 28, 29, 34 and 96) digested with EcoRI that releases the complete cp gene. c Southern blot analysis of total DNA extracted from the same transgenic lines digested with HindIII, which shows the copy number of the transgene integrated. The nylon membranes were probed with a digoxigenin-11-dUTP-labeled fragment of the cp coding region (CPV1-CPV2 probe). d Northern blot analysis of total RNA extracted from the transgenic lines (1, 18, 20, 21, 23, 28, 29, 34 and 96) and probed with [α-32P-CTP]-labeled DNA fragment of the cp gene (CPV1-CPV2 probe). e Total RNA extracted from the transgenic plants stained with ethidium bromide after gel electrophoresis showing RNA loads
Transformation and regeneration
Internodal stem segments from 6- to 12-month-old greenhouse-grown Pineapple sweet orange (Citrus sinensis L. Osbeck) seedlings were transformed by cocultivation with A. tumefaciens EHA 105 carrying the binary vector p35SGUSINT/CP. Selection of transformants was performed in a culture medium containing kanamycin (100 μg/ml) (Peña et al. 1995).
The basal part of the shoots regenerated from the stem segments was assayed for uidA gene expression (GUS) by overnight incubation in a 2 mM X-Gluc solution at 37°C (Cervera et al. 2005). When the reaction was positive, shoot tips were grafted in vitro on decapitated and etiolated seedlings of Troyer citrange as previously described (Peña et al. 1995). After 3–4 weeks, scions had developed several leaves which were then screened for the presence of the cp transgene by PCR. Positive transgenic plantlets were then side-grafted on vigorous 6-month-old seedlings of rough lemon (Citrus jambhiri Lush) and grown in a greenhouse maintained at 24–26°C, with an 18 h day length provided by a mixture of natural and supplemental light. The presence of the cp and uidA genes in the GUS positive transgenic lines was verified by PCR using the CPV1 (5′-GCTTCCTGGAAAAGCTGATG-3′) and the CPV2 (5′-TCTGTTTTTGTCAACACACTC-3′) primers designed from the cp gene (Barthe et al. 1998) and GUS1 (5′-GGTGGGAAAGCGCGTTACAAG-3′) and GUS2 (5′-TGGATCCCGGCATAGTTAAA-3′) from the uidA gene (Peña et al. 1995). A regenerated shoot without agro-infection was obtained and used as a regenerated non-transgenic control.
Southern and northern blot analyses
Southern blot assays were performed to analyze the integrity of the cp expression cassette, to estimate the number of copies of the cp gene in the transgenic plant genome and to analyze the possibility of transgene methylation.
In order to analyze the integrity and the number of copies of the cp transgene, DNA was extracted from 0.75 g of leaves according to Futterer et al. (1995), and aliquots of 20 μg were digested with EcoRI, which releases the cp expression cassette, or with HindIII which cuts outside of this cassette. The digestion products were fractioned in 0.8%-TAE agarose gel, and then blotted onto positively charged nylon membranes (ROCHE) by capillarity using 20× SSC buffer. After fixation, by baking at 120°C, membranes were incubated with a digoxigenin-11-dUTP-labeled fragment of the cp gene (CPV1-CPV2) prepared by PCR according to the manufacturer’s instructions (ROCHE Applied Science) or probed with a [α-32P-CTP]-labeled fragment from the cp gene (CPV1, CPV2).
For the analysis of transgene methylation, 20 μg of total DNA extracted from 0.75 g of leaves from lines 23, 96 and non-transformed controls were digested with the restriction enzyme EcoRI and the methylation-sensitive restriction enzyme DdeI. After digestion, DNA fragments were fractioned in 1.5%-TAE agarose gel, and then blotted onto positively charged nylon membrane (ROCHE). The membrane was incubated with two digoxigenin-11-dUTP-labeled fragments of the cp gene, prepared by PCR using the CPV1 and CPV2 primers and C2 (5′-TATTGAGCTCAGCATCTGGG-3′) and 48Kdir (5′-TAAGCTCGAGCAATGTCGATTCC-3′) primers.
Hybridization with the digoxigenin-11-dUTP-labeled probe was performed in 5× SSC, 2% Blocking Reagent (ROCHE), 0.1% N-lauroylsarcosine, 0.2% SDS and 50 mM sodium phosphate (pH7) overnight at 65°C. After three washing steps in 2× SSC, 0.2× SSC and 0.1× SSC plus 0.1% SDS at 65°C, the detection was done by a chemiluminescent substrate according to the manufacturer’s instructions (ROCHE) and exposed for 18 h to X-ray film. Hybridization with the [α-32P-CTP]-labeled probe was performed in 5× SSC, 5× Denhardt, 0.2% SDS and 500 μg/ml denatured herring sperm DNA (Invitrogen) overnight at 65°C. After three washing steps in 2× SSC, 1× SSC and 0.1× SSC plus 0.1% SDS at 65°C, membrane was exposed for 30 days to X-ray film.
For northern blot analysis, 40 μg of total RNA extracted from 1 g of citrus leaves were isolated according to Bekesiova et al. (1999). The samples were incubated for 5 min at 95°C in a mixture containing formamide and formaldehyde and then fractionated by electrophoresis in 1% agarose–formaldehyde denaturing gel in MOPS buffer. Nucleic acids were transferred to a positively charged nylon membrane (ROCHE) by capillarity using 20× SSC buffer. After fixation through UV exposure, the membrane was probed with a [α-32P-CTP]-labeled fragment from the cp gene by PCR with CPV1 and CPV2 primers. Hybridization was performed in 5× SSC, 2× Denhardt, 0.1% SDS, 300 μg/ml denatured herring sperm DNA (Invitrogen) and 3 mg/ml denatured yeast tRNA (SIGMA) overnight at 65°C. After three washing steps in 2× SSC, 0.2× SSC and 0.1× SSC plus 0.1% SDS at 65°C, membrane was exposed for 30 days to X-ray film.
Accumulation of CP in the transgenic lines
In order to determine the level of accumulation of the transgene-derived CP, triple-sandwich immunoassay (TAS-ELISA-HRP) was performed as previously described by Zanek et al. (2006). Briefly, total protein was extracted from 200 mg of leaf tissue by grinding with liquid nitrogen using PBS-Tween, 2% w/v polyvinyl pyrrolidone (PVP-40000), and 2.5% w/v of defatted milk powder. Plate coating, antibody incubations and washing conditions were performed as described previously, using 1/2,500 dilution of polyclonal anti-serum A376 (kindly provided by Pedro Moreno, IVIA, Spain) and 1/3,000 dilution of Goat Anti-Mouse IgG (H + L) Horseradish Peroxidase (PIERCE). The substrate was incubated for 30 min at room temperature and the reaction stopped with 0.7 M H2SO4. Optical density (OD) at 492 nm was measured in a Tecan Spectra (Austria).
Samples were taken from six to seven times for 4 months and the data were subjected to analysis of variance using StatSoft, Inc. (2003) STATISTICA (data analysis software system), version 6, to determine the significance of differences in the accumulation of CP in transgenic plants. Means were separated using the Fisher LSD test.
Propagation and graft-inoculation
Transgenic lines were propagated by bud grafting onto 6- to 8-month-old rough lemon seedlings as rootstocks, since rough lemon is vigorous and thus, ensures fast growth of the transgenic scion.
After 10 months, replicates of each line were graft-inoculated on the transgenic scion using pieces of infected bark tissue (bark pieces) carrying the CPV-4 isolate, from which cp transgene was generated.
In a first assay, 3–4 of the 6 replicates of the transgenic lines 21, 34, 28 and 96 were graft-inoculated with two bark pieces that remained in the plants throughout the challenge period. The plants were maintained in a light-controlled greenhouse at 22–24°C. They were observed for development of symptoms (rings, spots and flecking) in four successive flushes for 4.5 months. After each observation, leaf tissues were collected and analyzed by RT-PCR and TAS-ELISA as described below. Controls for these experiments were a non-transformed and a regenerated non-transgenic plant.
Two different conditions were tested in which the number of bark pieces and the time they remained as grafts (time of virus transmission) were varied to evaluate a lowest viral load sufficient to infect all plants. For this purpose, ten non-transgenic sweet orange seedlings were graft-inoculated with one infected bark piece carrying the CPV-4 isolate. After 10 days, the bark pieces were removed from four plants. In another four plants, the bark pieces were removed 16 days post inoculation (dpi). In the other two plants, considered as positive controls, the bark piece remained grafted throughout the experiment. Symptoms were observed in new flushes from all plants 25 dpi. To confirm the presence of the virus and since the plants were not transgenic, RT-PCR using CPV1 and CPV2 primers for detection of the cp gene (RNA 3) was applied on total RNA extracted from foliar tissue.
For a second challenge assay, buds from transgenic lines 28, 96 and 23 were propagated by grafting onto 6- to 8-month-old rough lemon seedlings. After 10 months and depending on the results of the viral load experiment, 6 of the 8 replicates were graft-inoculated on the scion with only one bark piece carrying the CPV-4 isolate and 16 days later the inoculum was removed. The plants, maintained in the greenhouse, were inspected individually for symptoms development in four successive flushes for 6 months and after 1 year.
Virus detection
RT-PCR using the primers designed from CPsV RNA 2 was used to test the viral infection in the transgenic lines. For amplification, the forward primer was A (5′ TAGATCCATGCTCAGTCACC 3′) and the reverse primer was B (5′ TTGACAATGATGGACACTGG 3′).
Total RNA was extracted from 100 mg of fresh tissue with TRIZOL (Invitrogen), following the manufacturer´s instructions. RNA samples were resuspended in 50 μl of RNase-free water. Total RNA weighing 1 μl was used for one-step RT-PCR which was conducted in a 15 μl reaction mixture containing 1 mM dNTPs, 2.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl pH 9.0 (at room temperature), 0.1% triton X-100, 1 μM of each primer, 16 units RNAsin® Ribonuclease Inhibitor (Promega), 12 units SuperScript™ III Reverse Transcriptase (Invitrogen) and 1 unit of Taq DNA polymerase (Promega). The thermocycling conditions included 30 min at 48°C for RT, 4 min at 94°C for inactivation of reverse transcriptase and initial denaturation, 36 cycles of 10 s at 94°C, 10 s at 50°C and 50 s at 72°C and a final elongation step of 4 min at 72°C in Perkin-Elmer 2400 equipment. PCR products were analyzed by electrophoresis on 2%-TAE agarose gel stained with ethidium bromide.
TAS-ELISA was the assay chosen to test the progress of the CPsV infection in the plants. Using this method for diagnosis, a sample is considered positive for CPsV when its OD value is ≥3 times the healthy control (Alioto et al. 1999).
Results
Production of transgenic sweet orange plants and their analysis
Twenty-one independent transgenic plants carrying the cp gene were obtained by A. tumefaciens-mediated transformation with the plasmid p35SGUSINT/CP. After performing GUS assays from small pieces of kanamycin-resistant regenerated shoots, positive scions were grafted in vitro on decapitated seedlings. Small pieces of leaf tissue of each line were tested by PCR in order to confirm the presence of the cp gene. Transformation efficiency (transgenic lines per inoculated explant) varied from 3.3 to 4.5%.
Figure 1b, c shows the integrity of the transgene and the number of copies inserted in each of nine transgenic lines, respectively, as determined by Southern blot. Most transformed plants had integrated one transgene copy (lines 1, 18, 20, 21, 28, 29 and 34). Lines 96 and 23 contained two and three copies, respectively.
Lines 18, 28, 21, 29, 34 and 96 showed higher transgene expression than lines 1, 23, and 20 in several northern blots. As examples, results from two northern blots revealing different levels of cp expression are shown in Fig. 1d.
There was no correlation between the transgene copy number and the expression of the cp mRNA in these nine lines. The seven lines with one copy showed different levels of mRNA. For example, lines 34 and 96 containing one and two copies of the transgene, respectively, expressed similar amount of cp mRNA. In line 23, cp mRNA detection was difficult, and in fact, only one of three northern blots showed a faint band (Fig. 1d).
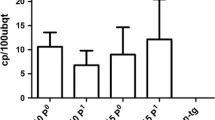
CP could not be detected by western blot in any of the lines using the available anti-sera (data not shown). However, TAS-ELISA-HRP, which is a more sensitive ELISA, was able to detect CP quantitatively. Figure 2 shows the optical density (OD), and the statistical values obtained in the nine lines and a non-transgenic plant used as control. In general, most lines accumulated low amounts of CP, but the amounts observed in lines 18, 20, 21, 28, 29, 34 (P < 0.01), and line 1 (P < 0.05) were significantly higher that in the non-transgenic control. All transgenic plants displayed normal growth and development, similar to that of non-transgenic controls.
TAS-ELISA-HRP for the detection of CPsV CP in transgenic citrus plants. The OD values refer to the mean optical densities for six to seven independent experiments. Small black boxes indicate the mean, the empty boxes the mean ± SE and the bars the mean ± SD. The OD values differences were analyzed according to the Fisher LSD test. a represents a statistically significant difference in OD value mean compared with non-transgenic line (healthy) at P < 0.01. b represents a statistically significant difference in OD value mean compared with non-transgenic line (healthy) at P < 0.05
Challenge of transgenic lines with the CPV-4 isolate
To investigate the response of the transgenic plants against viral infection, lines 21, 23, 28, 34, and 96 were challenged with CPV-4 by graft-inoculation. In these experiments, the non-transgenic and the regenerated non-transgenic plants were considered as the same control (C), since, when infected, both showed the characteristic symptoms of psorosis.
In the first experiment, four replicates of lines 21, 28, 34 and 96 were tested. In the first flush, the presence of the virus was determined by RT-PCR using the RNA 2 primers. TAS-ELISA was applied to measure virus multiplication quantitatively. The new flushes of the challenged plants had higher OD values than in the non-infected transgenic lines and were similar to those of infected controls. RT-PCR for detection of the viral RNA 2, TAS-ELISA, and symptom expression confirmed virus accumulation in the four lines throughout the 4 months of observation (data not shown). Some replicates of lines 28 and 96 died or grew slowly, for this reason they were analyzed again in a second experiment.
Since the two infected bark pieces used for grafting-inoculation might carry an excessive viral load, a less aggressive inoculum was tested in non-transgenic sweet orange plants before the start of the second challenge experiment. One bark piece infected with CPV-4 was grafted and removed either 10 or 16 dpi. Twenty-five days after inoculation, three of the four inoculated plants (10 dpi) and four of the four inoculated plants (16 dpi) developed symptoms of psorosis. The presence of the virus in the symptomatic plants was confirmed by RT-PCR.
Therefore, the second challenge was performed by grafting with one bark piece for 16 dpi. Lines 28, 96 and 23, containing 1, 2 and 3 copies of the cp gene, respectively, developed characteristic symptoms of psorosis (rings, spotting and flecking) for 1 year and no differences were noticed among lines or among replicates of the same line (Fig. 3). These plants were analyzed individually by RT-PCR in the first flush and the RNA 2 of CPsV was detected (data not shown). TAS-ELISA was applied for the three successive flushes after inoculation (4 months) to quantify the virus multiplication. As shown in Fig. 4, the three lines gave OD values several times higher than those of the non-infected transgenic plants and were similar to those of infected controls (C-I). Moreover, there was no recovery from symptoms several months later, indicating that plants were still infected.
Evaluation of transgenic lines challenged with CPsV. Analysis of virus accumulation of six replicates of lines 96, 23, 28 and the control plant (C) by TAS-ELISA-HRP. The OD values refer to mean optical densities for the six replicates infected with CPV-4 isolate for 96 (96-I), 23 (23-I) and 28 (28-I) transgenic lines in the three successive flushes for 4 months. The OD values for mean optical densities for two non-inoculated replicates of each transgenic line analyzed are included in the graph (96-NI, 23-NI and 28-NI). C-I refers to mean for infected control. C-NI refers to mean for non-infected control. Standard deviations are indicated by bars
Analysis of transgene methylation
Line 23 contained three copies of the transgene, but mRNA was barely detectable in this line, and CP accumulation was negative, suggesting that the cp transgene could be silenced in this line. For this reason, methylation of the transgene in line 23 was analyzed. Line 96 was included for comparison with line 23 because it had two copies of the transgene and expressed detectable CP mRNA and no CP.
Figure 5 shows the results of the Southern blot from total DNA digested with EcoRI to excise the cassette, and total DNA digested with DdeI, which is cytosine methylation-sensitive. Line 96 showed the same digestion pattern as the binary plasmid p35SGUSINT/CP, suggesting that the transgene was not methylated in this case. On the other hand, the pattern obtained in line 23 showed a band of about 2,400 nt corresponding to the fragment digested by EcoRI and not by the DdeI, indicating methylation of the ORF region. Another faint band of about 2,000 nt could be the product of partial promoter methylation.
Analysis of transgene methylation from CP lines. a Schematic representation of the T-DNA fragment containing the expression cassette of the cp gene showing the resulting fragments after digestion with the restriction enzyme EcoRI and the methylation-sensitive restriction enzyme DdeI. The relative sizes of cp probes and the expected hybridization products are indicated. b Southern blot hybridization of DNA from the binary plasmid p35SGUSINT/CP (lane 1), non-transgenic control (lane 2) and lines 23 and 96 (lanes 3 and 4, respectively) digested with EcoRI and DdeI. The nylon membrane was probed with digoxigenin-11-dUTP-labeled fragments of the cp coding region (CPV1-CPV2 and C2-48Kdir probes). The size of DNA markers are indicated in bp
Discussion
Twenty-one transgenic lines containing the cp gene of Citrus psorosis virus (CPV-4 isolate) were obtained and nine were evaluated. The integrity of the transgene and the mRNA expression were analyzed and showed that the transgene had been integrated and expressed in the plant genome.
Transgenic lines grew normally and were indistinguishable morphologically from non-transgenic Pineapple sweet orange controls. Thus, this result is an indication that the expression of cp gene was insufficient under the conditions tested to produce psorosis symptoms as described for transgenic citrus lines expressing the p23 protein of Citrus tristeza virus (CTV) (Ghorbel et al. 2001). It has also been reported that CP expression from the transgene was able to promote virus accumulation in some cases (Kouassi et al. 2006). In our case, in the first flush after inoculation, none of the lines showed enhanced symptoms over those produced by the non-transgenic controls, indicating there was no increase in susceptibility.
We used TAS-ELISA-HRP, which has the same sensitivity as RT-PCR (Zanek et al. 2006) to detect CP expressed in transgenic citrus plants. Applying this method, some variability in the expression of the transgene in the first 5 months of growth was observed, probably due to the sympodial growth pattern of citrus and the physiological condition of the plants. According to statistical analysis, all lines, except for lines 23 and 96, were low, but positive expressers, of the CP. Variable and non-detectable expression of the transgene has also been observed in transgenic citrus plants with the major CP of CTV (Domínguez et al. 2000) and with other proteins in some woody transgenic plants such as grapevines (Gambino et al. 2005; Maghuly et al. 2006).
Western blots failed to detect CP in transgenic citrus. However, in Nicotiana benthamiana plants transformed with the same construct, the CP was detected by western blot (data not shown) indicating that the whole protein is stable in the herbaceous plant and it could be probably synthesized intact in citrus plants.
In the first challenge experiment, lines 21 and 34 were chosen because they were considered high expressers of the CP, whereas lines 28 and 96 were low expressers. Lines 28 and 96 contained different number of copies of the transgene, and for this reason, they were included in the second experiment. Line 23 contained three copies of methylated transgene, expressed only small amounts of its mRNA, and accumulated negligible amounts of CP, suggesting PTGS of the cp transgene. Thus, it was a good candidate to be challenged since there is usually a correlation between PTGS of the transgene and virus resistance in other transgenic plant-virus combinations (Kalantidis et al. 2002; Scorza et al. 2001; Fagoaga et al. 2006).
In the challenge experiments, the virus multiplication was detected by RT-PCR using primers designed for the RNA2, but TAS-ELISA-HRP was used to follow the progress of the infection. The OD values of the non-infected transgenic plants were very low, and when infected, the OD increased during the experiment 5–10 times higher than the non-inoculated controls (Fig. 4).
Since CPsV is probably transmitted by a vector (Beñatena and Portillo 1984; Palle et al. 2004), the virus could take years to multiply in a tree and to manifest symptoms. Furthermore, previous experiments in the greenhouse indicated that graft-transmission of the psorosis agent can occur in a few days (P. Moreno and L. W. Timmer, personal communication). These observations suggested that under greenhouse conditions, the viral inoculum and transmission time could be reduced. The viral load assay revealed that one bark piece in 16 days was enough to transmit the virus and ensured 100% infection. For this reason, in the second challenge, the number of bark pieces and the time of virus transmission were reduced. However, we think that the viral inoculum dose was still too high, since the transgenic plants showed symptoms simultaneously and with the same severity as the controls in which the inoculum was not eliminated. An inherent difficulty in this assay is that as long the rootstock is susceptible to the CPsV, it can move to the rootstock and replicate there delivering virus to the scion. This problem cannot be overcome at the moment because there is no citrus or citrus relative rootstock resistant to CPsV.
None of the evaluated lines was resistant to CPsV or showed any delay in the symptom expression. Moreover, we have not found any significant differences in the response to virus challenge among the lines or among the replicates as reported in transgenic citrus expressing the major CP of CTV (Domínguez et al. 2002). In the CP-mediated PDR strategy, resistance might be provided either by transgene-derived CP over-accumulation or by PTGS induction triggered by transgene expression. If the mechanism of protection were protein mediated, the protein accumulation in the transgenic lines might not be sufficient to prevent viral infection under the conditions assayed (high viral load and young seedlings). If we propose a PTGS mechanism to protect the plants, line 23 would have been a good candidate since it contains three copies of the transgene, expresses a low level of mRNA and no CP, and the ORF is cytosine-methylated. Our results of transgene methylation revealed that this line could have partial promoter methylation. Thus the possibility of TGS induction cannot be discarded. Nevertheless, we speculate that PTGS could have been induced in this line since low level of CP mRNA could be detected.
PDR has been shown to be the means to control virus infection in some plant-virus systems. Nevertheless, most of the PDR has been reported for viruses of positive-stranded genome RNA and applied to transgenic herbaceous hosts. Transformation of citrus and other woody plants like grapevines and plums has been performed with positive-stranded virus genes, such as CTV (Domínguez et al. 2002; Fagoaga et al. 2006), GFLV (Gambino et al. 2005; Maghuly et al. 2006) and Plum pox virus (Ravelonandro et al. 1997; Hily et al. 2004). There are few reports of transformation with genes of negative-stranded genome viruses like Tomato spotted wilt virus, TSWV (Levin et al. 2005; Pang et al. 1992; Prins et al. 1995) and none on negative-stranded viruses in transgenic woody plants.
Several factors could contribute to the lack of protection observed in the transgenic lines evaluated here: (a) the high dose of virus delivered to plants by graft-inoculation could overcome the potential protection afforded by constitutive expression of the cp gene in transgenic cells, (b) the variable level of transgene expression during sympodial growth and development of transgenic plants could affect the PDR mechanism, and/or (c) the CPsV genome could encode proteins breaking putative PTGS at least in line 23.
Abbreviations
- CPsV:
-
Citrus psorosis virus
- CP:
-
Coat protein
- TAS-ELISA:
-
Triple sandwich immunoassay
- PDR:
-
Pathogen-derived resistance
- CaMV:
-
Cauliflower mosaic virus
- AMV:
-
Alfalfa mosaic virus
- nos :
-
Nopaline synthase gene
- OD:
-
Optical density
- dpi:
-
Days post inoculation
- C-I:
-
Infected control
- C-NI:
-
Non-infected control
- PTGS:
-
Post-transcriptional gene silencing
- CTV:
-
Citrus tristeza virus
- GFLV:
-
Grapevine fan-leaf virus
- TSWV:
-
Tomato spotted wilt virus
References
Alioto D, Gangemi M, Deaglio S, Sposato S, Noris E, Luisoni E, Milne RG (1999) Improved detection of citrus psorosis virus using polyclonal and monoclonal antibodies. Plant Pathol 48:735–741. doi:10.1046/j.1365-3059.1999.00410.x
Barthe GA, Ceccardi TL, Manjunath KL, Derrick KS (1998) Citrus psorosis virus: nucleotide sequencing of the coat protein gene and detection by hybridisation and RT-PCR. J Gen Virol 79:1531–1537
Baulcombe DC (1996) Mechanisms of pathogen-derived resistance to viruses in transgenic plants. Plant Cell 8:1833–1844. doi:10.1105/tpc.8.10.1833
Beachy RN, Loesch-Fries S, Tumer NE (1990) Coat protein-mediated resistance against virus infection. Annu Rev Phytopathol 28:451–474
Bekesiova I, Nap J-P, Mlynarova L (1999) Isolation of high Quality DNA and RNA from leaves of the carnivorous plant Drosera rotundifolia. Plant Mol Biol Rep 17:269–277
Beñatena HN, Portillo MM (1984) Natural spread of psorosis in sweet orange seedlings. In: Garnsey SM, Timmer LW and Dodds JA (eds) Proceedings of the 9th conference of the international organization of citrus virologists, IOCV, Riverside, pp 159–164
Cervera M (2005) Histochemical and fluorometric assays for uidA (GUS) gene detection. Methods Mol Biol 286:203–214
Danós E (1990) La psorosis de los cítricos: la epidemia en curso en Argentina y el desafio de su control. In: International Foundation for Science (IFS) e Instituto Nacional de Tecnologia Agropecuaria (INTA) (eds) Revista de Investigaciones Agropecuarias, vol 22, pp 265–277
Domínguez A, Guerri J, Cambra M, Navarro L, Moreno P, Peña L (2000) Efficient production of citrus transgenic plants expressing the coat protein gene of Citrus Tristeza Virus. Plant Cell Rep. 19:427–433. doi:10.1007/s002990050751
Domínguez A, Hermoso de Mendoza A, Guerri J, Cambra M, Navarro L, Moreno P, Peña L (2002) Pathogen-derived resistance to Citrus tristeza virus (CTV) in transgenic Mexican lime (Citrus aurantifolia (Christ. ) Swing.) plants expressing its p25 coat protein gene. Mol Breed 10:1–10. doi:10.1023/A:1020347415333
Fagoaga C, López C, Hermoso de Mendoza A, Moreno P, Navarro L, Flores R, Peña L (2006) Post-transcriptional gene silencing of the p23 silencing supresor of Citrus tristeza virus confers resistance to the virus in transgenic Mexican lime. Plant Mol Biol 60:153-165. doi:10.1007/s11103-005-3129-7
Futterer J, Gisel A, Iglesias V, Kloti A, Kost B, Mittelsten Sceid O, Neuhaus-Url G, Schrott M, Shillito R, Spangenber G, Wang ZY (1995) Standard molecular techniques for analysis of trangenic plants. In: Potrykus I, Spangenberg G (eds) Gene tranfer to plants, Springer Lab manual, vol 25, pp 215
Gambino G, Gribaudo I, Stephan L, Scharti A, Laimer M (2005) Molecular characterization of grapevine plants transformed with GFLV resistance genes:I. Plant Cell Rep 24:665–662. doi:10.1007/s00299-005-0006-4
García ML, Dal Bo E, Grau O, Milne R (1994) The closely related citrus ringspot and citrus psorosis viruses have particles of novel filamentous morphology. J Gen Virol 75:3585–3590
Garnsey SM, Timmer LW (1980) Mechanical transmissibility of citrus ringspot virus isolates from Florida, Texas, and California. In: Calavan EC, Garnsey SM, Timmer LW (eds) Proceedings of the Eighth conference of the international organization of citrus virologists, IOCV, Riverside, pp 174–179
Garnsey SM, Youtsey CO, Bridges GD, Burnett HC (1976) A necrotic ringspot-like virus found in a “Star Ruby” grapefruit tree imported without authorization from Texas. In: Proceedings of the Florida State Horticultural Society, vol 89, pp 63–67
Ghorbel R, Lopez C, Fagoaga C, Moreno P, Navarro L, Flores R, Peña L (2001) Transgenic citrus plants expressing the citrus tristeza virus p23 protein exhibit viral-like symptoms. Mol Plant Pathol 2:27–36
Goldbach R, Bucher, Prins M (2003) Resistance mechanisms to plant viruses: an overview. Virus Res 92:207–212. doi:10.1016/S0168-1702(02)00353-2
Hily JH, Scorza R, Malinowski T, Zawadzka B, Ravelonandro M (2004) Stability of gene silencing-based resistance to Plum pox virus in transgenic plum (Prunus domestica L.) under field conditions. Transgenic Res 13:427–436. doi:10.1007/s11248-004-8702-3
Kalantidis K, Psaradakis S, Tabler M, Tsagris M (2002) The occurrence of CMV-specific short RNAs in transgenic tobacco expressing virus-derived double-stranded RNA is indicative of resistence to the virus. Mol Plant Microbe Interact 15:826–833
Kouassi NK, Chen L, Siré C, Bangratz-Reyser M, Beachy RN, Fauquet CM, Brugidou C (2006) Expression of rice yellow mottle virus coat protein enhances virus infection in transgenic plants. Arch Virol 151:2111–2122. doi:10.1007/s00705-006-0802-3
Levin JS, Thompson WF, Csinos AS, Stephenson MG, Weissinger AK (2005) Matrix attachment regions increase the efficiency and stability of RNA-mediated resistance to Tomato Spotted Wilt Virus in transgenic tobacco. Transgenic Res 14:193–206. doi:10.1007/s11248-004-5413-8
Maghuly F, Stephan L, da Câmara Machado A, Borroto Fernandez E, Mahmood AK, Gambino G, Gribaudo I, Schartl A, Laimer M (2006) Molecular characterization of grapevine plants transformed with GFLV esistance genes: II. Plant Cell Rep 25:546–553. doi:10.1007/s00299-005-0087-0
Milne RG, Garcia ML, Moreno P (2003) Citrus psorosis virus. Association of applied biologists (AAB) descriptions of plant viruses. http://www.dpvweb.net/dpv/showdpv.php?dpvno=401
Naum-Ongania G, Gago-Zachert S, Pena E, Grau O, Garcia ML (2003) Citrus psorosis virus RNA 1 is of negative polarity and potentially encodes in its complementary strand a 24 K protein of unknown function and 280 K putative RNA dependent RNA polymerase. Virus Res 96:49–61. doi:10.1016/S0168-1702(03)00172-2
Palle SR, Miao H, Seyran M, Louzada ES, da Graça JV, Skaria M (2004) Preliminary evidence for natural transmission of citrus psorosis virus by an olpidium-like fungus. In: 16th conference of the international organization of citrus virologists, IOCV, Abstract conference, Mexico
Pang SZ, Nagpala P, Wang M, Slightom JL, Gonsalves D (1992) Resistance to heterologous isolates of tomato spotted wilt virus in transgenic tobacco expressing its nucleocapsid protein gene. Phytopathology 82:1223–1229
Peña L, Cervera M, Juárez J, Navarro A, Pina JA, Durán-Vila N, Navarro L (1995) Agrobacterium-mediated transformation of sweet orange and regeneration of transgenic plant. Plant Cell Rep 14:616–619. doi:10.1007/BF00232724
Powell-Abel P, Nelson RS, De B, Hoffmann N, Rogers SG, Fraley RT, Beachy RN (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 232:738–743. doi:10.1126/science.3457472
Prins M de Haan P, Luyten R, van Veller M, van Grinsven MQ, Goldbach R (1995) Broad resistance to tospoviruses in transgenic tobacco plants expressing three tospoviral nucleoprotein gene sequences. Mol Plant Microbe Interact 8:85–91
Ravelonandro M, Scorza R, Bachelier JC, Labonne G, Levy L, Damsteegt V (1997) Resistance of Prunus domestica L. to plum pox virus infection. Plant Dis 81:1231–1235
Roistacher CN (1991) Graft-transmissible diseases of citrus. In: Food and Agriculture Organization of the United Nations, FAO (ed) Handbook for detection and diagnosis, Rome, pp 115–126
Roistacher CN (1993) Psorosis-A review. In: Moreno P, da Graça JV, Timmer LW (eds) Proceedings of the 12th conference of the international organization of citrus virologists, IOCV, Riverside, pp 139–154
Sánchez de la Torre ME, Riva O, Zandomeni R, Grau O, García ML (1998) Mol Plant Pathol. Online http://www.bspp.org.uk/mppol/1998/1019sanchez
Sánchez de la Torre ME, López C, Grau O, García ML (2002) RNA2 of citrus psorosis virus is of negative polarity and has a single open reading frame in its complementary strand. J Gen Virol 83:1777–1781
Sanford JC, Johnston SA (1985) The concept of parasite-derived resistance—deriving resistance genes from the parasite’s own genome. J Theor Biol 113:395–405
Scorza R, Callahan A, Levy L, Damsteegt V, Webb K, Revelonandro M (2001) Post-transcriptional gene silencing in plum pox virus resistant transgenic European plum containing the plum potyvirus coat protein gene. Transgenic Res 10:201–209. doi:10.1023/A:1016644823203
Swingle WT, Webber HJ (1896) The principal diseases of citrus fruits in Florida. United States Department of Agriculture, Division of Vegetable Physiology and Pathology, Bulletin 8
Vancanneyt G, Schmidt R, O´Connor-Sanchez L, Willmitzer L, Rocha-Sosa M (1990) Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet 220:245–250
Zanek MC, Peña EJ, Reyes CA, Figueroa J, Stein B, Grau O, García ML (2006) Detection of Citrus psorosis virus in the northwestern citrus production area of Argentina by using an improved TAS-ELISA. J Virol Methods 137:245–251. doi:101016/jjviromet200606021
Acknowledgments
We thank Dr. L.W. Timmer for helpful discussion and manuscript revision, and Ing. G. Chiarrone and Pto. Agr. Fabián Ramos for greenhouse work. M. L. García, Eduardo J. Peña, Carina A. Reyes and M. C. Zanek belong to the staff of Facultad de Ciencias Exactas, UNLP, O.G. is recipient of the research career award from CICBA, and MLG from CONICET. Lic. Zanek and Reyes are fellows of CONICET. Lic. E.J. Peña is fellows of ANPCyT. This work was supported by grants from BID802 OC-AR PICT 6198 SECyT-CONICET, AECI, CICBA and INTA EEA Concordia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W. Harwood.
Rights and permissions
About this article
Cite this article
Zanek, M.C., Reyes, C.A., Cervera, M. et al. Genetic transformation of sweet orange with the coat protein gene of Citrus psorosis virus and evaluation of resistance against the virus. Plant Cell Rep 27, 57–66 (2008). https://doi.org/10.1007/s00299-007-0422-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-007-0422-8