Abstract
The β-glucosidase gene of maize (ZmGLU1) was suggested to hydrolyze cytokinin-conjugate and release free cytokinin during plant growth and development. A clone containing the upstream region of ZmGLU1 was isolated and sequenced from a maize genomic library. The full-length ZmGLU1 promoter and a series of its 5′ deletions were fused to the beta-glucuronidase (GUS) reporter gene and transferred into tobacco. The GUS activity of transgenic plants was assayed at various developmental stages. The results showed that ZmGLU1 promoter-driven GUS gene had the highest expression level in the roots and that the expression of GUS gene declined during seed maturation and down to the lowest level in mature seeds. The ZmGLU1 promoter-driven GUS expression increased during seed germination, reaching a peak on day 11. The results also showed that this promoter could be inhibited by 6-BA, trans-zeatin, and NAA, but was not affected by GA3, ABA, SA, cold, salt, drought, and submergence treatments. The histochemical staining revealed that GUS activity was located in vigorous cell division zones with dominant staining associated with vascular tissues. Deletion analysis showed that the promoter contained a putative leaf-specific and stem-specific negative regulative element and two putative enhancers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many plant genes resistant to disease have been discovered and successfully applied to crop improvement. They have mostly been promoted by heterogeneous constitutive promoters, such as the cauliflower mosaic virus (CaMV) 35S promoter, which has increased the burden on plant growth and the biosafety debate. In some cases, specific promoters from plant genomes would be more compatible (Punja 2001). In previous research, we used mRNA differential display to isolate seedling-specifically expressed genes from maize shoots and radicals (Zhang et al. 2000). Seven differential expressed fragments were cloned and sequenced after confirmation by reverse-Northern blotting analysis. Of these, a fragment coding β-glucosidase (ZmGLU1) exhibited especially high expression in both the shoots and radicles of maize seedlings (Zhang et al. 2000).
β-glucosidase (β-d-glucosidase glucohydrolase. EC 3.2.1.21) catalyzes the hydrolysis of various aryl and alkyl β-d-glucosides as well as short chain β-linked oligosaccharides (e.g., cellobiose and laminoribose, etc.). The enzyme occurs widely in both prokaryotes and eukaryotes. In plants, the β-glucosidases are associated with a variety of functions, including: (I) chemical defense against pathogen and other pests through the release of saponins, thiocyanates, HCN, or terpenes (Nisius 1988; Poulton 1990; Sahi et al. 1990); (II) phytohormone activation from their inactive glucoconjugation (Brzobohaty et al. 1993; Wiese and Grambow 1986); (III) hydrolysis of β-linked oligosaccharides resulting in cell wall degradation in endosperm during seed germination (Leah et al. 1995); and (IV) response to abiotic stresses (Gerardi et al. 2001; Seki et al. 2001; Thorlby et al. 2004).
In maize, ZmGLU1 is one of the β-glucosidases that has been suggested to hydrolysis cytokinin-O-glucosides to liberate free cytokinins (Brzobohaty et al. 1993). Although inactive cytokinin conjugate is abundant in plants, only a small amount of free cytokinins are available to stimulate and control plant growth (Letham and Palni 1983). The O-glucosides are the major mobilizable conjugated form of cytokinin from which active cytokinin can be released by ZmGLU1. As such, ZmGLU1 is one of the key enzymes controlling cytokinin homeostasis in maize.
Tissue-specific location analysis of ZmGLU1 by Northern blotting and in situ hybridization revealed high expression in the meristem (e.g., primordial leaves, shoot node, mesocotyl, and primary roots) of seedlings and female reproductive organs (e.g., ovule, silk, and husk) (Esen 1993). No expression was detected in plant parts that were terminally differentiated and matured (e.g., endosperm, ear axis, and rachis) (Kristoffersen et al. 2000). In some cases, low levels of enzyme activity can be detected in silk, anthotaxy, and mature leaves, suggesting a role in the release of toxins to defend against pathogens (Esen 1993). Other research findings suggest that the coleoptile has the highest enzyme activity, followed by roots and pistil (Esen 1993; Nikus and Jonsson 1999). All of these reports are consistent with the distribution of active cytokinin and indicates that β-glucosidase plays not only a critical role in plant growth and development, but also that its promoter may have tissue specific expression in intense cell division zones.
In order to understand more about the spatial and temporal expression of the ZmGLU1 and its response to phytohormones and abiotic stresses, we isolated a ZmGLU1 promoter (ZmGLU1P) and investigated its expression pattern on the basis of GUS reporter gene expression in transgenic tobacco. A series of 5′deletions were also examined to determine the putative regulatory cis-elements of the promoter.
Materials and methods
Genomic library screening and sequence analysis
Total RNA was extracted from 4-day-old maize seedlings of hybrid “Nongda 3138” following methods described by Logemann et al. (1987). mRNA was obtained using the PolyA-Track mRNA isolation system (Promega). In two separate reactions, 4 μg mRNA was reverse transcribed at 60°C using oligo d(T)18 as a primer. The 5′ fragment (580 bp) of the maize β-glucosidase cDNA was obtained by PCR using Probe-S: 5′-ACCTAGTAGGACCCAACAATGAGAG-3′ as a sense primer and Probe-R: 5′-CTCTTATCTAGGAAGCCGCCGTAC-3′ as a reverse primer (+62 to +86 and +618 to +641 related to the translation start codon ATG) (Fig. 1) and labeled with [α-32P]dCTP by random priming (Prime-A-Gene, Promega). A genomic library of maize inbred B73 in the phage EMBL-3 vector was purchased from CLONTECH (cat. FL1032D). The library was screened by plaque hybridization using the probe mentioned above. Nylon membrane replicas of independent plaques were prehybridized at 42°C for 5 h with 50% formamide/5 × Denhardt's solution/0.75 M NaCl/0.05 M NaPO4, pH 7.4/3 mM EDTA/0.1% SDS/0.1 mg/ml salmon sperm. Hybridization was carried out at 42°C overnight under the same conditions. Membranes were washed with buffer I (2 × SSC/0.5% SDS) at 42°C for 30 min followed by washings with buffer II (1 × SSC/0.5%SDS) and III (0.1 × SSC/0.1% SDS) at 65°C for 45 min each and then exposed to X-ray film (Fuji, Tokyo, Japan) at −70°C overnight. Positive plaques were isolated and rescreened. SalI/XbaI digested phage DNA hybridization generated a 4 kb positive fragment containing a 2 kb upstream fragment of ZmGLU1. This fragment was separately subcloned into the pGEM-3ZF(−) (Promega) vector and sequenced.
Primers and putative cis-elements on ZmGLU1 promoter region. Numbers indicate the positions relative to the transcription start site +1. The translation initiation codon is in shade. The putative TATA box, CAAT box, and other important putative cis-elements are boxed and labeled above it. Deletion primers, DRP primer, and Probe-S primer are underlined and labeled under it
Determination of transcription start site
The transcription start site was determined using 5′RACE technique following the Ambion ALM-RACE Kit protocol. Total RNA was extracted and cDNA obtained as described above. The PCR of cDNA was performed using Clontech adaptor primers (outer primer: 5′-GCTGATGGCGATGAATGAACACTG-3′ and inner primer: 5′-CGCGGATCCGAACACTGCGTTTCTGGCTTTGATG-3′), and ZmGLU1 cDNA specific outer (Probe-R) and inner (DRP: 5′-TCCATGGCCCCCCCTTTGCT-3′) primers (Fig. 1). The first PCR was carried out using the two outer primers with the cDNA as a template, followed by a second PCR using the two inner primers with the first PCR product as a template. The second PCR product was subcloned into a pGEM T-easy vector (Promega) and sequenced. RT-PCR was further performed to confirm the result with D8(-82) used as a forward primer, DRP as a reverse primer (Fig. 1), and the first strand cDNA reverse transcribed from the total RNA of the maize seedling as a template. The isolated positive plaque was used as positive template. The DNA template was omitted in the negative control.
Construction of the ZmGLU1 promoter and its deletions
The entire ZmGLU1P region (from −1677 to +64 bp, just include ATG), designated as D0 (−1677), and its eight deletions (designated as D1–D8) (Fig. 1) were created by PCR amplification using the following primers: D0(−1677) (D0: 5′-CCGTCTAGAGGTGGAAATATCTTCTCAAGC-3′), D1(−846) (D1: 5′-CGCTCTAGACAAAGCTGCCGCATTC-3′), D2(−798) (D2: 5′-CGGTCTAGAACGTTGACAAAAAAAATGAA-3′), D3(−758) (D3: 5′-GCGTCTAGACGGTCTAGATTATGGATATGGCGCGGT-3′), D4(−717) (D4: 5′-GCGTCTAGACTTTCCATAGGATTGTCCCAT-3′), D5(−679) (D5: 5′-GCGTCTAGATAAGTTTATCCCTGAATAACACG-3′), D6(− 530) (D6: 5′-CGGTCTAGAGAACGGTGTTAGGTAGTCATTG-3′), D7(−315) (D7: 5′-CCGTCTAGACCCACATGCATACGATGC-3′), D8(−82) (D8: 5′-GCGTCTAGAAGTGATAAACTATTGGGCAACC-3′) at the 5′end and deletion reverse primer (DRP) at the 3′end (Fig. 1). Every 5′end deletion primer coupled with a DRP primer produced a corresponding deletion. A XbaI restriction site was introduced into the forward primers and a NcoI restriction site introduced into the reverse primer. The amplified sequences were digested by XbaI and NcoI and then cloned into the XbaI and NcoI digested binary vector pCAMBIA3301 to replace the CaMV35S promoter in front of the beta-glucuronidase gene. The nine constructions were respectively described as D0(−1677), D1(−846), D2(−798), D3(−758), D4(−717), D5(−679), D6(−530), D7(−315), and D8(−82) (Fig. 1). For ease of understanding and illustration here, we refer to D0(−1677) as ZmGLU1P-GUS and pCAMBIA3301 as 35S-GUS.
Plant transformation, materials, and growth conditions
The nine plasmids described above and 35S-GUS were introduced into Agrobacterium tumefaciens stain LBA4404 using a freeze-thaw method. Single colonies were selected to transform tobacco (Nicotiana tabacum cv. Samsun NN) using the leaf disk infection method (Horsch et al. 1985). Transformed shoots were selected on a solidified MS medium (Murashige and Skoog 1962) containing 100 μg/ml phosphinothricin (ppt); and 250 μg/ml cefotaxim (cef). Tobacco plants were grown in a growth chamber at 28°C under fluorescent white light in a 16:8 h light/dark cycle.
Analysis of transgenic plants by Southern blotting
Genomic DNA was extracted from young leaves of T0 transgenic plants using CTAB DNA extraction system (Doyle and Doyle 1987). Twenty micrograms of DNA was then digested with EcoRI, fractionated by electrophoresis in 0.8% agarose gel and transferred to Hybond N+ Nylon membranes (Amersham Pharmacia) by capillary transfer. The DNA was fixed by baking at 80°C for 1 h. A fragment (590 bp) of GUS cleaved by EcoRV and BamHI from the pBI221 vector and labeled with [α-32P] dCTP using the prime-a-gene labeling system (Promega) was used as a probe. Membranes were hybridized with this probe at 65°C overnight in Church solution (Church and Gilbert 1984) (0.25 M NaPO4, pH 7.2/0.7%SDS/1%BSA/1 mM EDTA) after overnight prehybridization. The membranes were washed successively with 2 × SSC/0.5% SDS, 1 × SSC/0.5% SDS, and 0.5 × SSC/0.1% SDS at 65°C for 15 min each, then exposed to X-ray film at −70°C for a week.
Quantitative GUS activity assay
Five single-copy transgenic tobacco lines of ZmGLU1P-GUS and 35S-GUS were selected and their T2 homozygotes generated for subsequent assays. For the tissue-specific expression assay, leaves, petioles, stems, and roots were collected from each selected plant after growing in soil for ∼45 days when they had six to eight leaves. Flower organs were collected one day after the flower's opening and immature seeds collected about 15 days after flower opening. Developing fruits were collected on the 0, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, and 55 days after flower opening. For the germination assays, the surface-sterilized seeds of T2 transgenic plants were spread onto 20-mm-diameter discs with two layers of fiber filter paper (Whatman) moistened with water, and incubated in a growth chamber at 28°C. Aliquots were sampled 0, 1, 2, 3, 4, 5, 6, 7, 9, 11, 13, and 15 days after incubation. For the response of ZmGLU1 promoter to phytohormones and abiotic stresses assays, T2 seeds from the same five representative lines above were germinated on MS medium and grown in a growth chamber at 28°C. For phytohormone treatments, 14-day seedlings were transferred to new MS medium containing 10 μM 6-benzylaminopurine (6-BA), 10 μM trans-zeatin, 10 μM abscisic acid (ABA), 10 μM naphthalene-acetic acid (NAA), 10 μM gibberellin (GA3), 1 μM salicylic acid (SA), 100 μM NaCl, or 200 μM mannitol for 10 days, respectively. For cold treatment, 21-day seedlings were transferred to a cold chamber maintained at 4°C for 3 days. For submergence treatment, 14-day seedlings were transferred to a 150-ml Erlenmeyer flasks containing new MS medium. Seven days later distilled water was added into the flasks so that the seedlings were completely submerged in water, and incubated for 3 days. Twenty four-day-old seedlings growing in MS medium without any supplement or treatment were served as control. All parallel samples grew under the same conditions.
In the deletion analysis, shoots from tobacco calli were transferred to a solidified selective MS medium containing 100 μg/ml ppt and 250 μg/ml cef. After conditioning in a growth chamber for about 40 days, the leaves, stems, and roots of transgenic seedlings were selected for GUS activity assay.
Plant tissues were harvested and immediately homogenized by grinding in 0.7 ml protein extraction buffer: 0.05 M NaPO4, pH 7.0/0.1% SDS/10 mM EDTA/20% methanol/10 mM β-ME/Triton X-100. Centrifugation at 13,000×g 4°C for 15 min. Fifty microliters of supernatant was transferred into a microcentrifuge tube containing 250 μl GUS reaction buffer (protein extraction buffer containing 2 mM MUG) preincubated at 37°C and fully mixed. Fifty microliters of this mixture was immediately transferred into a 950 μl GUS stop buffer (0.2 M Na2CO3) to serve as a control. The other mixture was incubated at 37°C. Fifty microliters of aliquots were removed at 10, 20, 30, 40, and 60 min and mixed with 950 μl GUS stop buffer. Activity assays were performed in a Microfluor fluorometer (Model 450) with emission at 455 nm and excitation at 365 nm. Protein concentrations of the samples were determined using the Bradford protein assay (Bradford 1976). GUS enzyme activity was expressed as nmol 4-methylumbelliferone (MU) produced per minute per milligram protein.
Histochemical GUS staining
For histochemical GUS analysis, samples were immersed in staining solution (50 mM NaPO4, pH 7.0/10 mM EDTA/2mM 5-bromo-4-chloro-3-indoyl glucuronide/1 mM potassium ferrocyanide) as described by Jefferson (Jefferson 1987) at 37°C overnight after being fixed in 42.5% ethanol/5% acetic acid/3.7% formaldehyde. The samples were then cleared of chlorophyll using 70% ethanol and used for imaging.
Results
Isolation of genomic DNA sequence from maize
In our previous cDNA-differential display, an EST representing ZmGLU1 was detected showing especially high expression levels in both the shoot and radicle (Zhang et al. 2000). A 580 bp fragment from the 5′end of ZmGLU1 cDNA (GenBank accession number: X74217)—obtained by PCR using Probe-S and Probe-R primers (Fig. 1)—served as a probe to screen a lambda genomic library of maize inbred B73 and yielding a 2 kb positive sequence. A sequence blast in GenBank indicated that this 2 kb fragment contained 1904 bp upstream and 105 bp downstream sequences of ZmGLU1 related to the ATG codon, and that the 105 bp downstream sequence showed 100% identity with the ZmGLU1 and suggesting, therefore, that the 2 kb fragment contained the ZmGLU1 promoter region (GenBank accession number: DQ333310). Sequence analysis of the fragment also revealed some conservative sequence motifs. The nucleotide sequence of this fragment and results of its major putative cis-elements analysis are shown in Fig. 1.
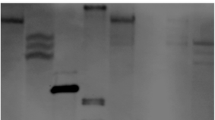
Determination of the transcription start site by 5′RACE. a 5′RACE products resolved on a 1.5% agarose gel. Adaptor primers and two nested ZmGLU1 cDNA specific primer, Probe-R, and DRP, were used to carry out 5′RACE. The reversed cDNA of maize hybrid “nongda3138” was used as template (M, DNA marker; 1, 2, 3, 4, four independent PCR from different cDNA as templates). b RT-PCR performed to solidify the transcription start site using DRP and D8(-82) primers (M, DNA marker; W, water as template; cDNA, cDNA from “nongda3138” as template; λDNA, isolated positive λ clone as template
Determination of transcription start site
A 5′RACE was performed to determine the transcription start site of the ZmGLU1 gene. A two-step PCR was performed after reverse transcription of total RNA extracted from 4-day-old seedlings of “Nongda 3138” with two nested reverse ZmGLU1 gene specific primers, Probe-R and DRP (Fig. 1), and yielding a ∼140 bp extended product (Fig. 2a). The amplified fragments were subsequently cloned and sequenced. Analysis of the fragment revealed that the site joined to the 3′end of the inner adaptor primer was the putative transcription start site, which was 58 bp upstream of the ATG codon. This result is also consistent with that predicted by http://www.fruitfly.org/cgi-bin/seq_tools/promoter.pl. RT-PCR was further performed to solidify this result. D8(−82) and DRP were select as RT-PCR primers (Fig. 1). First strand cDNA reverse transcribed from the total RNA of maize seedlings was used as a template. No production was obtained in the system or the negative control, while the isolated positive lambda DNA gave a product of ∼140 bp (Fig. 2b). This finding confirmed that the transcription start site was at the position within 140 nt upstream of the ATG translation start site, most probably at −58 bp. During inspection of the upstream sequences, a potential TATA box (TATAAAT) was identified at −25 to −31 nt upstream of the transcription start site (Fig. 1), which was consistent with the regular feature of eukaryotic promoters (Ke et al. 1997). A potential CAAT box (GCAAT) was also identified from −51 to −55 upstream of the transcription start, which is 20 nt upstream of the TATA box (Fig. 1). These additional findings support the results from the 5′RACE that the transcription start site was −58 bp from translation start site ATG.
GUS activity of different tissues and organs of transgenic tobacco plants carrying ZmGLU1P-GUS and 35S-GUS constructs. Leaves, petioles, stem, and roots were harvested about 45 days after budding when plant had six to eight true leaves. Reproductive tissues and expanded leaves were harvested after flower opening. Error bars represent standard error
Activity analysis of ZmGLU1 promoter in transgenic tobacco and comparison with CaMV35S promoter
The result showed that GUS activity driven by the ZmGLU1 promoter was detected in many plant tissues (Fig. 3), and that the activity was about 15 nM 4-MU/min/mg. Roots had the highest GUS activity, exhibiting nearly six times that of the most other tissues. This was followed by immature seeds and the pistol at about three to four times that of other tissues. The lowest level of activity was found in mature seeds and old leaves, which exhibited less than 2 nM 4-MU/min/mg enzyme activity while in the control, the 35S promoter showed no significant change in these organs. Histochemical staining revealed that GUS activity in the roots was found mainly in the elongation zone and not in the root tips (Fig. 4c and d). Enzyme activity in the meristematic region of protruding lateral root basement was remarkably higher than at any other location. Intense GUS staining was also observed in the vascular cylinder of roots (Fig. 4g). In the stem, GUS activity around the node was much stronger than that in the internode, and it was mainly present in the phloem and xylem (Fig. 4h–j). In the leaves, GUS activity was detected in petioles and veins, but little was found in the mesophyll (date not shown). The branching points of the shoot and the lateral root basement also displayed prominent GUS activity, in which high levels of expression were mostly associated with vascular tissues. In the flowers, the female reproductive organ had stronger GUS stain compared to the male organs (Fig. 4e and f). In the sepal and petal, GUS expression was found before flower opened, with little if any found afterward. In the developing anther, however, the opposite case was true with GUS activity nearly undetectable before the flower opened and then strongly stained afterward.
Histochemical localization of GUS gene expression driven by the ZmGLU1 full length promoter in transgenic tobacco. a, b Seed on 0, 2 day after germination. GUS signal was present in embryo but not in endosperm (em, embryo; en, endosperm). c 20 days plantlet grown on MS medium. GUS staining was found in root elongating zone but not in root tip. d Seedlings on day 7 after germination. High level of GUS activity represents in elongating zone, no GUS activity in root tip and cap (sc, seed coat; rt, root tip). e, f Flowers before (e) and after (f) flower opening. GUS staining exists mainly in female organs. g Longitudinal section of young root (lrb, lateral root basement). High GUS activity was shown in lateral root basement. h Longitudinal section of stem around the reciprocal second leaf of a 2-month-old tobacco. GUS activity was higher in node than internode (nd, node). i Transverse section of a young stem. GUS activity was present in phloem and xylem (25×). j Magnification of i (100×). (op, outer phloem; ip, internal phloem; x, xylem)
Comparing to the 35S promoter, ZmGLU1P activity was somewhat lower (about 1/3) in most tissues (i.e., leaves, stem, petiole, sepal, stamen, petal), but higher in the roots (nearly 3-fold), pistil (nearly 2-fold) and immature seeds (2-fold) (Fig. 3). GUS activity was much lower in mature seeds, about 1/20 that of 35S-GUS.
Activity analysis of ZmGLU1 promoter during fruit development and seed germination
Given the noticeable amount of GUS activity in the immature seeds of ZmGLU1P-GUS transgenic tobacco (20 times more than the mature seed (see Fig. 2)), we speculated that ZmGLU1 may play an important role in seed development and germination. To test our hypothesis, we examined the change in activity of ZmGLU1P between these two stages. However, because immature seeds are hard to separate, we only analyzed the activity change in the whole fruit.
Gus activity in fruits of transgenic tobacco harboring ZmGLU1P-GUS decreased during seed maturation, and reached a minimum when the seed was mature at 1/70 the GUS activity of pollination period (Fig. 5). After seed germination, however, expression of the GUS gene increased again (Fig. 6). GUS activity exhibited two major spurts after germination (day 2–3, and day 5–11), corresponding to the period of radicle and cotyledon emergence. On day 11 it reached a peak showing 300 times more GUS activity than dry seeds. However, Gus activity driven by the 35S promoter appears to present no significant changes during fruit development and seed germination (Figs. 5 and 6).
Histochemical staining indicated that the GUS activity of ZmGLU1P-GUS transgenic plants was diverse during fruit development and seed germination. During fruit development, strong GUS activity was detected in the fruit center column and fruit wall (data not shown). In early stages of seed germination, the staining showed mainly in the embryo with none in the endosperm (Fig. 4a and b). During the early seedling stage, the hypocotyl and radical were densely colored (Fig. 4d), with staining becoming more prominent in the root elongating zone, lateral root basement, and petiole later in the seedling (Fig. 4c).
Response of ZmGLU1 promoter to phytohormones and abiotic stress
Gus activity of tobacco seedlings containing ZmGLU1P-GUS and 35S-GUS constructions was analyzed under six familiar phytohormones and four major environmental stresses (Fig. 7). ZmGLU1P-GUS transgenic tobacco seedlings subjected to cold, salinity (NaCl), submergence, and drought (mannitol) did not display any significant induction or inhibition as measured by GUS expression. Responses to phytohormones, on the other hand, varied. There was no significant change in GUS activity following treatments with GA3, ABA, and SA. However, GUS activity decreased ∼70% when the seedlings were exposed to exogenous NAA, trans-zeatin, and 6-BA (Fig. 7).
Deletion analysis of the ZmGLU1 promoter
A series of 5′-deletions of the promoter region were created to characterize the promoter in greater detail. The entire ZmGLU1 promoter and corresponding deletion segments are illustrated in Fig. 1. We selected more than ten independent transgenic lines for each construct. Eight typical GUS positive independent transgenic lines for each construct were used in the final analysis. GUS activity was assayed in the leaves, stem, and roots of transgenic tobacco plants carrying the shortened ZmGLU1 promoter. As shown in Fig. 8, reducing the promoter sequence from D0(−1677) to D2(−798) had no significant effect on GUS activity. When the promoter was further reduced to D3(−758), there was a dramatic increase in GUS activity in the leaves and stem, but no significant change in the roots. This suggests that there might be an important cis-element for leaf-specific and stem-specific expression in the 40 bp sequence. When deleted from D4(−717) to D5(−679) and from D5(−679) to D6(−530), two distinct reductions in GUS activity were detected in all tissues. This suggests that there might be important enhancers capable of responding to high-level expression in these regions. The deletion from D7(−315) to D8(−82) produced an obvious reduction in GUS activity. Further, GUS activity driven by D8(−82) showed no difference from the wild type suggesting that the core-promoter of ZmGLU1 should be located within −315 bp.
The changes of GUS activities of ZmGLU1P-GUS and 35S-GUS transgenic tobacco under phytohormones and abiotic stresses. The GUS activity of the seedlings grown on MS medium without any supplement or treatment was set as 100% to calculate relative values of ZmGLU1P-GUS and 35S-GUS exposed to various phytohormones or abiotic stresses
Discussion
Our previous RNA differential assay indicated that ZmGLU1 had a high expression level in both the radicle and shoot (Zhang et al. 2000). However, the histochemical staining undertaken in this research revealed that GUS activity driven by ZmGLU1 promoter exhibited a high expression level in the radical, but low in the shoot (Fig. 4c and d). This difference may result from the heterogeneous environment for this promoter—the lack of a cis-element or cis-trans interaction necessary for shoot expression in tobacco, or from imprecise material separation in the RNA differential assay where some of the upper part of the radical may have separated into shoot.
Northern blotting by Esen (1993) showed that the highest ZmGLU1 activity was in the coleoptile, followed by the roots and pistil. In our experiment, the coleoptiles could not be separated from the tiny tobacco seeds so the roots showed the highest level of activity followed by the pistil and the immature seed (Fig. 3). However, in our histochemical staining of the germinating seeds, the staining existed mainly in the embryo (Fig. 4a and b). Histochemical staining of the seedlings suggested that the radical had the highest GUS activity (Fig. 4c and d). Tissue-specific expression of ZmGLU1 was observed mainly in the vascular bundle (Fig. 4g, i, and j). This finding supports somewhat Brzobohaty's speculation that ZmGLU1 guard the vascular bundle and liberate free cytokinins from the exogenous supply of cytokinin-O-glucosides arriving from other tissues, mainly the roots, and consequently maintain meristem activity (Brzobohaty et al. 1993). Several other phytohormone related promoters have also been discovered with high vascular tissue expression levels (Capone et al. 1994; Lee et al. 2003; Silverstone et al. 1997; Wong et al. 1996). This may suggest the close relationship between vascular tissue and the regulation of plant growth and development.
Promoter function and utilization are essential for endogenous and transgenic (exogenous) expression. Here we isolated a novel promoter (ZmGLU1P) from Zea mays and tested its temporal and spatial expression pattern using a GUS reporter gene system in transgenic tobacco. In comparison with the 35S promoter, ZmGLU1P is a strong and active promoter with somewhat root-specific traits. Its activity is about 1/3 that of the 35S promoter in most tissues, but much higher in roots (nearly 3-folds) (Fig. 3). Interestingly, its activity in mature seed is much lower, about 1/50 that in roots. If using this promoter in crop genetic engineering, it may reduce the transcription of exogenous gene in mature seeds, and in turn reduce protein accumulation in the seeds, the edible part of crops. However, the true outcomes also depend on the stability of the protein concerned. To our knowledge, it is the first reported promoter that could drive strong expression in all tissues except old tissues and matured seed.
A series of ZmGLU1P deletions were also fused to the GUS reporter gene and transformed into tobacco for analysis of important cis-elements. No significant change in GUS expression occurred with deletion from D0(−1677) to D2(−798). Further deletion from D2(−798) to D3(−758) resulted in dramatically increased activity in the leaves and stem, but no significant change in the roots (Fig. 8). This suggests there might be a potential suppressor binding site in the 40 bp region for leaf-specific and stem-specific expression. A search of the http://www.gene-regulation.com database for potential cis-elements in the 40 bp region revealed several reported cis-elements (Fig. 9), such as cgtgg: the conserve cis-element for anaerobic growth condition in maize ADH1 gene promoter (Ferl and Nick 1987); acgtg: the binding site of a bZIP transcription factor, TRAB1, in rice embryonic abundant protein EM1 and 2 (Hobo et al. 1999;Tokunori et al. 1999); tgaca: a elicitor response element in tomato PR10a-1 promoter (Desveaux et al. 2000); tcatttttt, matched eight of nine positions to the consensus sequence, tcacttttt, the binding site of the zinc finger transcription factor Dof2 in maize PEPC3 (Yanagisawa and Sheen 1998); ttgac, w-box of transcription factor werky18 in Arabidopsis (Du and Chen 2000); catttttt, matched seven of eight positions to the consensus sequence, catttttg, a cis-element for soybean embryo factor 4, SEF4 (Lessard et al. 1991). Although these binding sites were not reported for leaf-specific or stem-specific expression, it at least indicated that the 40 bp fragment was very active and important in the ZmGLU1 promoter. Deletion from D4(−717) to D5(−679) and D5(−679) to D6(−530) resulted in an obvious reduction in promoter activity (Fig. 8) and suggesting that at least two essential enhancers that confer high-level expression were imbedded within these fragments. Analysis of deleted DNA sequences revealed a GC-rich motif that has been shown to be important for basal and enhanced expression in soybean genes (Buzeli et al. 2002) and many AT-rich sequence-like enhancers (Boston et al. 1991) (Fig. 1). A reported vascular specific expression factor binding site (Torres-Schumann et al. 1996) was also found in regions from −508 to −500 (Fig. 1), which might account for the vascular specific expression (Fig. 4g, i, and j). When deleted from D7(−315) to D8(−82), no GUS activity could be detected indicating that the core promoter sequence might be located within −315 bp.
ZmGLU1 homologous genes from other species suggest a number of functions. Several β-gluconsidase conferring stress-responsive expression against stress tolerance have been reported (Gerardi et al. 2001; Seki et al. 2001; Thorlby et al. 2004). In this research, ZmGLU1P-GUS transgenic tobacco seedlings were exposed to four environment stresses, submergence, salinity, drought and cold, and six phytohormones, trans-zeatin, 6-BA, NAA, ABA, GA3, and SA and GUS activity changes monitored (Fig. 7). Expression of GUS driven by ZmGlU1P decreased to 30% when seedlings were exposed to 6-BA, trans-zeatin, and NAA. The changes under trans-zeatin and 6-BA treatments may involve in a cytokinin feedback mechanism. A substrate specific report showed that ZmGLU1 had a very restricted substrate (Brzobohaty et al. 1993). However, this report focused only on the glycosidic bond whereby ZmGLU1 could hydrolyze O-glucosides and N3-glucosides, but not N7-glucosides or N9-glucosides. A similar response between trans-zeatin and 6-BA in our research may indicate that ZmGLU1 has no cytokinin-specific hydrolysis. According to changes associated with NAA treatment, ZmGLU1P is not likely to be infected directly by NAA since the hydrolysis of IAA-glucosides could not be demonstrated (Brzobohaty et al. 1993). Other experiments may be needed to show up the details between ZmGLU1 promoter and auxin (NAA). Under other phytohormones and abiotic stresses, ZmGLU1 promoter activity exhibited no visible change indicating that the ZmGLU1 gene did not seem to function in stress tolerance and other phytohormone activation.
In summary, we have isolated ZmGLU1 promoter from maize inbred B73 genomic library and investigated its activity in transgenic tobacco. Our main conclusion was that the ZmGLU1 promoter has strong activity in most plant tissues and organs except for mature seeds and old leaves. These observations would seem to suggest that the ZmGLU1 promoter has a promising future in agribiotechnology engineering.
Abbreviations
- ABA:
-
Abscisic acid
- BA:
-
Benzylaminopurine
- CaMV:
-
Cauliflower mosaic virus
- cef:
-
Cefotaxim
- GA:
-
Gibberellin
- IAA:
-
Indole-3-acetic acid
- NAA:
-
Naphthalene-acetic acid
- ppt:
-
Phosphinothricin
- SA:
-
Salicylic acid
- ZmGLU1P :
-
ZmGLU1 promoter
References
Boston RS, Fontes EPB, Shank BB, Wrobel RL (1991) Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell 3:497–505
Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brzobohaty B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K (1993) Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 262:1051–1054
Buzeli RA, Cascardo JC, Rodrigues LA, Andrade MO, Almeida RS, Loureiro ME, Otoni WC, Fontes EP (2002) Tissue-specific regulation of BiP genes: a cis-acting regulatory domain is required for BiP promoter activity in plant meristems. Plant Mol Biol 50:757–771
Capone I, Frugis G, Costantino P, Cardarelli M (1994) Expression in different populations of cells of the root meristem is controlled by different domains of the rolB promoter. Plant Mol Biol 25:681–691
Church GM, Gilbert W (1984) Genomic sequencing. PNAS 81:1991–1995
Desveaux D, Despres C, Joyeux A, Subramaniam R, Brisson N (2000) PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12:1477–1490
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Du L, Chen Z (2000) Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J 24:837–847
Esen A (1993) Tissue-specific expression of beta-glucosidase. Maize Genetics Cooperation Newsletter 67
Ferl R, Nick H (1987) In vivo detection of regulatory factor binding sites in the 5′ flanking region of maize Adh1. J Biol Chem 262:7947–7950
Gerardi C, Blando F, Santino A, Zacheo G (2001) Purification and characterisation of a beta-glucosidase abundantly expressed in ripe sweet cherry (Prunus avium L.) fruit. Plant Sci 160:795–805
Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. PNAS 96:15348–15353
Horsch RB, Fry JE, Hoffman NL, Eicholtz D, Rogers SG, Fraley RT. (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report 5
Ke J, Choi JK, Smith M, Horner HT, Nikolau BJ, Wurtele ES (1997) Structure of the CAC1 gene and in situ characterization of its expression (the arabidopsis thaliana gene coding for the biotin-containing subunit of the plastidic acetyl-coenzyme a carboxylase). Plant Physiol 113:357–365
Kristoffersen P, Brzobohaty B, Hohfeld I, Bako L, Melkonian M, Palme K (2000) Developmental regulation of the maize Zm-p60.1 gene encoding a beta-glucosidase located to plastids. Planta 210:407–415
Leah R, Kigel J, Svendsen I, Mundy J (1995) Biochemical and molecular characterization of a barley seed beta-glucosidase. J Biol Chem 270:15789–15797
Lee JH, Takei K, Sakakibara H, Sun Cho H, Kim do M, Kim YS, Min SR, Kim WT, Sohn DY, Lim YP, Pai HS (2003) CHRK1, a chitinase-related receptor-like kinase, plays a role in plant development and cytokinin homeostasis in tobacco. Plant Mol Biol 53:877–890
Lessard P, Allen R, Bernier F, Crispino J, Fujiwara T, Beachy R (1991) Multiple nuclear factors interact with upstream sequences of differentially regulated beta-conglycinin genes. Plant Mol Biol 16:397–413
Letham DS, Palni LMS (1983) The biosynthesis and metabolism of cytokinins. Ann Rev Plant Physiol 34:163–197
Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163:16–20
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiologia Plantarum 15:473–497
Nikus J, Jonsson LMV (1999) Tissue localization of β-glucosidase in rye, maize and wheat seedlings. Physiologia Plantarum 107:373–378
Nisius A (1988) The stromacentre in Avena plastids: an aggregation of β-glucosidase responsible for the activation of oat-leaf saponins. Planta 173: 474
Poulton JE (1990) Cyanogenesis in plants. Plant Physiol 94:401
Punja ZK (2001) Genetic engineering of plants to enhance resistance to fungal pathogens—a review of progess and future prospects. Can J Plant Pathol 23:216–223
Sahi S, Chilton M, Chilton W (1990) Corn metabolites affect growth and virulence of agrobacterium tumefaciens. PNAS 87:3879–3883
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13:61–72
Silverstone AL, Chang C, Krol E, Sun TP (1997) Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12:9–19
Thorlby G, Fourrier N, Warren G (2004) The sensitive to freezing 2 gene, required for freezing tolerance in arabidopsis thaliana, encodes a {beta}-glucosidase. Plant Cell 16:2192–2203
Tokunori H, Mihoko A, Yasuo K, Tsukaho H (1999) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19:679
Torres-Schumann S, Ringli C, Heierli D, Amrhein N, Keller B (1996) In vitro binding of the tomato bZIP transcriptional activator VSF-1 to a regulatory element that controls xylem-specific gene expression. Plant J 9:283–296
Wiese G, Grambow H (1986) Indole-3-methanol-β-d-glc and indole-3-carboxylic acid-β-d-glc are products of indole-3-acetic acid degradation in wheat leaf segments. Phytochemistry 25:2451
Wong LM, Abel S, Shen N, de la Foata M, Mall Y, Theologis A (1996) Differential activation of the primary auxin response genes, PS-IAA4/5 and PS-IAA6, during early plant development. Plant J 9:587–599
Yanagisawa S, Sheen J (1998) Involvement of maize dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10:75–90
Zhang XH, Wang GY, Hu J, Zhao J (2000) The differences of gene expression in shoot and radicle of maize seedling and the cloning of differentially expressed cDNA fragments. J Agric Biotechnol 8:345
Acknowledgement
This research was supported by China High-tech program (863) and the Cultivation Fund of the Key Scientific and Technical Innovation Project, Ministry of Education of China (No.: 705009).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Puigdoménech
Rights and permissions
About this article
Cite this article
Gu, R., Zhao, L., Zhang, Y. et al. Isolation of a maize beta-glucosidase gene promoter and characterization of its activity in transgenic tobacco. Plant Cell Rep 25, 1157–1165 (2006). https://doi.org/10.1007/s00299-006-0177-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-006-0177-7