Abstract
Genes conferring resistance to kanamycin are frequently used to obtain transgenic plants as spontaneous resistance to kanamycin is not known to exist in higher plants. Nevertheless, mutations conferring kanamycin resistance have been identified in Chlamydomonas reinhardtii, raising the question as to why kanamycin-resistant mutants have not been found in higher plants. While attempting plastid transformation of alfalfa, we obtained non-transgenic but kanamycin-resistant somatic embryos following 2 months of culture in the presence of 50 mg l−1 kanamycin. Sequencing of the plastid DNA region corresponding to the decoding site of the 16S rRNA in ten independent resistant events revealed an A to C transversion at position 1357 of the 16S plastid rDNA, the same site at which an A to G conversion confers kanamycin resistance to C. reinhardtii by reducing the ability of the antibiotic to bind to its target site. All plants derived from the resistant embryos through additional cycles of somatic embryogenesis in the absence of kanamycin retained the mutant phenotype, suggesting that the mutation was homoplastomic. Resistant plants produced 85% less biomass than controls; their leaves were chlorotic during early development and over time slowly turned green. The absence of kanamycin- resistant mutants in higher plants might be explained by the requirement for a regeneration system capable of resulting in homoplastomic individuals, or it may be the result of the detrimental effect of the mutation on the phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Kanamycin is used frequently to select transgenic plants by transforming plant cells with the neomycin phosphotransferase II (nptII) gene from Escherichia coli (Bevan et al. 1983; Herrera-Estrella et al. 1983). Aminoglycoside antibiotics such as kanamycin bind to the bases in the A-site decoding region (helix 44) of the 16S rRNA. Specifically, aminoglycoside antibiotics bind to bases at positions 1408 and 1494 (base numbering according to Brosius et al. 1978) of the E. coli 16S rRNA (Moazed and Noller 1987, 1990; Woodcock et al. 1991). These bases are next to the 1409–1491 positions that form the base of a stem structure and are directly implicated in translation fidelity (Gregory and Dahlberg 1995). Binding of the antibiotic modifies the shape of the ribosome when a tRNA is present at the A site (Jerinic and Joseph 2000). As a result, the amino acid incorporation error rate is increased, and ribosome translocation is inhibited both in vitro (Davies et al. 1965) and in vivo (Edelmann and Gallant 1977).
Using E. coli base numbering, eukaryotic cytoplasmic ribosomes (Wilhelm et al. 1978) have a G at position 1408 of the 16S rRNA (Recth et al. 1999a), which reduces the binding affinity of aminoglycoside antibiotics. In contrast, prokaryotic ribosomes, including those of the plastid and mitochondrion, have an A at the 1408 position (Gutell 1994). Mutations in this region reduce the binding affinity of aminoglycoside antibiotics (Recth et al. 1999b) and provide resistance to the antibiotic. The A to G mutation at position 1408 in E. coli confers resistance to aminoglycoside antibiotics that have a 6′-NH2 group (Recth et al. 1999a). The same mutation results in resistance to kanamycin and apramycin in Mycobacterium (Alangaden et al. 1998). Methylation of the N1 position of A1408 confers resistance to the same antibiotics in Streptomyces tenjimariensis (Skeggs et al. 1985).
Spontaneous mutations giving rise to kanamycin resistance occur in Chlamydomonas reinhardtii. When kanamycin is used for the selection of transplastomic C. reinhardtii, 10% of the recovered cells are spontaneous kanamycin-resistant mutants (Bateman and Purton 2000). Kanamycin resistance in C. reinhardtii is caused by the A to G transition of base 1340 of the plastid 16S rRNA, which corresponds to the 1408 position of E. coli (Bartlett et al. 1979; Harris et al. 1989), or by the C to T transition at position 1341.
We describe herein the first kanamycin-resistant mutation reported in a higher plant, alfalfa (Medicago sativa L.). As in C. reinhardtii, it consists of a single base substitution in the plastid 16S rRNA. This mutation was obtained during the course of an alfalfa transformation project in which cell lines underwent extended selection on a medium supplemented with kanamycin.
Materials and methods
Plant material, cell culture, and recovery of resistant plants
An alfalfa (Medicago sativa L.) plant showing a high potential for somatic embryogenesis, designated RSY-1, was selected from seed of Regen-SY germplasm (Bingham 1991), using a regeneration protocol based on that of Austin et al. (1995). Young, fully expanded leaves from greenhouse-grown plants were rinsed for 20 s in 50% ethanol and then surface-sterilized in 0.6% (w/v) sodium hypochlorite for 20 min. Ten explants (0.7×0.7 cm) were excised and plated in the center of a 10-cm petri dish containing B5H medium (Brown and Atanassov 1985), resulting in a target approximately 5 cm2 in size. In addition, cotyledon-stage somatic embryos, obtained as described by Austin et al. (1995) but without Agrobacterium co-cultivation and antibiotics, were plated in the same way.
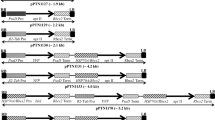
The vector, pMSCP1, was used for the transformation trials (Fig. 1). This vector was designed for alfalfa plastid transformation and contains the expression cassette from pSKKmR, which uses the aphA-6 gene from Acinetobacter baumannii as the selectable marker (Bateman and Purton 2000). The aphA-6 gene has been used for plastome transformation in combination with kanamycin selection in C. reinhardtii (Bateman and Purton 2000) and tobacco (Huang et al. 2002). DNA preparation, precipitation onto gold particles, and bombardment were as described by Stewart et al. (1996), except that 0.6-μm gold particles were used as microprojectiles.
Eighteen petri dishes containing leaf explants were bombarded 2 days after plating using the PDS 1000/He (BioRad, Hercules, Calif.) apparatus. Two days after bombardment, leaf explants were spread on the surface of fresh B5H medium containing 50 mg l−1 kanamycin and cultured at 24–26°C under a 23/1-h (light/dark) photoperiod with light provided by cool-white fluorescent tubes at a photon flux of approximately 10 μmol m−2 s−1. The tissues were subcultured every 2 weeks to fresh selection medium over a period of up to 3 months.
Green somatic embryos were isolated 2–3 months after bombardment and matured for approximately 2 weeks on B5H0 medium (Brown and Atanassov 1985) supplemented with 50 mg l−1 kanamycin. The embryos were placed for a second time on kanamycin-supplemented B5H medium to induce a second round of somatic embryogenesis, which enabled the recovery of multiple embryos from each primary embryo. In total, three to four such regeneration cycles were performed. After desiccating the embryos for 2 days, half of the somatic embryos from the second regeneration cycle were converted into plants by transferring them to petri dishes containing half-strength MS basal medium (Murashige and Skoog 1962) with or without 50 mg l−1 of kanamycin. Plants from seven independently recovered kanamycin-resistant embryos were transferred to GA-7 boxes (Magenta, Chicago, Ill.) containing the same medium for plant development and then to sterile soil in plastic pots placed in coupled GA-7 boxes. The plants were acclimatized and transferred to a greenhouse.
Plant characterization
To assess the effect of kanamycin resistance on fitness, we sampled fully expanded leaves of plants germinated from kanamycin-resistant embryos that had been grown in a greenhouse for about 4 months. Two 10-cm petri plates (replications) containing B5H with 50 mg l−1 kanamycin and two containing kanamycin-free-B5H medium were prepared with five leaf explants each per genotype. Two plates were prepared with control leaves on kanamycin-free medium and ten on kanamycin-supplemented medium. Callus and embryo production were scored after 2 weeks and 5 weeks, respectively.
Embryos obtained on kanamycin-free medium were converted into plants as described above and used to assess the stability of resistance. Leaf explants were excised from chlorotic and green leaves of in vitro-grown resistant plants obtained on antibiotic-free medium, and a new regeneration cycle was performed as described using both kanamycin-supplemented (50 mg l−1) and kanamycin-free media, with two replications per regeneration event and leaf color.
Five plants (replications) of the same age from each of four regeneration events and the control, all germinated on antibiotic-free medium, were removed from culture 3 weeks after their transfer to GA-7 boxes. These were desiccated at 100°C for 24 h and weighed to measure dry matter. Shoot dry matter accumulation was calculated by harvesting the top-growth of greenhouse-grown resistant (six events) and control plants after 4 weeks, when the control plants began flowering. The fresh material was dried and weighed as described. After verification of the normality of the data, weights were subjected to analysis of variance.
Molecular analyses
Genomic DNA was extracted using a CTAB protocol (Doyle and Doyle 1990) from the leaves or somatic embryos of ten independently recovered kanamycin-resistant plants and from the susceptible control. Two primer pairs targeted to the aphA-6 coding sequence (AphA-1F: 5′-TTTTCAGGATGAGCAGTTTG and AphA-1R: 5′-TAACCCAGCACGACCAAG; AphA-160F: 5′-TACAGTGTCTCTCGTGAAGCG and AphA-680R: 5′-AGGCAACGTTCAACAAAGG) were used for amplification.
The Medicago truncatula plastid 16S rRNA plastome sequence (GenBank AC093544) was used to design two primers (16S-236F: 5′-TAGTTTGGAACCCTGAAC; 16S-609R: 5′-GCTACCTTGTTACGACTTC) used for sequencing the alfalfa plastome region orthologous to the site of the C. reinhardtii kanamycin resistance mutation (Harris et al. 1989). The amplicons obtained from ten independently derived resistant plants and from the susceptible plant were sequenced twice using the primer MSCP-2F (5′-CGTCATAACTTCCCTCTAGACC).
To ensure that the kanamycin-resistant phenotype was not due to the insertion of aphA-6 into the plastome of resistant plants, we amplified the predicted insertion site of the alfalfa plastome from five independently derived resistant plants using primers MSCP-2F and MSCP-2R (5′-AAAAAGATTGATGAGAGAAACG) and sequenced it. To check for nuclear integration of aphA-6, 10 μg of genomic DNA from resistant and control plants were digested with SphI and used for Southern blotting according to a standard protocol (Sambrook et al. 1989). The probe was obtained by PCR amplification of aphA-6 using primers AphA-160F and AphA-680R.
In all cases, PCR was performed in a 20-μl reaction volume using: 1× PCR buffer, 0.25 μM of each primer, 0.2 mM dNTP, 1–10 ng μl−1 DNA, and 1 U Taq polymerase (Pharmacia, Piscataway, N.J.). Thermal cycling consisted of one cycle at 94°C for 4 min; 40 cycles at 94°C for 45 s, 50°C for 45 s, 72°C for 45 s; a final extension at 72°C for 10 min.
Progeny analyses
Kanamycin-resistant plants were maintained in a greenhouse until they flowered, at which point reciprocal crosses were carried out with wild-type plants. Seeds were harvested, surface-sterilized as described previously for leaf explants, and germinated in petri dishes containing half-strength MS basal medium with 50 mg l−1 of kanamycin. Susceptible plants were transferred to kanamycin-free medium and allowed to recover. All surviving plants were acclimatized and transferred to the greenhouse as described. Leaf tissue was collected and DNA extracted and sequenced as described previously.
Results and discussion
Thirty-seven green embryos were isolated within a 3-month period following bombardment, all from the leaf explants. Twenty-seven of these embryos regenerated in the presence of kanamycin during a second regeneration cycle, thereby confirming their resistance to kanamycin.
The first indication that kanamycin resistance was due to a mutation and not the integration of the transgene was the failure of PCR to produce the expected amplicon, along with the lack of aphA-6 hybridization signals from Southern blotting. Sequencing of the putative kanamycin resistance site of the 16S plastid rDNA in ten resistant events and in the control revealed a mutation at position 1357 of the 16S plastid rDNA, corresponding to the 1340 position of C. reinhardtii and to the 1408 position of E. coli (Table 1). In all of the resistant events sequenced, the alfalfa mutation was an A to C transversion. The same mutation was found in the three kanamycin-resistant events obtained from non-bombarded leaf explants cultured on kanamycin for 4 months, which showed that the mutation was not attributable to the bombardment.
The reason why this mutation occurred repeatedly and consistently—to the exclusion of other mutations in the sequenced region—is unknown. In a similar case, point mutations in the 16S ribosomal RNA gene of Nicotiana plumbaginifolia provide streptomycin resistance (Yeh et al. 1994). Nevertheless, while the authors identified three different point mutations in 11 resistant mutants, one mutation (C to T at position 912) accounted for 9 of the 11 mutants.
Phenotype of resistant plants
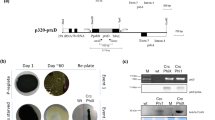
All the plants from independent mutation events had the same phenotype, and on both kanamycin and kanamycin-free media they developed more slowly than did the control plants on kanamycin-free medium. The leaves were pink when they initially appeared (Fig. 2a) and gradually turned white during leaf expansion (Fig. 2c, e, f). Green veins were present in incompletely expanded leaves. Greening occurred after leaf expansion, starting from the distal end of each leaflet (Fig. 2d). After about 2 weeks, the whole leaf had become green but was still paler than leaves from control plants. The growth of kanamycin-resistant plants regenerated in the absence of antibiotic was much lower than that of control plants, both in vitro (66% reduction) and in the greenhouse (85% reduction) (Table 2). Resistant plants displayed the chlorotic phenotype, grew much less than control plants, and had thinner stems (Fig. 2g).
Phenotype of kanamycin-resistant alfalfa plants. a Plantlet obtained from the conversion of a resistant somatic embryo, b plantlet obtained from the conversion of a control somatic embryo, c 3- week-old resistant plant, d greening of the mutant plant leaves compared with a control leaf (lower right control leaf), e, f plantlets from two resistance genotypes germinating on kanamycin-free (e) and kanamycin-supplemented media (f), g greenhouse-grown kanamycin-resistant and control (center) plants
Even though all of the leaf explants obtained from mutant plants were able to regenerate a second time in the presence of kanamycin, embryo production in the absence of the antibiotic was significantly higher than in the presence of the antibiotic (Table 3), and the average time for regeneration was 52 days on kanamycin versus 36 days in the absence of kanamycin. Together, these observations suggest that kanamycin resistance is not complete. This is consistent with the effect of the kanamycin resistance mutation in C. reinhardtii, in which the amino acid incorporation rate by the mutant ribosomes in the presence of the antibiotic was estimated to be reduced by 13–22% (Bartlett et al. 1979). Likewise, the 1408 A to G mutation in E. coli results in a longer doubling time in the presence of the antibiotics (Recht et al. 1999a).
Somatic stability of resistance
For stable maintenance of a plastome-encoded trait, homoplastomy is necessary; that is, all of the plastome copies within all plastids of all cells should be genetically identical. No difference in callus or embryo production was evident between green and chlorotic leaf explants, both on kanamycin and kanamycin-free medium (Table 3), suggesting that leaf greening was not due to the accumulation of wild-type plastids that may have remained in the cells (i.e., heteroplastomy). Of the 408 somatic embryos that were obtained from the mutant tissue, 194 germinated on kanamycin-free medium. All of the shoots displayed the chlorotic phenotype, green plants were never regenerated from them, and sequencing of their DNA revealed the presence of the mutation. Heteroplasty would have resulted in a double peak at the mutation site in the electropherograms (See et al. 2000), and no such double peaks were ever obtained in our investigation. Homoplastomy was expected for two reasons. First, the plants were evaluated after four to five regeneration cycles on kanamycin, during which time wild-type plastids were never recovered. Secondly, antibiotic resistance based on reduced binding affinity with the ribosome is recessive; that is, homoplastomy may be required for the expression of resistance and regeneration (Dix and Kavanagh 1995).
Progeny analysis
Both kanamycin-resistant and susceptible progeny plants were obtained. All resistant progeny plants had the chlorotic phenotype and the original mutation. Specifically, crossing susceptible and resistant plants led to the recovery of both resistant and susceptible progeny, suggesting that the resistant progeny inherited their plastids from the paternal parent, thereby confirming the observation of Smith et al. (1986) that there can be paternal transmission of plastids in alfalfa. The reciprocal cross also led to the recovery of both resistant and susceptible progeny. In this case, the susceptible progeny would have obtained their wild-type plastids from the male parent.
The question remains as to why other kanamycin-resistant plants have not been recovered previously, given that kanamycin has been used for plant transformation studies for 20 years. One possibility is that a longer selection period is required to obtain kanamycin-resistant mutants than is used by most transformation protocols. In the transformation of alfalfa, the first transgenic embryos appear after 3–4 weeks following co-cultivation. In contrast, because the original goal of this work was to obtain plastome transformation, leaf explants were cultured on selection medium for over 3 months. Secondly, it is possible that mutants may have occurred many times in other laboratories during transformation experiments, but the resulting plants were discarded as “escapes” or unwanted somaclonal variants.
References
Alangaden GJ, Kreiswirth BN, Aouad A, Khetarpal M, Igno FR, Moghazeh SL, Manavathu EK, Lerner SA (1998) Mechanism of resistance to amikacin and kanamycin in Mycobacterium tubercolosis. Antimicrob Agents Chemother 42:1295–1297
Austin S, Bingham ET, Matthews BF, Shahan MN, Will J, Burgess RR (1995) Production and field performance of transgenic alfalfa (Medicago sativa L.) expressing alpha-amylase and manganese-dependent lignin peroxidase. Euphytica 85:381–393
Bartlett SG, Harris EH, Grabowy CT, Gillham NW, Boynton JE (1979) Ribosomal subunits affected by antibiotic resistance mutations at seven chloroplast loci in Chlamydomonas reinhardtii. Mol Gen Genet 176:199–208
Bateman JM, Purton S (2000) Tools for chloroplast transformation in Chlamydomonas: expression vectors and a new dominant selectable marker. Mol Gen Genet 263:404–410
Bevan MW, Flavell RB, Chilton MD (1983) A chimaeric antibiotic resistance gene as a selectable marker for plant cell transformation. Nature 304:184–187
Bingham ET (1991) Registration of alfalfa hybrid Regen-SY germplasm for tissue culture and transformation research. Crop Sci 31:1098
Brosius J, Palmer LM, Kennedy PJ, Noller F (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA 75:4801–4805
Brown DCW, Atanassov A (1985) Role of genetic background in somatic embryogenesis in Medicago. Plant Cell Tissue Organ Cult 4:111-122
Davies J, Gorini L, Davis BD (1965) Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol Pharmacol 1:93–106
Dix PJ, Kavanagh TA (1995) Transforming the plastome-genetic-markers and DNA delivery systems. Euphytica 85:29–34
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dron M, Rahire M, Rochaix JD (1982) Sequence of the chloroplast 16S rRNA gene and its surrounding regions of Chlamydomonas reinhardtii. Nucleic Acids Res 10:7609–7620
Edelmann P, Gallant J (1977) Mistranslation in E. coli. Cell 10:131–137
Gregory ST, Dahlberg AE (1995) Nonsense suppressor and antisuppressor mutations at the 1409–1491 base-pair in the decoding region of Escherichia coli 16S ribosomal RNA. Nucleic Acids Res 23:4234–4238
Gutell RR (1994) Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res 22:3502–3507
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281–292
Herrera-Estrella L, De Block M, Messen E, Hernalstens JP, Van Montagu M, Schell J (1983) Chimeric genes as dominant selectable markers in plant cells. EMBO J 2:987–995
Huang FC, Klaus SMJ. Herz S, Zou Z, Koop HU, Golds TJ (2002) Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Mol Genet Genomics 268:19–27
Jerinic O, Joseph S (2000) Conformational changes in the ribosome induced by translational miscoding agents. J Mol Biol 304:707–713
Moazed D, Noller HF (1987) Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389–394
Moazed D, Noller HF (1990) Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J Mol Biol 211:135–145
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–597
Recth MI, Douthwaite S, Puglisi JD (1999a) Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J 18:3133–3138
Recth MI, Douthwaite S, Dahlquist KD, Puglisi JD (1999b) Effect of mutations in the A site of 16S rRNA on aminoglycoside antibiotic-ribosome interaction. J Mol Biol 286:33–43
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
See D, Kanazin V, Talbert H, Blake T (2000) Gel-based detection of single nucleotide polymorphisms. Biotechniques 28:710–716
Skeggs PA, Thompson J, Cundliffe E (1985) Methylation of 16S ribosomal RNA and resistance to aminoglycoside antibiotics in clones of Streptomyces lividans carrying DNA from Streptomyces tenjimariensis. Mol Gen Genet 200:415–421
Smith SE, Bingham ET, Fulton RW (1986) Transmission of chlorophyll deficiencies in Medicago sativa—evidence for biparental inheritance of plastids. J Hered 77:35–38
Stewart CN Jr, Adang MJ, All JN, Boerma HR, Cardineau G, Tucker D, Parrott WA (1996) Genetic transformation, recovery, and characterization of fertile soybean [Glycine max (L.) Merrill] transgenic for a synthetic Bacillus thuringiensis CRY IA(c) gene. Plant Physiol 112:121–129
Wilhelm JM, Pettitt SE, Jessop JJ (1978) Aminoglycoside antibiotics and eukaryotic protein synthesis: structure-function relationships in the stimulation of misreading with a wheat embryo system. Biochemistry 17:1143–1149
Woodcock J, Moazed D, Cannon M, Davies J, Noller HF (1991) Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J 10:3099–3103
Yeh KC, To KY, Sun SW, Wu MC, Lin TY, Chen CC (1994) Point mutations in the chloroplast 16S ribosomal RNA gene confer streptomycin resistance in Nicotiana plumbaginifolia. Curr Genet 26:132–135
Acknowledgements
Support for this research was provided by State and Hatch money allocated to the Georgia Agricultural Experiment Station. D.R. acknowledges NATO Advanced Fellowship Programme 1999, grant no. 215.32 that supported his stay at The University of Georgia. The assistance of Gabriella Petesse and Greg Martin is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C.F. Quiros
Rights and permissions
About this article
Cite this article
Rosellini, D., LaFayette, P.R., Barone, P. et al. Kanamycin-resistant alfalfa has a point mutation in the 16S plastid rRNA. Plant Cell Rep 22, 774–779 (2004). https://doi.org/10.1007/s00299-004-0757-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-004-0757-3