Abstract
The plant hormone auxin plays a major role in a variety of growth and developmental responses, even in the more ancient plants—for example, cell differentiation in mosses. Nevertheless, almost nothing is known about the distribution of auxin during moss development. To address this question, we characterised auxin distribution in the moss Physcomitrella patens using auxin-inducible reporter gene systems. Stable transgenic Physcomitrella plants were produced expressing the β-glucuronidase (GUS) gene driven by the auxin-inducible promoters GH3 and DR5, respectively. Both fusions showed remarkable differences with respect to auxin-induced promoter strength and expression kinetics. A detailed characterisation of the GUS expression pattern in different developmental stages revealed that the highest auxin concentrations were in dividing and ontogenetic young cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The moss Physcomitrella patens is unique among land plants as targeted mutagenesis by homologous recombination is feasible (Reski 1998a). EST data covering more than 95% of the Physcomitrella transcriptome is available (Rensing et al. 2002), and two different approaches of targeted mutagenesis via transposon tagging have been performed (Nishiyama et al. 2000; Egener et al. 2002). Gene content, expression and regulation as well as codon usage are comparable to those found in higher plants (Reski 1998b; Rensing et al. 2002). Consequently, Physcomitrella is particularly suitable as a model organism for the study of molecular and cellular plant processes.
The comparatively simple Physcomitrella developmental cycle comprises only a few differentiation steps, which are induced by phytohormones (Reski 1998b; Bopp 2000). Following germination of the moss haplospore, the filamentous protonema with apical growth is formed. Protonema filaments consist of two different cell types: the older chloronema—short cells with many plastids separated by perpendicular cell walls—and the younger caulonema, characterised by longer cells with fewer plastids and oblique cell walls. While the physiological importance of auxin for caulonema formation can be shown (Johri and Desai 1973; Bopp 2000), almost nothing is known about auxin distribution, spatial influences of auxin with respect to moss development and its changes during the developmental progression. The presumption is that auxin builds up a gradient in the moss protonema (Bopp and Atzorn 1992a, 1992b). The highest auxin concentration seems to be in the tip cell and then to gradually decline to the basal cells.
As auxin is involved in a wide array of growth and developmental processes, the control of auxin distribution is crucial for plant development. In seed plants, auxin is produced mainly in the young parts of the plant shoot, i.e. the apical meristem, lateral buds and leaves, and is transported to the roots (Friml and Palme 2002). In mosses, auxin flow remains a poorly characterised phenomenon. A three-faced cell that results in bud formation and, subsequently, the growth of the adult gametophore with leaflets and rhizoid cells initiates the next developmental step.
Our aim was to examine the distribution of auxin in Physcomitrella by means of a molecular approach. This approach uses an auxin-inducible reporter gene that represents a molecular marker for monitoring changes either in auxin concentration or in cellular sensitivity to auxin. For this purpose, Physcomitrella protoplasts were stably transformed with a construct containing the auxin-inducible GH3 promoter from soybean in front of the GUS-encoding uidA gene (Hagen et al. 1991). The usage of the GH3::GUS fusion for transient expression analysis in Physcomitrella protoplasts was demonstrated by Imaizumi et al. (2002). In seed plants, the GH3 promoter is rapidly induced only by active auxins (Hagen et al. 1984, 1991; Li et al. 1999). It contains at least three auxin-response elements (AuxRE) occupied by ARF proteins after auxin induction that activate GH3 expression (Liu et al. 1994; Ulmasov et al. 1995). In addition to the native GH3 promoter, the synthetic DR5 promoter is widely used to monitor auxin responses in higher plants (Guilfoyle 1999). DR5 comprises tandem repeats of the auxin-responsive TGTCTC element (Ulmasov et al. 1997a, 1997b). In seed plants, this synthetic promoter may possess a stronger auxin responsiveness than the native GH3 promoter itself (Guilfoyle 1999), but to date the functionality and auxin-inducibility of the GH3 promoter and the DR5 promoter has never been compared within one plant.
In the investigation reported here, transgenic Physcomitrella plants containing two different auxin-inducible reporter gene fusions—GH3::GUS and DR5::GUS—were produced and molecularly characterised. Both constructs were functional in Physcomitrella but displayed differences with respect to promoter strength and induction kinetics. We also describe GH3::GUS and DR5::GUS expression patterns during the different developmental stages of Physcomitrella. The GUS-transgenic plants will provide an excellent platform for studying the involvement of auxin during various growth and developmental events.
Materials and methods
Plant material and growth conditions
Physcomitrella patens (Hedw.) B.S.G. has been characterised by Reski et al. (1994). Plants were grown axenically under standard conditions (in agitated liquid Knop medium, 250 mg/l KH2PO4, 250 mg/l MgSO4×7 H2O, 250 mg/l KCl, 1,000 mg/l Ca(NO3)2×4 H2O, 12.5 mg/l FeSO4×7 H2O, pH 5.8, or on solid Knop medium with 1.2% (w/v) agar) in a growth chamber under controlled conditions [25±1°C; light provided from above by two fluorescent tubes (Philips TL-D 36 W/25, light flux of 55 µmol s-1 m-2 outside the flasks), light-dark regime of 16:8 h). The plants were subcultured at 7-day intervals.
Protonema material for performing the GUS assays was obtained from liquid cultures grown in Knop medium with a density of 100 mg/l dry weight, and induction was always carried out directly after subculturing. GUS staining was analysed for up to 7 days after induction with 10 µM NAA and 5 µM cytokinin [6-(γ,γ-Dimethylallyl-amino)purine], respectively. GH3 protonema was treated with increasing NAA concentrations (0, 0.1, 1, 10, 100 µM), and GUS staining was performed 1 day after induction. In the case of treatment with 2-NAA (10 µM), GUS staining was analysed 1 day after induction for GH3 plants and 7 days after induction for DR5 plants. Buds were taken from protonema cultures grown on plates with solidified Knop medium covered with a sheet of cellophane, and gametophores were obtained from moss cultures grown on Knop plates. Physcomitrella buds and gametophores were incubated in 2 ml liquid Knop medium supplemented with 10 µM NAA and 5 µM cytokinin, respectively, for up to 7 days.
Molecular cloning
The original plasmids containing the GH3::GUS and DR5::GUS elements were kindly provided by Tom Guilfoyle (University of Missouri). The plasmids were digested with EcoRI or EcoRI and SalI to cut out the GH3::GUS and the DR5::GUS fusion, respectively. The two fragments were inserted into the pRT99 vector (Töpfer et al. 1988) that provides a neomycin phosphotransferase II gene (nptII) selection cassette against the antibiotic G418, yielding the new constructs pRTGH3::GUS and pRTDR5::GUS. Before transfection of the Physcomitrella protoplasts, the pRTGH3::GUS plasmid was linearised with BamHI.
Transfection of Physcomitrella protoplasts
Protoplasts were isolated, and 3×105 cells were transfected with 30 µg of plasmid DNA and regenerated in regeneration medium [Knop medium, 3% (w/v) mannitol, 5% (w/v) glucose, pH 5.8] as described previously (Strepp et al. 1998). Three rounds of selection with the transgenic plants were performed against 50 µg/ml G418.
Flow cytometric analysis
Flow cytometric analysis was performed as described by Hohe et al. (2001).
DNA isolation
Moss protonemata were ground to a fine powder under liquid nitrogen and incubated with agitation at 60°C for 1 h in 1 volume CTAB buffer [2% (w/v) CTAB, 1.4 M NaCl, 20 mM EDTA, 100 mM Tris-HCl, pH 8.0] with 2% (v/v) β-mercaptoethanol, 100 µg/ml Proteinase K and 0.1 volume N-laurylsarcosine buffer [10% (w/v) N-laurylsarcosine, 20 mM EDTA, 100 mM Tris-HCl, pH 8.0], the buffer/plant ratio being at least 2 ml/g. The homogenate was extracted twice with chloroform/isoamylalcohol, and nucleic acids were then precipitated with isopropanol. The pellet was resuspended in 10 mM Tris, 1 mM EDTA and precipitated with 1.2 M NaCl and ethanol. The pellets were washed with 70% (v/v) ethanol, resuspended in H2O and incubated with 50 µg/ml RNase A overnight at 4°C and subsequently for 1 h at 37°C.
Southern blot analysis
The digested genomic DNA (10 µg) was separated on a 0.7% (w/v) agarose gel and transferred to Hybond-N+ nylon membranes (Amersham Biosciences, Freiburg, Germany) according to standard procedures (Sambrook and Russell 2001). The ECL random prime labelling and detection system (Amersham Biosciences) was used for probe generation and detection according to the manufacturer´s instructions.
RNA isolation
For RNA isolation, 7-day-old protonema was taken from liquid culture, ground to a fine powder under liquid nitrogen, mixed with 3 ml extraction buffer (0.15 M NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 9.0) and 0.7 ml 10% (w/v) sodium dodecyl sulfate (SDS) per gram moss material. The suspension was incubated with 7 ml phenol for 15 min on a rotating platform. After centrifugation at 1,600 g for 10 min, the supernatant was again extracted twice with 1 volume phenol/chloroform/isoamylalcohol (25:24:1) and once with chloroform. The supernatant was mixed carefully with 1 volume 5 M LiCl and RNA was precipitated overnight at 4°C. After centrifugation at 4,500 g, the pellet was washed twice with 70% (v/v) ethanol and resuspended in H20. All solutions were prepared with DEPC-treated H2O.
Northern blot analysis
Fifteen micrograms of total RNA were separated on a formaldehyde-agarose gel, transferred to Hybond-N+ nylon membranes (Amersham Biosciences) and hybridised with [32P]-labelled DNA probes for the uidA and L21 gene, respectively. The RediprimeTMII kit (Amersham Biosciences) was used for generation of the probes according to the manufacturer's instructions. The membrane was washed twice in 2× SSC, 0.1% (w/v) SDS at 64°C and exposed to X-ray film (BioMaxTM MS, Kodak, USA) using a BioMax Transcreen HE Intensifying Screen (Kodak) at −80°C overnight.
Histochemical assay for GUS enzyme activity
Plant tissues were incubated in 100 µl X-gluc solution (1 mM 5-bromo-4-chloro-3-indolyl-β-glucoronide, 100 mM sodium phosphate, pH 7.0) at 37°C for 12–16 h. Fixation was done in 200 µl 5% (v/v) formalin for 10 min followed by an incubation for 10 min in 200 µl 5% (v/v) acetic acid. Pigments in the tissues were removed by serial incubations in 30%, 50%, 70% and finally 96% (v/v) ethanol.
Results
Generation and molecular analysis of auxin-inducible GUS-expressing Physcomitrella plants
To obtain stable auxin-inducible GUS-expressing Physcomitrella plants, we cloned the GH3::GUS and DR5::GUS fusions from the original vector into the pRT99 plasmid (Töpfer et al. 1988), which offers the possibility of antibiotic selection of the transgenic plants. Physcomitrella was transformed with the new vectors pRTGH3::GUS and pRTDR5::GUS (Fig. 1), and stable plants were obtained via selection with G418. Flow cytometric analysis confirmed the maintenance of the original haploid state for all of the transgenic plants analysed (data not shown).
Map of the pRTGH3::GUS and pRTDR5::GUS constructs. A Plasmid pRTGH3::GUS containing the 592-bp promoter fragment of the auxin-inducible gh3 gene from soybean upstream of the GUS reporter gene. B Plasmid pRTDR5::GUS containing the synthetic auxin-inducible DR5 promoter in front of the reporter gene. GH3 GH3 promoter, uidA β-glucuronidase, nptII neomycin phosphotransferase II, bla β-lactamase
The transgenic plants were screened for wild-type-like phenotype. After the first GUS staining experiments of 14 GH3 plants, five GH3::GUS-expressing plants (G1–G5) showing different staining intensities were chosen for further characterisation as were five DR5 plants (D1–D5) obtained after the selection process. Southern blot analyses were performed to confirm the stable integration of the constructs into the Physcomitrella genome and to estimate the copy number of the transgenes (Fig. 2). All of the analysed plants expressing GH3::GUS showed stable integration of the pRTGH3::GUS construct into their genome. Transgenic plants G1 and G3 contained between three and ten copies of the pRTGH3::GUS construct (Fig. 2A, lanes 2, 3), G5 fewer than 20 copies and G2 and G4 more than 50 copies (data not shown). Stable integration of the pRTDR5::GUS construct could also be demonstrated for the five DR5::GUS-expressing plants. Three of the transformants (D1, D2 and D3) carried between 20 and 50 copies, with plants D4 and D5 showing high numbers (>50) of pRTDR5::GUS integration (Fig. 2B, lanes 1–5).
Southern blot analysis of transgenic Physcomitrella plants. The DNA was cleaved with HindIII, separated on 0.7% (w/v) agarose, blotted onto Hybond-N+ nylon membrane and hybridised with a probe derived from the uidA gene. The restriction enzyme HindIII cuts in both constructs, liberating an approximate 6.2-kb GUS-vector fragment. For estimation of transgene copy numbers, we added a dilution series of the pRTGH3::GUS and pRTDR5::GUS vector, respectively, corresponding to different copy numbers. A Analysis of GH3 plants G1 (lane 2) and G3 (lane 3). Lane 1 Wild type, lanes 4–9 dilution series of the pRTGH3::GUS vector corresponding to 1, 3, 5, 10, 50 and 100 copies, respectively. B Analysis of DR5 plants D1–D5 (lanes 1–5). Lane 6 Wild type, lanes 7–9 dilution series of pRTDR5::GUS vector corresponding to 10, 50 and 100 copies, respectively
Expression analyses by Northern blotting were performed with 7-day-old protonema grown in liquid culture. The analyses confirmed weak expression for GH3 plants G1 and G3 and strong expression for plants G2, G4 and G5 (Fig. 3A). Surprisingly, GUS expression levels in the DR5 plants were very weak compared to the numbers of integrated copies (Fig. 3B). Only plants D4 and D5 containing the highest copy numbers showed detectable signals within the Northern blot analysis. Thus, expression analyses reflected the GUS-gene integration numbers. Northern blot analysis of auxin-treated plants (10 µM NAA) confirmed auxin inducibility of both constructs (data not shown).
Northern blot analysis of transgenic Physcomitrella plants. RNA was separated on 1.25% (w/v) agarose in the presence of formaldehyde, blotted onto Hybond-N+ nylon membrane and hybridised with [32P]-labelled cDNA fragments of GUS, with the expectation of obtaining a transcript size of about 2 kb. As a loading control, the transcript of ribosomal protein L21 from Physcomitrella (about 900 bp) was used, which is expressed constitutively under our conditions (bottom). A Analysis of transgenic GH3 plants G1–G5, B analysis of transgenic DR5 plants D1–D5. WT Wild type
GH3::GUS and DR5::GUS expression during Physcomitrella development
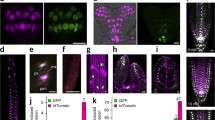
Following the production and molecular characterisation of stable GH3 and DR5 plants, we investigated the GUS staining patterns during different developmental stages. Experiments were done using the molecularly characterised GH3 plants G1–G5 and DR5 plants D1–D5, with special emphasis on the plants with a low copy number (plants G1, G3, G5, D1, D2, D3). The GUS staining patterns of different plants with similar copy numbers were always compared with each other to exclude any positional effects of construct integration on GUS expression. Different moss tissues (protonema, buds, gametophores) were analysed with and without application of exogenous phytohormones (auxin and cytokinin). Treatment with cytokinin was chosen because of the known close relationship between the two phytohormones auxin and cytokinin in the Physcomitrella protonema (Reutter et al. 1998). While treatment with auxin induced specific responses in the transgenic GH3 and DR5 plants, the application of exogenous cytokinin showed no further influence on GUS expression (data not shown). To confirm the specificity of GUS expression in response only to active auxins, the inactive auxin analogue 2-NAA was applied to protonema of GH3 and DR5 plants, respectively. GUS staining was faintly enhanced compared to that found in non-stimulated plants (Fig. 4C). The GUS staining patterns of the analysed GH3 and DR5 plants corresponded to the results of the molecular analyses. Signals were detected with and without auxin induction in the GH3 plants, but DR5 plants showed hardly any staining without auxin application. In principle, low copy numbers resulted in a weak GUS staining and high copy numbers in a strong GUS staining. The spatial staining patterns in all of the analysed GH3 and DR5 plants, respectively, were comparable with each other.
Staining kinetics of auxin-treated GH3 and DR5 protonema. Pictures were taken 1, 3 and 7 days after tissue disruption and auxin induction (10 µM NAA). A Upper row Untreated GH3 G1 culture, lower row auxin-treated GH3 G1 culture. B Upper row Untreated DR5 D2 culture, lower row auxin-treated DR5 D2 culture. C Protonema filaments treated with 10 µM NAA and 10 µM 2-NAA, respectively, 1 day (GH3 plants) and 7 days (DR5 plants) after induction. Bar: 100 µm
Protonema
In order to analyse GUS expression in protonema tissue, we incubated liquid cultures with a density of 100 mg/l dry weight with and without the active synthetic auxin NAA (10 µM) directly after subculturing for 7 days. GUS staining was visible in the GH3 plants even without auxin induction (Fig. 4A). After 1 day, a blue colour was detectable in outgrowing protonema cells, but this staining pattern changed over time, and at day 7, it was evenly distributed within the protonema filaments. This gradient was even more apparent in auxin-treated protonema. Conversely, DR5 plants showed no GUS activity in the protonema without auxin induction (Fig. 4B). Application of exogenous auxin resulted in an induction of GUS expression in the DR5 plants that was initially detectable after 3 days and which increased in intensity during the following days. After 7 days, GUS staining was evenly distributed throughout the DR5 protonema.
Protonema from GH3 plants grown in liquid culture was treated with increasing auxin concentrations to assure that the GUS staining reaction was operating within a reasonable range of auxin concentrations. As low a concentration as 0.1 µM NAA was enough to induce GUS expression in GH3 protonema 1 day after auxin treatment. At all of the auxin concentrations tested there was a difference in staining intensity between younger and older protonema cells (data not shown).
Buds
Analysis of the GUS expression pattern in buds was performed on moss plants cultivated on agar plates. A strong GUS staining could be observed in buds of GH3 plants (Fig. 5A). The outgrowing leaves of these buds showed no GUS activity. Following auxin induction, the leaves were also coloured (Fig. 5A). In contrast, the buds of DR5 plants showed no GUS staining without auxin induction (Fig. 5B); after induction with 10 µM NAA, the DR5 plants were also stained blue (Fig. 5B). GUS expression within outgrowing leaves could be induced by auxin as well (data not shown). The staining kinetics for GH3 and DR5 plants were the same as those already described for protonema tissue. The application of cytokinin greatly increased the number of buds (Reski and Abel 1985), but GUS expression pattern for the buds was the same as that in untreated transgenic plants: GUS staining in GH3 buds and no detectable staining in DR5 plants.
GUS expression in the buds of transgenic plants. Overview (left) of GH3 and DR5 protonema with buds (middle). Protonema and buds were incubated with 10 µM NAA, and pictures were taken 1 day after induction for the GH3 plants and 3 days after induction for the DR5 plants (right). As an example, GH3 plant G1 and DR5 plant D1 is shown. A GH3 plants display GUS staining in buds but not in outgrowing leaves. B There is no detectable GUS expression in uninduced DR5 buds. Bar: 50 µm
Gametophores
Plants taken from solid Knop medium were used to examine the GUS staining pattern of GH3 and DR5 gametophores. The analysis revealed a centre of GUS activity in the stem apex for both GH3 and DR5 gametophores (Fig. 6). In detail, gametophores of GH3 plants showed GUS staining of the whole stem with two maxima, one in the apex and the other one in the base of the stem (Fig. 6A). The leaves were never stained. Induction of GH3::GUS expression could be triggered by the application of exogenous auxin within 1 day and resulted in the staining of the whole gametophore (Fig. 6A). With respect to the DR5 plants, the stem apex of the gametophores was the only tissue within the plant developmental cycle in which GUS expression could be observed without auxin induction (Fig. 6B). This DR5::GUS staining is comparable to the apical staining maximum of the GH3 plants. The leaves of the gametophores were never stained without auxin induction. The addition of auxin induced an increase in GUS expression that commenced after 3 days and resulted in GUS expression nearly throughout the whole gametophore (Fig. 6B).
Gametophores of GH3 and DR5 plants. In GH3::GUS and DR5::GUS expressing plants, a staining maximum is located in the stem apex. GH3 plants show an additional GUS maximum at the base of the stem. Additionally, gametophores were incubated with 10 µM NAA. Pictures were taken 1 day after NAA induction for GH3 plants and 3 days after NAA induction for the DR5 plants. A GH3 gametophore G5, B DR5 gametophore D2. Bar: 100 µm
Discussion
GH3::GUS and DR5::GUS fusions are functional in Physcomitrella
The GH3::GUS and DR5::GUS fusions are molecular instruments that enable auxin concentration or auxin sensitivity to be measured at the cellular level (Li et al. 1999; Friml and Palme 2002). Both fusions are widely used and well investigated in higher plants such as tobacco, white clover (GH3), and Arabidopsis (DR5; Hagen et al. 1991; Mathesius et al. 1998; Li et al. 1999; Sabatini et al. 1999). In the present study, the functionality of the auxin-inducible GH3::GUS and DR5::GUS fusions was analysed in the ancient land plant Physcomitrella patens. Physcomitrella has a phylogenetic position near the base of extant land plants (Theissen et al. 2001), and it is this position that makes it an interesting tool for studying the evolution of plant gene regulatory mechanisms.
Stable transgenic GH3 and DR5 plants were produced. GUS staining was observed in different tissues of the transgenic plants but with differences between GH3::GUS- and DR5::GUS-expressing transformants with respect to staining intensities. The transgene copy numbers and the expression levels of the characterised plants without auxin application correlated well with their GUS staining behaviour.
A main feature of the GH3::GUS and DR5::GUS fusions is their inducibility by active auxins (Hagen et al. 1991; Ulmasov et al. 1997b). This auxin inducibility was tested for in the GH3 and DR5 plants by treating different tissues with the active auxin NAA. Both fusions, GH3::GUS and DR5::GUS, were able to respond to the application of exogenous auxin in all of the different moss tissues analysed. A further induction of GUS expression by cytokinin could not be shown. These divergent induction properties of auxin and cytokinin are in agreement with results obtained for GH3::GUS and DR5::GUS fusions in higher plants (Hagen et al. 1991). Thus, these promoter elements might be evolutionary conserved and used in the same way in Physcomitrella as in seed plants. The auxin-inducible elements comprising the GH3 and synthetic DR5 promoters were quite obviously detected by the moss P. patens. Therefore, it seems likely that the corresponding binding transcription factors are conserved as well. A search of our EST database (Rensing et al. 2002) revealed the existence of all known gene families related to auxin-induced gene expression according to Hagen and Guilfoyle (2002), such as Aux/IAA, ARF, Saur-like and GH3-like proteins (data not shown): the moss seems to contain the whole set of known transcription factors recognising auxin-inducible elements. The binding of ARF transcription factors to the TGTCTC element within the GH3 and DR5 promoters was demonstrated by Ulmasov et al. (1997a, 1999). ARF transcription factors should form homo- and heterodimers (Ulmasov et al. 1999) and additionally heterodimers with Aux/IAA proteins to regulate the expression of primary auxin-responsive genes by binding the auxin-response elements within their promoters (Hagen and Guilfoyle 2002). This system may also be present in the moss Physcomitrella.
The addition of auxin resulted in the induction of GUS staining in nearly all of the developmental stages of the transgenic plants that were examined (protonema, buds, gametophores). Hence, we conclude, that every Physcomitrella cell has the competence to respond to auxin and that the staining pattern in the transgenic plants is dependent on auxin concentration. The concentration of auxin in unstained parts of the plant restricts the level of GUS gene expression, indicating that both reporter systems actually measure differences in auxin concentration in the plant rather than the auxin sensitivity of cells. However, different staining intensities between younger and older cells in auxin-treated protonema suggest an additional influence of auxin sensitivity on GUS expression.
GH3::GUS and DR5::GUS fusions display differential auxin responsibility in the moss P. patens
Our investigation is the first reported attempt to compare the GH3::GUS and DR5::GUS fusions in the ancient land plant P. patens. The GH3 promoter exhibited a broad GUS-staining pattern in different developmental stages of Physcomitrella without the application of auxin. In contrast, the DR5 promoter showed a limited GUS-staining pattern without exogenous auxin. There were also differences in the behaviour of the two promoters with respect to the time course of induction. The GH3 promoter immediately responded within the first day of auxin application, whereas the DR5 promoter needed a longer time to respond: Only after 3 days of auxin application was the first induction of additional GUS expression detectable with this promoter. Thus, the two elements display wide differences concerning promoter strength and the kinetics of promoter induction by auxin. Obviously, the DR5 promoter is less sensitive than the GH3 promoter—at least in the moss Physcomitrella. In contrast to this, for seed plants the assumption is that the DR5 promoter should be more active than the GH3 promoter (Ulmasov et al. 1997b; Hagen and Guilfoyle 2002). This difference between the two promoters in Physcomitrella might be due to their divergent composition. DR5 is a synthetic promoter consisting of a sevenfold repeat of the same element (Ulmasov et al. 1997b). The GH3 promoter forms part (592 bp) of the naturally occurring soybean GH3 promoter (Hagen et al. 1991) in which at least three different auxin-response elements can function independently of one another (Liu et al. 1994), thereby providing more flexibility for putative binding proteins. While the GH3 system may find broad application in Physcomitrella, the lower sensitivity of the DR5 promoter makes it most useful for specific studies when enhanced auxin concentrations are expected.
Auxin concentration in moss gametophores is highest in dividing/ontogenetic young cells
Very little is known about auxin distribution during different developmental stages in Physcomitrella, especially the situation in the gametophore. Bopp and Atzorn (1992b) were only able to demonstrate the presence of high auxin concentration in the tip cell of protonema. The transgenic moss reporter plants now available offer the means to observe auxin distribution during different stages of Physcomitrella development.
In the transgenic plants, especially in the GH3 plants, the highest GUS expression was always visible in actively growing young cells. In the case of protonema, we were able to observe a faint GUS staining in the outgrowing protonema filaments, which gradually declined in the older cells. Our results are in agreement with those of Bopp and Atzorn (1992b), who detected a peak of endogenous auxin in the tip cells of protonema filaments by immunofluorescence. Auxin plays an important role in the differentiation step from the chloronema to the caulonema stage of protonema (Johri and Desai 1973) and is also important for the maintenance of the caulonema state (Bopp 2000). As the protonema filaments grew older, the GUS expression pattern changed to an even distribution throughout the protonema. The reason for this change might be that older protonema cells react to the auxin accumulation in the medium. Physcomitrella excrete most of the auxin produced into the medium (Reutter et al. 1998), and after medium exchange the auxin concentration could rise again.
The next step in moss development, bud formation, can be stimulated by the application of exogenous cytokinins (Gorton and Eakin 1957). However, higher auxin concentrations inhibit normal bud development (Reski 1998b), and there are hints for a participation of auxin together with cytokinins in bud development (Cove and Ashton 1984). GUS expression in the developing buds demonstrates indirectly the presence of auxin in this developmental stage of Physcomitrella; therefore, a possible role of auxin in bud development must be assumed. For seed plants, high concentrations of auxin in the leaf primordia have been described (Avsian-Kretchmer et al. 2002). Analogue structures, although of gametophytic origin, are the outgrowing leaves of buds. In contrast to seed plants, no GUS expression—i.e. high auxin concentrations—were detectable in this functional tissue equivalent.
While investigators had made a few assumptions about the physiological role and the distribution of auxin in the protonema and the buds of Physcomitrella, previous to this study almost nothing was known of these topics in the leafy gametophore. The GUS expression pattern defines two main locations of high auxin occurrence in GH3 plants: the apical part and the basal part of the gametophore stem. It is likely that the stem apex provides a functional gametophytic analogue to the shoot apical meristem of seed plants, which is known to be correlated with high auxin concentrations (Avsian-Kretchmer et al. 2002). Another indication for the presence of a meristematic analogue is the occurrence of moss homologues for genes controlling the generation of the shoot apical meristem (Theissen et al. 2001). As the leaves of GH3 plants showed no GUS staining, they may contain no or undetectably low concentrations of auxin. In higher plants, the situation in leaves is different. Avsian-Kretchmer et al. (2002) analysed auxin distribution in developing leaves of Arabidopsis seedlings using the DR5::GUS fusion. They found that young leaf primordia showed high auxin concentrations, which decreased when the leaves grew older. GUS activity was concentrated at the distal end of the leaves and was also visible in the leaf vasculature.
In higher plants, the highest levels of auxin are found in regions of active cell division or ontogenetic young tissue, such as the apical meristems, young leaves (Avsian-Kretchmer et al. 2002) and the root meristem (Sabatini et al. 1999). As we demonstrated by indirectly measuring auxin concentration with the help of auxin-inducible reporter gene constructs, this was also true for the moss Physcomitrella as outgrowing protonema cells, buds and the apex of gametophores are locations with high levels of cell division or meristematic activity.
The Physcomitrella plants expressing the auxin-inducible reporter genes will be used for studying auxin transport processes and the involvement of auxin in many growth and developmental events such as tropism or tissue differentiation. Additionally, the transgenic plants will be used to characterise the function of genes involved in auxin transport or signal transduction using the unique possibility of Physcomitrella: targeted gene knockout by homologous recombination against the transgenic background.
Abbreviations
- DR5:
-
Synthetic auxin-inducible promoter
- EST:
-
Expressed sequence tag
- GH3:
-
Promoter of the soybean gh3 gene
- GUS:
-
β-Glucuronidase
- NAA:
-
1-Naphthaleneacetic acid
- 2-NAA:
-
2-Naphthaleneacetic acid
References
Avsian-Kretchmer O, Cheng J-C, Chen L, Moctezuma E, Sung ZR (2002) Indoleacetic acid distribution coincides with vascular differentiation pattern during Arabidopsis leaf ontogeny. Plant Physiol 130:199–209
Bopp M (2000) 50 years of the moss story. In: Progress in botany, vol 61. Springer, Berlin Heidelberg New York, pp 1–34
Bopp M, Atzorn R (1992a) Hormonelle Regulation der Moosentwicklung. Naturwissenschaften 79:337–346
Bopp M, Atzorn R (1992b) The morphogenetic system of the moss protonema. Cryptogam Bot 3:3–10
Cove DJ, Ashton NW (1984) The hormonal regulation of gametophytic development in bryophytes. In: Dyer AF, Duckett JG (eds) The experimental biology of bryophytes. Academic Press, New York, pp 177–201
Egener T, Granado J, Guitton M-C, Hohe A, Holtorf H, Lucht JM, Rensing S, Schlink K, Schulte J, Schween G, Zimmermann S, Duwenig E, Rak B, Reski R (2002) High frequency of phenotypic deviations in Physcomitrella patens plants transformed with a gene-disruption library. BMC Plant Biol 2:6
Friml L, Palme K (2002) Polar auxin transport—old questions and new concepts? Plant Mol Biol 49:273–284
Gorton BS, Eakin RE (1957) Development of the gametophyte in the moss Tortella caespitosa. Bot Gaz 119:31–38
Guilfoyle TJ (1999) Auxin-regulated genes and promoters. In: Hooykaas PPJ, Hall MA, Libbenga KR (eds) Biochemistry and molecular biology of plant hormones. Elsevier, New York, pp 423–459
Hagen G, Guilfoyle TJ (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Hagen G, Kleinschmidt A, Guilfoyle TJ (1984) Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 162:147–153
Hagen G, Martin G, Li Y, Guilfoyle TJ (1991) Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol Biol 17:567–579
Hohe A, Schween G, Reski R (2001) Establishment of a semicontinous bioreactor culture of Physcomitrella patens for mass production of protoplasts. Acta Hortic 560:425–427
Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14:373–386
Johri MM, Desai S (1973) Auxin regulation of caulonema formation in moss protonema. Nat New Biol 245:223–224
Li Y, Wu YH, Hagen G, Guilfoyle TJ (1999) Expression of the auxin-inducible GH3 promoter/GUS fusion gene as a useful molecular marker for auxin physiology. Plant Cell Physiol 40:675–682
Liu Z, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6:645–657
Mathesius U, Schlaman HRM, Spaink HP, Sautter C, Rolfe BG, Djordjevic MA (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14:23–34
Nishiyama T, Hiwatashi Y, Sakakibara K, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens, by shuttle mutagenesis. DNA Res 7:9-17
Rensing SA, Rombauts S, Van de Peer Y, Reski R (2002) Moss transcriptome and beyond. Trends Plant Sci 7:535–538
Reski R (1998a) Physcomitrella and Arabidopsis: the David and Goliath of reverse genetics. Trends Plant Sci 3:209–210
Reski R (1998b) Development, genetics and molecular biology of mosses. Bot Acta 111:1-15
Reski R, Abel WO (1985) Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 165:354–358
Reski R, Faust M, Wang XH, Wehe M, Abel WO (1994) Genome analysis of the moss Physcomitrella patens (Hedw.) B.S.G. Mol Gen Genet 244:352–359
Reutter K, Atzorn R, Hadeler B, Schmülling T, Reski R (1998) Expression of the bacterial ipt gene in Physcomitrella rescues mutations in budding and in plastid division. Planta 206:196–203
Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle TJ, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99:463–472
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein Ftsz, an ancestral tubulin. Proc Natl Acad Sci USA 95:4368–4373
Theissen G, Münster T, Henschel K (2001) Why don´t mosses flower? New Phytol 150:1-5
Töpfer R, Schell J, Steinbiss H-H (1988) Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res 16:8725
Ulmasov T, Liu Z, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7:1611–1623
Ulmasov T, Hagen G, Guilfoyle TJ (1997a) ARF1, a transcription factor that binds to auxin response elements. Science 276:1865–1868
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971
Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19:309–319
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft via a concerted programme on molecular analysis of phytohormone action (DFG RE 837/6). We thank Tom Guilfoyle and Jane Murfett for kindly providing the GH3::GUS- and the DR5::GUS-containing plasmids and Dr. G. Schween for carrying out the flow cytometric analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Lörz
Rights and permissions
About this article
Cite this article
Bierfreund, N.M., Reski, R. & Decker, E.L. Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens . Plant Cell Rep 21, 1143–1152 (2003). https://doi.org/10.1007/s00299-003-0646-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-003-0646-1