Abstract
In mammals, enteric salmonellas can use tetrathionate (ttr), formed as a by-product from the inflammatory process in the intestine, as electron acceptor in anaerobic respiration, and it can fuel its energy metabolism by degrading the microbial fermentation product 1,2-propanediol. However, recent studies have shown that this mechanism is not important for Salmonella infection in the intestine of poultry, while it prolongs the persistence of Salmonella at systemic sites in this species. In the current study, we show that ΔttrApduA strains of Salmonella enterica have lower net survival within chicken-derived HD-11 macrophages, as CFU was only 2.3% (S. Enteritidis ΔttrApduA), 2.3% (S. Heidelberg ΔttrApduA), and 3.0% (S. Typhimurium ΔttrApduA) compared to wild-type strains after 24 h inside HD-11 macrophage cells. The difference was not related to increased lysis of macrophages, and deletion of ttrA and pduA did not impair the ability of the strains to grow anaerobically. Further studies are indicated to determine the reason why Salmonella ΔttrApduA strains survive less well inside macrophage cell lines.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salmonella enterica is a major foodborne pathogen, contributing to an estimated 150 million illnesses and 60,000 deaths annually worldwide (Plumb et al. 2023). Poultry products contaminated with non-typhoid serovars, such as Salmonella Enteritidis (SE), Salmonella Heidelberg (SH), and Salmonella Typhimurium (STM) are of particular public health concern (CDC 2013, 2018; ECDC and EFSA 2021; Kipper et al. 2022).
Salmonella are highly specialized pathogens expressing a set of virulence factors involved in attachment and invasion of the intestinal epithelium, and subsequently evading the host immune response (Ménard et al. 2022), as well as a set of virulence factors, which allows the bacterium to survive and replicate within macrophages. The latter ability is essential for the systemic form of Salmonella infection (Barrow et al. 1994). In addition to the ability of Salmonella to overcome the intestinal mucosa epithelium barrier, the bacteria must compete with the existing microbiota in an anaerobic environment of the intestine. To this end, Salmonella spp. can utilize tetrathionate as an electron acceptor in anaerobic respiration through transcription of the ttr operon (Winter et al. 2010) and use 1,2-propanediol as an energy source through the expression of the pdu gene cluster, as first demonstrated with STM infection in murine experimental models (Lawley et al. 2008; Winter et al. 2010; Thiennimitr et al. 2011; Rivera-Chávez et al. 2013; Faber et al. 2017). For this reason, ttrA and pduA genes are considered central players during intestinal infection of mammals (Lawley et al. 2008).
The role of ttr and pdu operons in infection of chicken is less clear. Harvey et al. (Harvey et al. 2011) demonstrated that deleting the pdu operon from the STM genome reduced the bacterial shedding in chicken feces. A more recent report, however, showed that the absence of ttrA and pduA did not impair bacterial shedding after infection of chicken with SE, STM, and SH, but rather suggested it was highest (Saraiva et al. 2021; Góes et al. 2022). Thus, unlike the diverse set of factors that influence Salmonella pathogenicity (Winter et al. 2010; Harvey et al. 2011), such as fae, shdA, misL, sipABC genes related to fimbrial adherence, nonfimbrial adherence, invasion, and type III secretion system, respectively (Brown et al. 2021; Vinueza-Burgos et al. 2023; Monte et al. 2024), ttrA and pduA does not seem to be fundamental for intestinal infection in chicken. In systemic infection of chicken, no major differences in pathogenicity depending on the presence of these genes were likewise observed. However, the SE and STM ttrA and pduA mutants grew to higher numbers in the liver and spleen of both broiler and layer chickens (Saraiva et al. 2021), suggesting a different interaction with the host during systemic infections. Given the importance of macrophage survival for the propagation of Salmonella at systemic sites, we hypothesized that ttrA and pduA gene clusters affect Salmonella survival in the intracellular environment of chicken-derived macrophages, and the current study aimed to determine the ability of SE, STM and SH strains with defective ttrA and pduA genes to survive and grow inside chicken derived HD-11 macrophage-like cells.
Materials and methods
Bacterial strains
Deletion mutants in ttrA and pduA in SE, SH and STM (SEΔttrApduA, SHΔttrApduA, and STMΔttrApduA) were obtained from a previous study (Saraiva et al. 2021). The wild-type strains that provided the genetic background for the mutant constructions (SE P125109; SH a field-isolate; STM 98) (Saraiva et al. 2021) were used as positive controls in experiments.
Whole genome sequence analysis
To rule out changes in gene sequences in mutants, in addition to the designed deletions of ttrA and pduA, whole genome sequencing was carried out. Bacterial strains were cultured overnight in Lysogeny Broth (LB, Becton Dickinson, USA). Subsequently, 1.5 mL of each was centrifuged, and the resulting pellet was used for the genomic DNA extraction using a commercial kit (QIAmp Fast DNA Tissue, Qiagen, USA). Following DNA extraction, the integrity of DNA samples was evaluated using electrophoresis in a 1% agarose gel, and quantification was carried out using a spectrophotometer (Colibri, Titertek-Berthold, Germany). Subsequently, genomic libraries were prepared using Nextera XT (Illumina, USA), according to the manufacturer’s guidelines. Paired-end sequencing was performed in the MiSeq platform (Illumina) using a 2 × 250 cycle V2 kit (Illumina). Subsequent bioinformatic analyses, including removal of low-quality reads and adapters, genome assembly, and annotation, were performed using Geneious software (v. 2023.1.2).
Tetrathionate utilization assay
Following confirmation of gene knockout, an in vitro assessment of tetrathionate use by mutant and wild-type strains was carried out. Initially, all strains were adjusted to approximately 5 × 103 CFU/mL by inoculation into M9 minimal salt medium broth. In this M9 medium, propylene glycol (1,2 propanediol) is replaced by glucose in similar amounts found in the tetrathionate broth and thiosulfate sodium. Subsequently, iodine was added to half of the medium volume to oxidize thiosulfate to tetrathionate. Finally, both mutant and wild-type strains of each serovar were inoculated in M9 (thiosulfate or tetrathionate) in the presence or absence of oxygen. The inoculated samples were then incubated at 37 °C for 24 h under aerobic conditions and 48 h in an anaerobic jar with an anaerobic environment generator (Anaerobac®, Probac, Brazil). A detailed composition of the M9 medium used is described in Table S1.
Chicken-derived macrophage-like cell line HD-11 cultivation
The infection of chicken macrophage-like cell line HD-11 followed the protocol outlined by Huang et al. (Huang et al. 2019) and Rodrigues Alves et al. (Rodrigues Alves et al. 2024), with slight modifications. Initially, nitrogen-stocked cells were defrosted and transferred into 40 mL of RPMI (RPMI Medium 1640 + GlutaMAX™, ThermoFisher Scientific, USA) supplemented with 5% of inactivated Chicken Serum (Chicken Serum, Sigma-Aldrich, USA), 5% Fetal Bovine Serum (FBS EU Origin, Biowest, France), and 25 µg/mL gentamicin was added (Gentamicin 50 mg/mL, ThermoFisher Scientific, USA). The cell suspension was evenly transferred into 75 cm2 tissue culture flasks (TPPⓇ, Switzerland) and conditioned at 37 °C in a 5% CO2 incubator. The media were replaced as needed, based on cell morphology and multiplication. For media replacement, cells were washed with 20 mL of Dulbecco’s Phosphate Buffered Saline (DPBS [1x], Gibco, ThermoFisher Scientific, USA) and the same volume of supplemented fresh RPMI was added. Subsequently, the flasks were returned to the same conditions as before.
Upon reaching 80% of confluence, the cells were washed twice with 20 mL of DPBS. Subsequently, the monolayer cells were detached from the bottom of the flask using a sterile scraper on the final resuspension. The cell suspension was then transferred into a 50 mL Falcon tube and centrifuged at 5000 x g for 8 min. Following the centrifugation, supernatant was discarded, and 3 mL of RPMI was added. The cells were gently resuspended with a pipette and the supplemented medium was added until a final volume of 10 mL was reached. Then, 2 µL of 0.4% Trypan Blue (ThermoFisher Scientific, USA) was added to 18 µL of cell culture and 10 µL of the mixture was transferred into a hemocytometer chamber with a glass coverslip. The cells were counted in the two large squares diagonal from each other, and the density was calculated as follows: cells per mL = Average number of cells x 10,000. The concentration was adjusted to 5 × 105 cells/mL and 1 mL of the suspension was added to each well and incubated in the same conditions as previously described.
Macrophage infection
Wild-type (SE, SH, and STM) and mutant strains (SEΔttrApduA, SHΔttrApduA, and STMΔttrApduA) were recovered from – 80 ºC ultrafreezer and streaked on Lysogeny Agar (LA, Becton Dickinson, USA), which were incubated for 24 h at 37 ºC. The pre-inoculum was prepared on the day before the infection by transferring a single colony of each strain from LA into 10 mL of sterile LB. These cultures were then incubated under the previously mentioned shaking conditions.
On the infection day, 0.5 mL of the overnight cultures were transferred to 9.5 mL of fresh LB and incubated at 37 ºC under 180 rpm for approximately 4 h. Then, the inoculum was centrifuged at 5000 x g per 5 min and resuspended with 10 mL of DPBS supplemented with 20% of inactivated Salmonella antibody-free chicken serum, and then statically incubated at 37 ºC for 45 min to opsonize the bacteria. During this incubation period, the media of the wells were replaced by freshly RPMI without gentamicin, and an extra well was scraped to enumerate the number of cells per mL.
The cultures were washed three times with 0.9% NaCl solution by centrifugation at 5000 x g per 5 min and resuspended in 10 mL of supplemented RPMI without gentamicin. The optimal density (OD600) was measured using a spectrophotometer (Eppendorf BioPhotometer, Germany), and the cultures were adjusted to a concentration of 107 CFU/mL according to the number of cells. Subsequently, 0.1 mL of bacterial cultures were added to the corresponding wells, considering a Multiplicity of Infection (MOI) of 10:1. The inoculum concentrations were verified by decimal serial dilution on LA plates.
To facilitate the bacterial uptake by HD-11 and monocyte-derived primary macrophages, the 24-well plates were centrifuged at 455.59 x g for 5 min at 25 ºC immediately after inoculation. Subsequently, the tissue culture plates were incubated at 37 ºC in a 5% CO2 incubator. After 30 min post-inoculation, the media was pipette out, and 1 mL of RPMI supplemented with 100 µg/mL of gentamicin was added to eliminate extracellular bacteria (uptake hereafter). After 30 min of incubation, the wells were washed three times with 1 mL of pre-warmed 0.9% NaCl. Then, 1 mL of 0.1% Triton-X was added to the plate used to enumerate the bacterial uptake. To ensure efficient lysis, the bottom of the wells was gently scraped with sterile pipette tips, and after a 10-minute incubation period, bacterial counts were determined by performing ten-fold dilutions plated on LA.
To evaluate the intracellular survival, the supernatants of the other plates were removed, followed by the triple cell wash with 0.9% NaCl. Subsequently, 1 mL of RPMI-25 was added to each well. At one, four-, and 24 h post-cell infection, the triple washing, cell disruption, and decimal serial dilution steps were performed as previously described. Each assay was conducted in triplicates.
Bacterial uptake, fold-net replication, and survival from HD-11 infection
All the bacterial counts after infection of macrophage-like cell line were logarithmically transformed, and then calculated as follows: uptake was the proportion between CFU/mL from HD-11 cells (after initial lysis – 30 min) and CFU/mL of initial inoculum (used for infection). The fold-net replication was obtained by the proportion between CFU/mL of bacterial uptake and CFU/mL of CFU/mL from HD-11 cells (after 1 h lysis); these proportions were repeated for fold-net replication 4 h/1 h and 20 h/4 h. Ultimately, the survival of mutant strains was calculated by percentage concerning the wild-type survival: individual survival from mutant or wild-type strains was the proportion between CFU/mL from HD-11 cells (after 20 h lysis) and CFU/mL of bacterial uptake.
Macrophage cytotoxicity assay
To assess the cytotoxicity of infected macrophages, 50 µL supernatant from each well were transferred in duplicates at four- and 24-hours post-infection into 96-well microtiter plates. Then cytosolic lactate dehydrogenase (LDH) activity was measured according to the manufacturer´s instructions using the colorimetric CytoTox 96Ⓡ Non-Radioactive Cytotoxicity Assay (Promega Corporation, USA). The reactions were read under Optical Densitometry of 490 nm (OD490) and scored according to the following formula: Cytotoxicity % = (infected cell release – uninfected cell release x 100) / maximum release – uninfected cell release. Three biological replicates were done.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 10.0.0 for macOS (GraphPad Software, La Jolla, California, USA). Bacterial survival and fold-net replication data from macrophage infection assays, cytotoxicity assays, and bacterial enumeration in M9 medium were performed using two-way ANOVA followed by Bonferroni multiple comparison test. Inoculum and uptake were analyzed using an Unpaired T-test (Wilcoxon matched-pairs signed rank test). P-values lower than 0.05 (P < 0.05) were considered statistically significant.
Results and discussion
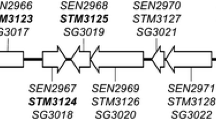
The study aimed to assess the survival capabilities of strains of SE, STM, and SH carrying deletion in ttrA and pduA during infection of HD-11 macrophage-like cells. Our initial focus was on confirming the mutations and their genetic context. To achieve this, we sequenced the entire genome of the mutant strains and compared them to the wild-type genome. Local alignment analyses (BLAST) showed proper mutation, leading to a 2981 bp deletion in ttrA and 201 bp deletion in pduA in all three serovars. A 90 bp-scar sequence in each gene was identified, but the relative length of the scar in comparison to the entire gene sequence (3,071 nt for ttrA; 291 nt for pduA) was unlikely to allow the proper protein expression, mainly considering the presence of stop codons at the end of the remaining sequence (Fig. 1A and B) (Baba et al. 2006; Fels et al. 2020). In both genes, promoter regions were removed without affecting genes located up and/or downstream of the depletion (Fig. 1A and B). Furthermore, no additional deletions or point mutations were found in any of the three mutant strains. The complete genome data of SEΔttrApduA, SHΔttrApduA, and STMΔttrApduA were deposited in DDBJ/ENA/GenBank with Accession No. SAMN38927547, SAMN38927628, and SAMN38927550, respectively.
Interestingly, a recent report has demonstrated that double deletion of ttrA and pduA enhances the immune cell response in chickens, especially by prominent infiltration of macrophages (Cabrera et al. 2023). Based on these findings, we conducted macrophage infection assays with SEΔttrApduA, SHΔttrApduA, and STMΔttrApduA and their respective wild-type strains to compare the intracellular survival of mutant and wild-type strains within HD-11 cells. There was a trend towards lower uptake of mutant strains than wild-type strains for all three serovars, however, none of the differences in bacterial uptake by HD-11 cells were significant (Fig. 2A and C, and 2E). Intracellular survival of the wild-type strains was higher at 4 h (STM) and 24 h (SE, SH, and STM) post-infection compared to their respective double mutant strains (Fig. 2B and D, and 2F), i.e., the lack of ttrA and pduA, reduced bacterial survival inside HD-11 to 2.3% (SE), 2.3% (SH), nd 3.0% (STM), respectively (Fig. 3B, D and F). CFU determined in gentamicin protection assays over time reflects the sum of intracellular survival and intracellular multiplication. Expressed as net-replication, the deletion of ttrA and pduA impaired the net replication rates in the intracellular environment of all three serovars (Fig. 3A and C, and 3E).
Fold-net replication of SEΔttrApduA and SE (A), SHΔttrApduA and SH (C), and STMΔttrApduA and STM (E) and replication/survival of mutants expressed as percentage of wild-type strains (B, D, F) within HD-11 cell-like macrophages 24 h post-challenge. ns: non-significant (P > 0.05); *, P < 0.05; **, P < 0.01
Results of studies of in vitro growth of Salmonella have suggested that tetrathionate-based respiration is the most important system for Salmonella growth under anaerobic condition (Hensel et al. 1999). To determine whether the lower net replication of mutant strains in the intracellular environment of macrophages was related to the inability to grow under anaerobic conditions, we compared the ability of wild-type and mutant strains to grow under conditions with and without oxygen and with and without the possibility to use tetrathionate as electron acceptor. This demonstrated no significant differences in growth between the ΔttrApduA mutants and their respective wild-type strain at both aerobic and anaerobic conditions (Fig. 4), suggesting that lack of the genes would not impair the ability of the strains to grow under anaerobic conditions. To rule out that differences in CFUs after 24 h were due to increased lysis of macrophages, which would subject the bacteria to the killing activity of gentamicin, a measurement of macrophage cytotoxicity was further carried out. No difference between the cytotoxicity of mutant and wild-type strains was observed for any of the serovars (Supplemental material Figure S1).
Enumeration of wild-type and mutant strains during growth with and without oxygen and with and without the possibility to use tetrathionate as electron acceptor. Tetrathionate + O2: growth in M9 broth supplemented with tetrathionate under aerobic condition; Thiosulfate + O2: growth in M9 broth supplemented with thiosulfate under aerobic condition; Tetrathionate + Ana: growth in M9 broth supplemented with tetrathionate under anaerobic condition; Thiosulfate + Ana = inoculum in M9 broth supplemented with thiosulfate in anaerobioses condition
Previous studies have predominantly focused on the role of ttrA and pduA genes in infections of the intestine under anaerobic conditions (Lawley et al. 2008), while metabolic pathways commonly investigated in relation to macrophage infection have centred around mgtC gene, which is essential to ATP homeostasis and persistence of Salmonella spp. within macrophages (Lee et al. 2013). Research has shown that a deletion of this gene in S. Gallinarum reduced proliferation inside HD-11 chicken derived macrophages when compared to the wild-type strain (Rodrigues Alves et al. 2024). From the current study, it appears that deletion of ttrA and pduA genes also leads to reduced proliferation inside this cell line. Interestingly, a significantly lower viable cell count was mostly observed at a late stage of infection (24 h post-infection - hpi). This contrasts with the effect of deletion of mgtC in S. Typhi (Retamal Patricio and Castillo-Ruiz 2009) and S. Typhimurium (Smith et al. 1998; Retamal Patricio and Castillo-Ruiz 2009), where lower bacterial loads were observed at the early stage of infections (between 2 and 4 h post-infection).
The current study was carried out with double deletion mutants, and it is not possible to conclude which of the genes was important for the observed phenotypes. Our observation is that pdu may be important for intracellular growth in macrophages; this gene deletion has already been associated with the accumulation of toxic intermediates such as propionaldehyde (Cheng et al. 2011), which could possibly influence intramacrophage survival. Furthermore, previous observations, for instance, proteomic analysis of S. Typhimurium grown in a magnesium-depleted minimal medium, which partially mimics the environment of the Salmonella-containing vacuole, indicates that proteins from the pdu operon are expressed, especially in the virulent strain ATCC 14,028 (Adkins et al. 2006), and according to Klumpp and Fuchs (2007), the cob-cbi-pdu gene cluster increases the intracellular fitness of S. Typhimurium in mouse-derived J774A.1 macrophages. Additionally, during the infection, the cell undergoes metabolic reprogramming that favors glycolysis, which is an anaerobic process (Jiang et al. 2021). Moreover, the availability of nutrients and reduced antimicrobial activity are essential for intracellular multiplication (Liss et al. 2017). Further studies are needed to elucidate how and why pduA and ttrA affect intracellular survival, including whether the absence of those genes can lead to higher levels of reactive oxygen species (ROS) which would impair bacterial survival after being ingested by macrophages (West et al. 2011).
The use of HD-11 cells in the current study had certain advantages: they are easy to cultivate and eliminate the need for primary extraction of macrophages from chicken, serving as an easy in vitro alternative to avian macrophages. However, it is important to note that despite their ease of cultivation, HD-11 cells may not fully replicate normal cell functionality and can express unique gene patterns not found in vivo (Beug et al. 1979; Carter and Shieh 2015).
Conclusion
Our findings show that the ttrA and pduA genes affect bacterial growth/survival within chicken-derived HD-11 macrophages. Further studies are needed to understand why this is the case, since our in vitro growth assays indicate that Salmonella grows equally well with and without these genes at anaerobic conditions.
Data availability
Sequence data that support the findings of this study have been deposited in DDBJ/ENA/GenBank with Accession No. SAMN38927547, SAMN38927628, and SAMN38927550.
References
Adkins JN, Mottaz HM, Norbeck AD et al (2006) Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol Cell Proteomics 5:1450–1461. https://doi.org/10.1074/mcp.M600139-MCP200
Baba T, Ara T, Hasegawa M et al (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2. https://doi.org/10.1038/msb4100050
Barrow PA, Huggins MB, Lovellt MA (1994) Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system
Beug H, von Kirchbach A, Döderlein G et al (1979) Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375–390. https://doi.org/10.1016/0092-8674(79)90057-6
Brown EW, Bell R, Zhang G et al (2021) Salmonella Genomics in Public Health and Food Safety. EcoSal Plus. https://doi.org/10.1128/ecosalplus.ESP-0008-2020. 9:eESP-0008-2020
Cabrera JM, Saraiva MMS, Rodrigues Alves LB et al (2023) Salmonella enterica serovars in absence of ttrA and pduA genes enhance the cell immune response during chick infections. Sci Rep 13. https://doi.org/10.1038/s41598-023-27741-x
Carter M, Shieh J (2015) Cell Culture Techniques. In: Carter M, Shieh J (eds) Guide to Research Techniques in Neuroscience. Academic Press, pp 295–310
Centers for Disease Control and Prevention – CDC (2018) Salmonella Infections Linked to Chicken Salad. https://www.cdc.gov/salmonella/typhimurium-02-18/index.html. Accessed 15 August 2023
Centers for Disease Control and Prevention – CDC (2013) Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Foster Farms Brand Chicken (Final Update). https://www.cdc.gov/salmonella/heidelberg-10-13/index.html. Accessed 15 August 2023
Cheng S, Sinha S, Fan C et al (2011) Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J Bacteriol 193(6):1385–1392. https://doi.org/10.1128/JB.01473-10
European Centre for Disease Prevention and Control – ECDC, European Food Safety Authority – EFSA (2021) Multi-country outbreak of Salmonella Enteritidis sequence type (ST)11 infections linked to poultry products in the EU/EEA and the United Kingdom. https://doi.org/10.2903/sp.efsa.2021.en-6486. EFSA Supporting Publications 18:
Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, Andrews-Polymenis HL, Winter SE, Bäumler AJ (2017) Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13:1–19. https://doi.org/10.1371/journal.ppat.1006129
Fels U, Gevaert K, Van Damme P (2020) Bacterial Genetic Engineering by means of Recombineering for Reverse Genetics. Front Microbiol 11
Góes V, Monte DFM, Saraiva M, de MS et al (2022) Salmonella Heidelberg side-step gene loss of respiratory requirements in chicken infection model. Microb Pathog 171. https://doi.org/10.1016/j.micpath.2022.105725
Harvey PC, Watson M, Hulme S et al (2011) Salmonella enterica serovar typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun 79:4105–4121. https://doi.org/10.1128/IAI.01390-10
Hensel M, Hinsley AP, Nikolaus T et al (1999) The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol 32:275–287. https://doi.org/10.1046/j.1365-2958.1999.01345.x
Huang K, Herrero-Fresno A, Thøfner I et al (2019) Interaction differences of the avian host-specific Salmonella enterica Serovar Gallinarum, the host-generalist S. Typhimurium, and the cattle host-adapted S. Dublin with chicken primary macrophage. Infect Immun 87:101128iai00552–101128iai00519. https://doi.org/10.1128/iai.00552-19
Jiang L, Wang P, Song X et al (2021) Salmonella Typhimurium reprograms macrophage metabolism via T3SS effector SopE2 to promote intracellular replication and virulence. Nat Commun 12. https://doi.org/10.1038/s41467-021-21186-4
Kipper D, Mascitti AK, De Carli S et al (2022) Emergence, dissemination and Antimicrobial Resistance of the Main Poultry-Associated Salmonella Serovars in Brazil. Vet Sci 9
Klumpp J, Fuchs TM (2007) Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiol (N Y) 153:1207–1220. https://doi.org/10.1099/mic.0.2006/004747-0
Lawley TD, Bouley DM, Hoy YE et al (2008) Host transmission of Salmonella enterica Serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun 76:403–416. https://doi.org/10.1128/iai.01189-07
Lee E-J, Pontes MH, Groisman EA (2013) A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell 154:146–156. https://doi.org/10.1016/j.cell.2013.06.004
Liss V, Swart AL, Kehl A et al (2017) Salmonella enterica remodels the host cell endosomal system for efficient Intravacuolar Nutrition. Cell Host Microbe 21:390–402. https://doi.org/10.1016/j.chom.2017.02.005
Ménard S, Lacroix-Lamandé S, Ehrhardt K et al (2022) Cross-talk between the intestinal epithelium and Salmonella Typhimurium. Front Microbiol 13
Monte DFM, Saraiva MMS, Cabrera JM et al (2024) Unravelling the role of anaerobic metabolism (pta-ackA) and virulence (misL and ssa) genes in Salmonella Heidelberg shedding using chicken infection model. Brazilian J Microbiol. https://doi.org/10.1007/s42770-023-01241-6
Plumb I, Fields P (Patti), Bruce B (eds) (2023) Salmonellosis, Nontyphoidal. In: CDC Yellow Book 2024: Health Information for International Travel
Retamal Patricio AND, Castillo-Ruiz MANDMGC (2009) Characterization of MgtC, a virulence factor of Salmonella enterica Serovar Typhi. PLoS ONE 4:1–6. https://doi.org/10.1371/journal.pone.0005551
Rivera-Chávez F, Winter SE, Lopez CA et al (2013) Salmonella Uses Energy Taxis to benefit from intestinal inflammation. PLoS Pathog 9. https://doi.org/10.1371/journal.ppat.1003267
Rodrigues Alves LB, de Freitas Neto OC, de Mesquita Souza Saraiva M et al (2024) Salmonella Gallinarum mgtC mutant shows a delayed fowl typhoid progression in chicken. Gene 892:147827. https://doi.org/10.1016/j.gene.2023.147827
Saraiva MMS, Rodrigues Alves LB, Monte DFM et al (2021) Deciphering the role of ttrA and pduA genes for Salmonella enterica serovars in a chicken infection model. Avian Pathol 50:257–268. https://doi.org/10.1080/03079457.2021.1909703
Smith RL, Kaczmarek MT, Kucharski LM, Maguire ME (1998) Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtCB during invasion of epithelial and macrophage cells. Microbiol (N Y) 144:1835–1843. https://doi.org/10.1099/00221287-144-7-1835
Thiennimitr P, Winter SE, Winter MG et al (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci 108:17480–17485. https://doi.org/10.1073/pnas.1107857108
Vinueza-Burgos C, Medina-Santana J, Maldonado R et al (2023) Evaluation of virulence of Salmonella Infantis and Salmonella Enteritidis with in vitro and in vivo models. Foodborne Pathog Dis 20:484–491. https://doi.org/10.1089/fpd.2023.0060
West AP, Brodsky IE, Rahner C et al (2011) TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472:476–480. https://doi.org/10.1038/nature09973
Winter SE, Thiennimitr P, Winter MG et al (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. https://doi.org/10.1038/nature09415
Acknowledgements
We are in debt to Xiao Fei for the valuable support in the cell culture assay.
Funding
This work was supported by The São Paulo Research Foundation (FAPESP) [grant numbers 2018/03189-0, 2018/21301-2, and 2020/0741-0].
Author information
Authors and Affiliations
Contributions
Mauro M. S. Saraiva: Writing – original draft, review & editing, Visualization, Validation, Supervision, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Valdinete P. Benevides: Writing – original draft, review & editing, Visualization, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Priscila R. Guerra: Writing – review & editing, Visualization, Validation, Methodology, Investigation, Formal analysis. Adriana Maria de Almeida: Methodology, Formal analysis. Isabella C. Campos: Writing – original draft, Methodology, Formal analysis. Lucas B. Rodrigues Alves: Methodology, Investigation, Formal analysis, Data curation. Jacqueline B. Paiva: Methodology, Formal analysis, Data curation. Lauanda M. Muniz: Methodology, Formal analysis. Adriana M. Almeida: Methodology, Formal analysis, Data curation. Oliveiro C. Freitas Neto: Writing – review & editing, Visualization, Validation, Supervision, Investigation, Data curation, Conceptualization. John E. Olsen: Writing – original draft, review & editing, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Angelo Berchieri Junior: Writing – review & editing, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saraiva, M., Benevides, V.P., Guerra, P.R. et al. Deletions of ttrA and pduA genes in Salmonella enterica affect survival within chicken-derived HD-11 macrophages. Curr Genet 70, 14 (2024). https://doi.org/10.1007/s00294-024-01299-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00294-024-01299-1