Abstract
RNA/DNA hybrids are processed by RNases H1 and H2, while single ribonucleoside-monophosphates (rNMPs) embedded in genomic DNA are removed by the error-free, RNase H2-dependent ribonucleotide excision repair (RER) pathway. In the absence of RER, however, topoisomerase 1 (Top1) can cleave single genomic rNMPs in a mutagenic manner. In RNase H2-deficient mice, the accumulation of genomic rNMPs above a threshold of tolerance leads to catastrophic genomic instability that causes embryonic lethality. In humans, deficiencies in RNase H2 induce the autoimmune disorders Aicardi–Goutières syndrome and systemic lupus erythematosus, and cause skin and intestinal cancers. Recently, we reported that in Saccharomyces cerevisiae, the depletion of Rnr1, the major catalytic subunit of ribonucleotide reductase (RNR), which converts ribonucleotides to deoxyribonucleotides, leads to cell lethality in absence of RNases H1 and H2. We hypothesized that under replicative stress and compromised DNA repair that are elicited by an insufficient supply of deoxyribonucleoside-triphosphates (dNTPs), cells cannot survive the accumulation of persistent RNA/DNA hybrids. Remarkably, we found that cells lacking RNase H2 accumulate ~ 5-fold more genomic rNMPs in absence than in presence of Rnr1. When the load of genomic rNMPs is further increased in the presence of a replicative DNA polymerase variant that over-incorporates rNMPs in leading or lagging strand, cells missing both Rnr1 and RNase H2 suffer from severe growth defects. These are reversed in absence of Top1. Thus, in cells lacking RNase H2 and containing a limiting supply of dNTPs, there is a threshold of tolerance for the accumulation of genomic ribonucleotides that is tightly associated with Top1-mediated DNA damage. In this mini-review, we describe the implications of the loss of RNase H2, or RNases H1 and H2, on the integrity of the nuclear genome and viability of budding yeast cells that are challenged with a critically low supply of dNTPs. We further propose that our findings in budding yeast could pave the way for the study of the potential role of mammalian RNR in RNase H2-related diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
R-loops accumulating in absence of RNases H1 and H2 lead to cell lethality when the supply of dNTPs is limiting for DNA synthesis
Cellular deoxyribonucleoside-triphosphates (dNTPs), the building blocks of DNA, are utilized by the three major replicative DNA polymerases (Pol) α, δ and ε for the duplication of genomic DNA (i.e. nuclear genome) during S-phase in eukaryotic cells (for a review, see e.g. Burgers and Kunkel 2017). Pol α initiates DNA synthesis at origins of replication in both leading and lagging strands, and at Okazaki fragments in lagging strand. Pol δ and Pol ε synthesize the bulk of the lagging and leading strands, respectively. Recent reports suggest that Pol δ participates in the initiation and termination of leading strand replication (e.g., Garbacz et al. 2020; Yeeles et al. 2017; Zhou et al. 2019).
For accurate and timely DNA replication, the levels and balance of the four dNTPs need tight regulation during S-phase (e.g., Kumar et al. 2010; Poli et al. 2012). Notably, dNTP pools are subject to a Goldilocks-like effect, that requires just the right concentration; low pools induce replicative stress (i.e. replication fork stalling), DNA mutagenesis, DNA damage and viral restriction, whereas high and imbalanced dNTP pools facilitate DNA mutagenesis, cancer growth and viral replication (for a review, see e.g. Aye et al. 2015; Coggins et al. 2020; Ganai and Johansson 2016; Mathews 2015; Pai and Kearsey 2017; Techer et al. 2017).
Ribonucleotide reductase (RNR) complex catalyses the rate-limiting step in dNTP synthesis and plays an essential role in both DNA replication and repair. In Saccharomyces cerevisiae, RNR complex is formed of a homodimer of Rnr1, which contains the regulatory and catalytic sites, and a heterodimer of Rnr2 and Rnr4 (for a review, see e.g. Sanvisens et al. 2013). Rnr3 is a homologue of Rnr1, but with much lower catalytic activity (Domkin et al. 2002). Rnr3 is not detectably expressed in unperturbed conditions, but is highly induced following replicative or genotoxic stress (e.g., Cerritelli et al. 2020; Elledge and Davis 1990; Gupta et al. 2013; Li et al. 2019b; Maicher et al. 2017). The absence of Rnr1 is merely tolerated in the S. cerevisiae BY4741 background (Cerritelli et al. 2020; Giaever et al. 2002; Gupta et al. 2013; Maicher et al. 2017), and leads to cell lethality in other budding yeast backgrounds (e.g., Elledge and Davis 1990; Gupta et al. 2013; and A.E.H. Unpublished observations). We and others (Cerritelli et al. 2020; Gupta et al. 2013; Maicher et al. 2017) found that the loss of Rnr1 in BY4741 background decreases cellular dNTP pools > 3-fold, particularly the levels of dGTPs, as compared to wild-type cells. Hence, the ratios of dNTPs to ribonucleoside-triphosphates (rNTPs) in cells lacking Rnr1 are much lower than those in wild-type cells, which naturally have several-fold higher concentrations of rNTPs than dNTPs (e.g., Balachander et al. 2020) (compare Fig. 1b with Fig. 1a). Moreover, we and others (Cerritelli et al. 2020; Gupta et al. 2013; Maicher et al. 2017) found that the absence of Rnr1 in BY4741 background modestly induces the expression of Rnr3, due to mild activation of the S-phase checkpoint-signaling pathway Mec1-Rad53-Dun1. Thus, by providing dNTPs, the Rnr3-containing-RNR complexes would support DNA synthesis in the absence of Rnr1, albeit at a reduced pace. This would create acute replicative stress that significantly slows down cell growth in S-phase. Consistent with these ideas, we (Cerritelli et al. 2020) found that Dun1 is essential for the viability of single mutant depleted of Rnr1. This is presumably due to total loss of RNR activity elicited by the absence of both Rnr1 and Rnr3, and to the ensuing arrest of DNA synthesis. Furthermore, our unpublished observations show that single mutant lacking Dun1, which grows like the wild-type strain in unperturbed conditions, is hypersensitive to medium doses (25 mM) of the RNR inhibitor hydroxyurea (HU), as previously reported (e.g., Li et al. 2019b). In this case, the hypersensitivity to HU of cells lacking Dun1 could be due to acute replicative stress triggered by a critically low supply of dNTPs.
Limiting supply of dNTPs leads to lethal R-loop-mediated replicative stress and DNA damage in the absence of RNases H1 and H2, and to catastrophic Top1-mediated DNA damage in the presence of a high load of unrepaired single genomic rNMPs. a Wild-type budding yeast cell. In unperturbed S-phase, the cytoplasmic Rnr1-containing-RNR complexes, which are hetero-tetramers comprised of two Rnr1 subunits (small, dark-green ovals) and one subunit each of Rnr2 (small, grey sphere) and Rnr4 (small, yellow sphere), provide dNTPs (blue dots) for the duplication and repair of nuclear DNA. The total cellular concentrations of rNTPs (pink dots) are several-fold higher than those of dNTPs. A replication fork is represented in the nucleus. Pol ε (labelled ε and colored in purple) continuously synthesizes the leading strand (one horizontal, blue arrow). Pol δ (labelled δ and colored in brown) discontinuously synthesizes Okazaki fragments, which form the lagging strand (three horizontal, blue arrows). Pol α, which initiates the synthesis of leading strand and Okazaki fragments, is omitted for clarity. Parental/template DNA strands for replication are represented by black horizontal lines. Pol ε and Pol δ incorporate rNMPs (small, pink dashes) in nascent DNA strands at low frequency. Pol ε naturally has ~ 3-fold lower discrimination against the utilization of rNTPs than Pol δ. RNase H2 (labelled H2), which is a hetero-trimeric enzymatic complex (three spheres colored pale-yellow, light-green and red), incises at the 5′-end of the rNMP, thereby initiating error-free removal of the rNMP by the cellular RER pathway. The replicative helicase complex CMG (Cdc45-MCM-GINS; large, red-crimson oval) unwinds the double-stranded DNA duplex ahead of the replication fork. The RNA (pink line) extruding from the RNA polymerase (large, dark-blue oval) could hybridize with the transcribed strand (bottom, black line), thereby forming a tripartite R-loop structure, which is comprised of an RNA/DNA hybrid and an unpaired non-transcribed strand (top, black line). The RNA moiety of the RNA/DNA hybrid could be cleaved by RNase H1 (purple sphere labelled H1) or RNase H2. The colors of the nascent DNA strands and the nascent RNA match the colors of their corresponding building blocks; i.e. blue for deoxyribonucleoside-monophosphate and pink for rNMP, respectively. The directions of replication and transcription are represented by small, horizontal, black arrows. The nucleus and cytoplasm are not drawn to scale. b Budding yeast cell depleted of Rnr1 and lacking RNases H1 and H2. In S-phase in absence of Rnr1, Rnr3 is modestly expressed, due to mild activation of the Mec1-Rad53-Dun1-dependent-S-phase checkpoint. The cytoplasmic Rnr3-containing-RNR complexes, which are hetero-tetramers comprised of two Rnr3 subunits (brown ovals) and one subunit each of Rnr2 and Rnr4, provide the cell with ~ 3-fold lower dNTP concentrations, as compared to Rnr1-containing-RNR complexes in panel a. This significantly slows down the replication fork and concomitantly increases (by ~ 5-fold) the incorporation of rNMPs by replicative Pols in newly synthesized DNA. Pol ε would frequently hand over to Pol δ at the nascent leading strand (labelled ε/δ), because of acute replicative stress. DNA repair could also be compromised by low dNTP pools. As RNase H2 is absent, rNMPs accumulate in the newly synthesized DNA, particularly the leading strand, and Top1 (orange sphere) incises at the 3′-end of some embedded rNMPs. Top1-mediated cleavages would lead to DNA mutagenesis and/or genome instability. The absence of RNases H1 and H2 would lead to a persistent R-loop. This could block the advancement of the replication fork (red, lightning signal), thereby triggering irreversible fork collapse and breakage, and ultimately leading to cell lethality. Note that RNA-polymerase-associated-R-loops can be co-directional or head-on with respect to the direction of the replication fork, and head-on collisions are likely to be more deleterious than co-directional collisions. Other details are as in panel a. c Budding yeast cell that bears an rNTP-permissive form of Pol ε, lacks RNase H2, and is also depleted of Rnr1. In S-phase in absence of Rnr1, the rNTP-permissive form of Pol ε, which is encoded by pol2-M644G allele (labelled ε*), excessively incorporates rNMPs in nascent leading strand. Processing of rNMPs by Top1 in absence of RNase H2, in the nascent leading strand, would induce single-strand breaks or DSBs. Repair of these DNA lesions (e.g. of DSBs by Rad51/Rad52-dependent-homologous recombination) might be compromised, ultimately leading to severe growth defects. Pol ε* would frequently hand over to Pol δ at the nascent leading strand (labelled ε*/δ). Other details are as in panels a and b. d Budding yeast cell that bears an rNTP-permissive form of Pol δ, lacks RNase H2, and is also depleted of Rnr1. In S-phase in absence of Rnr1, the rNTP-permissive form of Pol δ, which is encoded by pol3-L612M allele (labelled δ* and colored in brown), incorporates high loads of rNMPs in both nascent leading and lagging strands. Pol ε would frequently hand over to Pol δ* at the nascent leading strand (labelled ε/δ*). Processing of rNMPs by Top1 in absence of RNase H2, in both nascent DNA strands, would induce single-strand breaks or DSBs. Other details are as in panels a–c
It was recently reported (Forey et al. 2020) that wild-type budding yeast cells enter S-phase with a low supply of dNTPs, which is sufficient for the activation of hundreds of early origins (i.e. for the synthesis of ~ 5 Kb DNA on each side of the origin, which is ~ 10–15% of the genome; Poli, et al. 2012), but which is insufficient to sustain DNA synthesis from these origins. This leads to replication fork pausing. Consequently, Mec1 transiently activates Rad53, presumably through its mediator Mrc1 (for a review, see e.g. Moriel-Carretero et al. 2019), thereby ensuring replication fork stability and limiting DNA damage. Activation of Rad53 also triggers Dun1-mediated-degradation of Sml1, the natural inhibitor of Rnr1 (Zhao et al. 2001), therefore upregulating dNTP synthesis. Based on this model (Forey et al. 2020), and other published observations (e.g., Devbhandari and Remus 2020; Gan et al. 2017; Katou et al. 2003; Tercero et al. 2003; Zhao et al. 2001), we propose that Mec1 and Rad53, in addition to other roles in S-phase (for a review, see e.g. Corcoles-Saez et al. 2019; Giannattasio and Branzei 2017; Pardo et al. 2017), protect stalled replication forks from breakage and promote dNTP production for DNA synthesis, in both single mutant lacking Rnr1, and single mutant missing Dun1 and treated with HU. Because these two mutants are likely to experience acute replicative stress due to a critically low supply of dNTPs for DNA synthesis, the activation of Rad53 by Mec1 is presumably maintained throughout S-phase by a cross-talk between Mrc1 and Rad9, which is the other mediator of Mec1 (for a review, see e.g. Moriel-Carretero et al. 2019).
We (Cerritelli et al. 2020; and our unpublished observations) found that the triple mutant depleted of Rnr1 and lacking RNases H1 and H2, and the triple mutant lacking together Dun1 and RNases H1 and H2 and treated with HU (25 mM), are both non-viable. What could be causing the lethality in these triple mutants? RNase H1 is active throughout the cell cycle, while RNase H2 processes its substrates in S- and -G2/M phases of the cell cycle (Arudchandran et al. 2000; Lockhart et al. 2019). RNases H1 and H2 can cleave the RNA moiety in RNA/DNA hybrids (Fig. 1a) (e.g., El Hage et al. 2010, 2014; for a review, see e.g. Cerritelli and Crouch 2009; Hyjek et al. 2019). However, RNase H2 can also incise single rNMPs embedded in nuclear DNA at their 5′-end (Fig. 1a), thereby initiating the error-free, ribonucleotide excision repair (RER) pathway (Sparks et al. 2012; for a review, see e.g. Cerritelli and Crouch 2016; Hyjek et al. 2019; Williams et al. 2016). RNA/DNA hybrids can be found in the cell as part of R-loops, which are formed during transcription when the RNA extruding from the RNA polymerase hybridizes to the template strand, thereby leaving the non-template strand unpaired (Fig. 1a) (for a review, see e.g. Aguilera and Garcia-Muse 2012; Drolet 2006). In budding yeast, R-loops are highly enriched at frequently transcribed genes, and R-loop accumulation is increased in absence of RNases H1 and H2 (e.g., Chan et al. 2014; El Hage et al. 2014; Wahba et al. 2016). Furthermore, R-loops can block replication fork progression and induce genomic instability (for a review, see e.g. Gomez-Gonzalez and Aguilera 2019; Mikolaskova et al. 2018) (Fig. 1b). Accumulation of single rNMPs in genomic DNA in the absence of RNase H2 can also lead to genomic instability (for a review, see e.g. Williams et al. 2016). Remarkably, we (Cerritelli et al. 2020) found that depletion of Rnr1 in budding yeast cells significantly increases the utilization of rNTPs by replicative Pols, due to critically low [dNTP]/[rNTP] ratios, as compared to wild-type cells. Thus, yeast double mutant that lacks both RNase H2 and Rnr1 accumulate much more (~ 5-fold) genomic rNMPs than single mutant that only lacks RNase H2. Taken together, it would be reasonable to expect that the lethality of triple mutant depleted of Rnr1 and lacking RNases H1 and H2, and the lethality of triple mutant lacking together Dun1 and RNases H1 and H2 and treated with HU are both caused by a combination of persistent genomic R-loops and single genomic rNMPs. However, we (Cerritelli et al. 2020; and our unpublished data) found that the growth defects in both of these two triple mutants are rescued almost completely by the expression of a mutant variant of RNase H2 that cleaves RNA/DNA hybrids but that is RER-deficient (Chon et al. 2013). These results lead us thus to conclude that persistent genomic RNA/DNA hybrids (likely R-loops), and not single genomic rNMPs, are the main factor causing lethality in these triple mutants.
At the start of S-phase in wild-type budding yeasts, spontaneous replicative stress at early replicating regions is not caused by replication-transcription conflicts but rather by the suboptimal supply of dNTPs (Forey et al. 2020). Moreover, an earlier report showed that R-loops associated with highly transcribed tRNA genes in wild-type budding yeast cells do not induce replication fork pausing (Osmundson et al. 2017). Nonetheless, tRNA gene-associated-R-loops (Chan et al. 2014; El Hage et al. 2014; Wahba et al. 2016) can cause DNA damage independently of replication fork pausing, and can also enhance DNA mutagenesis (Saini et al. 2017; Tran et al. 2017). Other studies in budding yeast have reported R-loop-associated genome instability at the highly transcribed ribosomal RNA genes (Amon and Koshland 2016; El Hage et al. 2010; Stuckey et al. 2015). We propose that the lethality of triple mutant depleted of Rnr1 and lacking RNases H1 and H2, and the lethality of triple mutant lacking together Dun1 and RNases H1 and H2 and treated with HU are both caused by deleterious R-loop-associated-transcription-replication conflicts, particularly at sites of highly transcribed genes (Fig. 1b). In this case, the activation of the Mec1-Rad53-Dun1-dependent-S-phase checkpoint would be insufficient to protect replication forks against the harmful impact of R-loops, presumably because arrested forks at R-loop sites are not able to restart due to the limiting supply of dNTPs, thereby leading to their collapse and breakage (e.g., Morafraile et al. 2015; Poli et al. 2012; Zhao et al. 2018). Furthermore, damage repair of broken forks (for a review, see e.g. Ait Saada et al. 2018) could be compromised due to the insufficient supply of dNTPs for DNA synthesis. Another non-mutually exclusive explanation that we propose for the lethality of triple mutant depleted of Rnr1 and lacking RNases H1 and H2, and the lethality of triple mutant lacking together Dun1 and RNases H1 and H2 and treated with HU, is that R-loop-associated-DNA damage, particularly at highly transcribed genes, occurs in a replication-independent manner, outside S-phase (for a review, see e.g. Kim and Jinks-Robertson 2012). In this case, repair by DNA synthesis of R-loop-mediated-DNA damage would also be compromised due to a critically low supply of dNTPs (Owiti, et al. 2018, 2019). Finally, it is possible that during the repair process of R-loop-mediated-DNA damage, within or outside S-phase in both of these two triple mutants, there is increased accumulation of genomic rNMPs, due to the absence of both RNase H2 and Rnr1, which would further aggravate the genome integrity defects in these mutants.
High load of unrepaired genomic rNMPs leads to catastrophic Top1-mediated DNA damage when the supply of dNTPs is limiting for DNA synthesis
The major role of Topoisomerase 1 (Top1) is to relieve DNA torsional stress generated during the cellular transcription and replication processes (e.g., Bermejo et al. 2007; El Hage et al. 2010; French et al. 2011). This could be facilitated by the fact that Top1 is directly associated with both of these machineries (e.g., Baranello, et al. 2016; Gambus et al. 2006). However, Top1 can be irreversibly trapped while relieving torsional stress, which could lead to DNA mutagenesis and/or threaten genome stability (e.g., Jakobsen et al. 2019a, b; Lippert et al. 2011; Stingele et al. 2014; Takahashi et al. 2011; for a review, see e.g. Cho and Jinks-Robertson 2018). Additionally, in the absence of RNase H2, Top1 would cleave at the 3′-end of a single genomic rNMP (Fig. 1b). This could produce a non-ligatable, single-strand nick that compromises genome integrity (for a review, see e.g. Cho and Jinks-Robertson 2017, 2018; Williams et al. 2016). For instance, in budding yeast, Top1-mediated incisions within short tandem repeats at sites of unrepaired single genomic ribonucleotides (i.e. at sites of single rNMPs accumulating in absence of RNase H2) have been associated with a signature of 2–5 bp deletion (e.g., Kim et al. 2011; Nick McElhinny et al. 2010; Potenski et al. 2014). Another form of Top1-mediated lesion at unrepaired rNMP sites are double-strand breaks (DSB), which can be a serious threat to genome integrity and cell viability if not productively repaired by the cellular Rad51/Rad52-homologous recombination machinery (Huang et al. 2017).
In principle, Top1 could incise genomic rNMPs in wild-type cells, but this would be an extremely rare event, as embedded rNMPs are expected to be very transient due to their efficient removal by RER (e.g. Balachander et al. 2020; Sparks and Burgers 2015; Sparks et al. 2012). Furthermore, only a small fraction of unrepaired rNMPs would be incised by Top1. Moreover, as most of the Top1-mediated incisions at rNMP sites would be religated in the presence of Top1 (Huang et al. 2015; Sparks and Burgers 2015), only a subset of unrepaired rNMPs that are incised by Top1 would be removed in an error-free manner by the cellular DNA repair pathways (e.g., Li et al. 2019a; Potenski et al. 2014; Sparks and Burgers 2015), or on the contrary removed in an error-prone and/or genome-destabilizing manner. The detection of Top1 activity at genomic rNMP sites would, therefore, be dictated by the combination of all these factors. To enhance detection of Top1 activity at rNMP sites in cells lacking RNase H2, it is possible to increase rNMP incorporation in genomic DNA by using a replicative rNTP-permissive Pol variant that has a relaxed selectivity against rNTPs, as compared to its wild-type parent enzyme (for a review, see e.g. Brown and Suo 2011; Williams et al. 2016). Indeed, this approach has been extensively used to study Top1-mediated DNA damage at unrepaired rNMP sites in budding yeast (e.g., Cho et al. 2015; Huang et al. 2017; Li et al. 2019a; Potenski et al. 2014; Williams, et al. 2013, 2015). For instance, analyses of the infrequently transcribed URA3 reporter revealed that Top1-mediated short deletions at rNMP sites are strongly associated with the newly synthesized leading strand, in the double mutant lacking RNase H2 and bearing an rNTP-permissive form of Pol ε, which is encoded by the allele pol2-M644G (Pol2 is the catalytic subunit of Pol ε) (Williams et al. 2013). On the contrary, however, Top1-mediated short deletions at rNMP sites in the URA3 reporter were found to be weakly associated with the newly synthesized lagging strand, in the two double mutants that lack RNase H2 and bear an rNTP-permissive form of Pol α or Pol δ, which is encoded by the allele pol1-L868M or pol3-L612M, respectively (Pol1 and Pol3 are the catalytic subunits of Pol α and Pol δ, respectively) (Williams et al. 2015). The asymmetry of Top1 nicking at unrepaired rNMP sites with regard to the two strands could reflect the need for Top1 to relieve torsional stress in the newly synthesized leading strand; however, torsional stress may not accumulate in the newly synthesized lagging strand due to its discontinuous nature, thereby excluding Top1 from this strand, as previously suggested (Williams et al. 2015). Another non-mutually exclusive possibility to explain the asymmetry is the higher number of unrepaired rNMPs in the leading strand vs. the lagging strand (Williams et al. 2015). This is because the rNTP-permissive form of Pol ε encoded by the allele pol2-M644G utilizes ~ 3-fold more rNTPs than the rNTP-permissive form of Pol δ encoded by the allele pol3-L612M (Nick McElhinny et al. 2010; Williams et al. 2015). Moreover, most of the rNMPs incorporated in nascent lagging strand by the rNTP-permissive form of Pol α encoded by the allele pol1-L868M, which utilizes ~ 2.5-fold more rNTPs than the rNTP-permissive form of Pol ε encoded by the allele pol2-M644G (Nick McElhinny et al. 2010; Williams et al. 2015), are likely to be removed during the maturation of Okazaki fragments (e.g., Reijns et al. 2015).
It is worth noting that the asymmetry of Top1 nicking at unrepaired single rNMP sites with regard to the nascent leading and lagging strands is lost in highly transcribed genes. Indeed, a previous report showed that under high transcription conditions the rates of Top1-mediated short deletions at unrepaired single rNMP sites are greatly increased in either nascent leading or lagging strand, independently of the direction of replication, and that the short deletions are only associated with the non-transcribed strand (Cho et al. 2015; reviewed in Cho and Jinks-Robertson 2017).
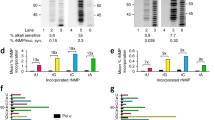
Recently, we (Cerritelli et al. 2020) showed that the combination of a replicative rNTP-permissive Pol variant with the depletion of Rnr1 and the absence of RNase H2 in budding yeast cells increases excessively the load of rNMPs in their genomic DNA. We (Cerritelli et al. 2020) also performed Southern blotting to analyze Top1 activity at rNMP sites in the infrequently transcribed gene AGP1 of budding yeast mutants that bear an rNTP-permissive form of Pol ε, Pol α or Pol δ and that also lack both Rnr1 and RNase H2, in presence/absence of Top1. We tested in parallel the viability of these strains. We found that the triple mutant bearing the pol2-M644G allele and lacking both Rnr1 and RNase H2 accumulates much more rNMPs and shows higher Top1 activity at rNMP sites in AGP1-leading strand (Fig. 1c), compared to the double mutant that bears the pol2-M644G allele and lacks only RNase H2. Strikingly, the triple mutant bearing the pol2-M644G allele and lacking both Rnr1 and RNase H2 suffered from severe growth defects, but these were reversed in the absence of Top1. It is possible that in this triple mutant, acute replicative stress and compromised DNA repair that are elicited by the insufficient supply of dNTPs highly exacerbate the impact of Top1-mediated damage on genome stability and cell growth. We also found that Top1 activity at rNMP sites is associated with both AGP1-leading and -lagging strands in the two triple mutants that bear the pol1-L868M or pol3-L612M allele and lack both Rnr1 and RNase H2 (for triple mutant bearing the pol3-L612M allele, see Fig. 1d). Additionally, we found that the triple mutant bearing the pol3-L612M allele has more rNMPs and a higher level of Top1 activity at rNMP sites in both AGP1-leading and -lagging strands, compared to its triple mutant counterpart bearing the pol1-L868M allele. As in the case of the triple mutant bearing the pol2-M644G allele, and presumably for similar reasons, the triple mutant bearing the pol3-L612M allele suffered from severe growth defects in the presence, but not in the absence, of Top1. Finally, it is worth emphasizing that we showed that the severe growth defects in the two triple mutants that bear the pol2-M644G or pol3-L612M allele and that also lack both Rnr1 and RNase H2 are not rescued by the expression of a mutant variant of RNase H2 that cleaves RNA/DNA hybrids but that is RER-deficient (Chon, et al. 2013). This result strongly indicates that single genomic rNMPs, and not genomic RNA/DNA hybrids, are the main factor causing severe growth defects in these strains.
Together, our data (Cerritelli et al. 2020) lead us to conclude that in the absence of RNase H2, Top1 incises at rNMP sites in both newly synthesized leading and lagging strands. We propose that the more rNMPs accumulate in nascent leading or lagging strands, the higher is the chance that Top1 incises at rNMP sites. Thus, there is a threshold for the detection of Top1-mediated incisions at rNMP sites (and the associated DNA damage) in both nascent leading and lagging strands that depends on the number of rNMPs, rather than a selectivity of Top1 for a strand over the other. It would be interesting to analyze on a whole-genome scale the distribution of embedded rNMPs, and the associated Top1-mediated DNA damage, in budding yeast mutants that lack Rnr1 and RNase H2 and also bear an rNTP-permissive replicative Pol.
Could RNR play a role in the etiology of RNase H2-deficient diseases?
The expansion of dNTP pools in S-phase of proliferating cells promotes high-fidelity DNA synthesis during replication and repair of genomic DNA (for a review, see e.g. Ganai and Johansson 2016; Pai and Kearsey 2017). Our recent findings in budding yeast (Cerritelli et al. 2020) support the idea that the increase in cellular [dNTP]/[rNTP] ratios during unperturbed S-phase (Chabes et al. 2003) would limit the incorporation of rNMPs in genomic DNA by replicative Pols, thereby preventing ribose-associated DNA damage, as previously suggested (Cerritelli and Crouch 2016). Consistent with this model, it was reported that ribose accumulation is increased in genomic DNA of mouse embryonic RNaseH2null fibroblast cells treated for 48 h with a low dose of HU (Reijns et al. 2012). This suggests that increased RNR activity during S-phase counteracts ribonucleotide incorporation in nuclear DNA of proliferating mammalian cells. Contrary to the situation in S-phase, however, it is possible that the lower cellular [dNTP]/[rNTP] ratios outside S-phase in wild-type budding yeast (Chabes et al. 2003) favor the incorporation of ribonucleotides by Pols in genomic DNA during the repair process of endogenous DNA damage. This could occur at highly transcribed genes, which are more prone to damage than the infrequently transcribed ones (for a review, see e.g. Kim and Jinks-Robertson 2012). Consistent with this idea, it was reported that in G1- or G2-phase of the cell cycle in budding yeast, high cellular [dUTP]/[dTTP] ratio favours the incorporation of dUMPs by Pols in DNA during the repair of lesions at frequently transcribed genes. This increases uracil-associated mutagenesis (Owiti, et al. 2018, 2019).
Elevated levels of genomic ribonucleotides are not tolerated during embryonic development in RNase H2-deficient mice, as they lead to catastrophic DNA damage that elicits a lethal p53-response (Hiller et al. 2012; Reijns et al. 2012; Uehara et al. 2018). Indeed, mice mutants expressing an RNase H2 variant that cleaves RNA/DNA hybrids but that is RER-deficient are embryonically lethal, strongly suggesting that single genomic rNMPs, and not RNA/DNA hybrids, are the main factor causing the death of these mice embryos (Uehara et al. 2018). In budding yeast, additional defects are needed to drive a sufficiently high density of unrepaired genomic ribonucleotides to generate a severe growth defect; e.g. in triple mutants that are depleted of Rnr1, lack RNase H2 and also bear an rNTP-permissive form of Pol ε or Pol δ (Cerritelli et al. 2020). As previously suggested (Uehara et al. 2018), we herein postulate that very fast cell division in early mice embryos (for a review, see e.g. Kojima et al. 2014) does not allow for the repair of the DNA damage (e.g., Ahuja et al. 2016; for a review, see e.g. Tichy and Stambrook 2008) that is caused by the accumulation of genomic ribonucleotides in the absence of RNase H2. Notably, it was recently reported (Zimmermann et al. 2018) that loss of RNase H2 sensitizes human cells to poly (ADP-ribose) polymerase (PARP) inhibition. This is because functional PARP1 is required to resolve DNA lesions created by TOP1-cleavages at sites of genomic rNMPs in RNaseH2null cells (Zimmermann et al. 2018). It is, therefore, possible that unresolved TOP1-mediated lesions at single rNMP sites lead to an early embryonic arrest in RNase H2-deficient mice. Unravelling the lethal role of TOP1 in RNase H2-deficient mice embryos may not be trivial, as TOP1 has ubiquitous roles in other important cellular functions (for a review, see e.g. Pommier et al. 2016). As RNR activity is a limiting factor in ribose incorporation during the synthesis of genomic DNA (Cerritelli et al. 2020; Reijns et al. 2012), it would be thus interesting to study the potential involvement of mammalian RNR in accumulation of genomic rNMPs, and the associated TOP1-mediated DNA damage (Zimmermann et al. 2018), during embryonic development of RNase H2-deficient mice. By modulating dNTP levels, one could perhaps exacerbate, or mitigate, ribonucleotide-associated-genome instability defects; e.g. by treating mice embryos with drugs that inhibit RNR activity (for a review, see e.g. Aye et al. 2015), or by supplementing embryos with nucleosides (e.g., Bester et al. 2011), respectively.
RNR plays important roles in cancer development by supplying dNTPs for the expansion of transformed cells. Notably, R2, the small subunit of mammalian RNR, is overexpressed in many types of cancer (for a review, see e.g. Aye et al. 2015). Moreover, several inhibitors of RNR, such as HU, are currently used as chemotherapeutic drugs to treat cancer (for a review, see e.g. Aye et al. 2015). Loss of RNase H2 has been recently implicated in skin and intestinal cancers, suggesting a correlation between rNMP incorporation in genomic DNA, DNA damage and unrestricted proliferation (Aden et al. 2018; Hiller et al. 2018). Reducing RNR activity in these RNase H2-deficient cells, e.g. by targeting the co-chaperones of heat shock protein 70 or 90, which normally stabilize RNR subunits (for a review, see e.g. Knighton et al. 2019), may increase the load of rNMPs in genomic DNA, and presumably also the associated TOP1-mediated DNA damage (Zimmermann et al. 2018), thereby selectively killing cancerous cells.
Modulation of RNR activity might also play a role in the prevention and treatment of two RNase H2-associated-autoimmune diseases, Aicardi-Goutières syndrome (AGS) (e.g., Crow et al. 2006, 2015), and systemic lupus erythematosus (e.g., Gunther et al. 2015; Pendergraft and Means 2015). AGS is a neuro-inflammatory autoimmune disorder that phenotypically resembles viral infection, and more than half of AGS patients have hypomorphic mutations in one of the three genes that encode the heterotrimeric RNase H2 complex. Either cytosolic RNA/DNA hybrids, or genomic rNMPs, or genomic R-loops, or a combination of these nucleic acids, may cause the auto-immune response in RNase H2-related-AGS patients. RNA/DNA reverse transcription intermediates from retroelements, which constitute about 40% of the human genome and which are mostly comprised of LINE-1 and Alu elements, could trigger the innate immune response in patients with RNase H2-related-AGS (e.g. Rice et al. 2018; for a review, see e.g. Ablasser and Hur 2020; Crow et al. 2020; Volkman and Stetson 2014). This may be unlikely, however, as two recent studies reported that loss of RNase H2 in mammalian cells abolishes rather than facilitates retrotransposition of LINE-1 and Alu elements (Bartsch et al. 2017; Benitez-Guijarro et al. 2018). It was recently reported that proliferating mammalian RNase H2-deficient cells have a high frequency of cytosolic DNA aggregates resembling micronuclei (Bartsch et al. 2017; Mackenzie et al. 2017). These can be formed by missegregation of chromosomal DNA during mitosis due to incomplete DNA replication and/or DNA damage. Remarkably, sensing of cytosolic micronuclei DNA by cGAS in proliferating RNase H2-deficient cells activates the cGAS-STING-IRF3 signaling cascade and the ensuing interferon-induced innate immune response (Bartsch et al. 2017; Mackenzie et al. 2017; for a review, see e.g. Ablasser and Hur 2020). These findings raise the intriguing possibility that chronic nuclear DNA defects in proliferating RNase H2-deficient cells, which are likely to be triggered by the accumulation of unrepaired genomic rNMPs and/or persistent genomic R-loops (e.g. Lim et al. 2015; Mackenzie et al. 2016; Pizzi et al. 2015; Zimmermann et al. 2018), lead to autoimmunity in patients with RNase H2-related-AGS. Because RNR activity is likely to play a role in ribose-associated- and R-loop-associated-genomic instability in RNase H2-deficient cells (Cerritelli et al. 2020), we hypothesize that the severity of the symptoms in some patients with RNase H2-related AGS could be innately reduced by increased endogenous RNR activity, or on the contrary enhanced by low endogenous RNR activity.
Deficiency of SAMHD1, which is a dNTPase that maintains balanced dNTP pools in mammals (for a review, see e.g. Coggins et al. 2020), is also associated with AGS (Rice et al. 2009). Recently, it was reported that nuclear DNA damage in SAMHD1-deficient proliferating mammalian cells induces the cGAS-STING-IRF3 signaling cascade and the ensuing innate immune response (Coquel et al. 2018). This is reminiscent of chronic damage in genomic DNA of proliferating RNase H2-deficient cells inducing the innate immune response (Bartsch et al. 2017; Mackenzie et al. 2017). However, while RNR activity could play a role in the etiology of RNase H2-related-AGS, this might not be the case for SAMHD1-related-AGS, as the protective role of SAMHD1 during replication of genomic DNA is likely to be independent of its dNTPase activity (Coquel et al. 2018).
Finally, our work in budding yeast unveiling the importance of RNR in mitigating the effects of persistent genomic R-loops and preventing the incorporation of ribonucleotides in genomic DNA might help in designing therapies for RNase H2-related-AGS and -cancer, two devastating human conditions.
References
Ablasser A, Hur S (2020) Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat Immunol 21:17–29. https://doi.org/10.1038/s41590-019-0556-1
Aden K, Bartsch K, Dahl J, Reijns MAM, Esser D, Sheibani-Tezerji R, Sinha A, Wottawa F, Ito G, Mishra N, Knittler K, Burkholder A, Welz L, van Es J, Tran F, Lipinski S, Kakavand N, Boeger C, Lucius R, von Schoenfels W, Schafmayer C, Lenk L, Chalaris A, Clevers H, Rocken C, Kaleta C, Rose-John S, Schreiber S, Kunkel T, Rabe B, Rosenstiel P (2018) Epithelial RNase H2 maintains genome integrity and prevents intestinal tumorigenesis in mice. Gastroenterology. https://doi.org/10.1053/j.gastro.2018.09.047
Aguilera A, Garcia-Muse T (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46:115–124. https://doi.org/10.1016/j.molcel.2012.04.009
Ahuja AK, Jodkowska K, Teloni F, Bizard AH, Zellweger R, Herrador R, Ortega S, Hickson ID, Altmeyer M, Mendez J, Lopes M (2016) A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat Commun 7:10660. https://doi.org/10.1038/ncomms10660
Ait Saada A, Lambert SAE, Carr AM (2018) Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair (Amst). https://doi.org/10.1016/j.dnarep.2018.08.017
Amon JD, Koshland D (2016) RNase H enables efficient repair of R-loop induced DNA damage. eLife. https://doi.org/10.7554/eLife.20533
Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ (2000) The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells 5:789–802
Aye Y, Li M, Long MJ, Weiss RS (2015) Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 34:2011–2021. https://doi.org/10.1038/onc.2014.155
Balachander S, Gombolay AL, Yang T, Xu P, Newnam G, Keskin H, El-Sayed WMM, Bryksin AV, Tao S, Bowen NE, Schinazi RF, Kim B, Koh KD, Vannberg FO, Storici F (2020) Ribonucleotide incorporation in yeast genomic DNA shows preference for cytosine and guanosine preceded by deoxyadenosine. Nat Commun 11:2447. https://doi.org/10.1038/s41467-020-16152-5
Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY, Guha R, Wilson K, Zhang X, Zhang H, Piotrowski J, Thomas CJ, Singer DS, Pugh BF, Pommier Y, Przytycka TM, Kouzine F, Lewis BA, Zhao K, Levens D (2016) RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165:357–371. https://doi.org/10.1016/j.cell.2016.02.036
Bartsch K, Knittler K, Borowski C, Rudnik S, Damme M, Aden K, Spehlmann ME, Frey N, Saftig P, Chalaris A, Rabe B (2017) Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet 26:3960–3972. https://doi.org/10.1093/hmg/ddx283
Benitez-Guijarro M, Lopez-Ruiz C, Tarnauskaite Z, Murina O, Mian Mohammad M, Williams TC, Fluteau A, Sanchez L, Vilar-Astasio R, Garcia-Canadas M, Cano D, Kempen MH, Sanchez-Pozo A, Heras SR, Jackson AP, Reijns MA, Garcia-Perez JL (2018) RNase H2, mutated in Aicardi-Goutieres syndrome, promotes LINE-1 retrotransposition. EMBO J. https://doi.org/10.15252/embj.201798506
Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M (2007) Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev 21:1921–1936. https://doi.org/10.1101/gad.432107
Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145:435–446. https://doi.org/10.1016/j.cell.2011.03.044
Brown JA, Suo Z (2011) Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 50:1135–1142. https://doi.org/10.1021/bi101915z
Burgers PMJ, Kunkel TA (2017) Eukaryotic DNA replication fork. Annu Rev Biochem 86:417–438. https://doi.org/10.1146/annurev-biochem-061516-044709
Cerritelli SM, Crouch RJ (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS J 276:1494–1505. https://doi.org/10.1111/j.1742-4658.2009.06908.x
Cerritelli SM, Crouch RJ (2016) The balancing act of ribonucleotides in DNA. Trends Biochem Sci 41:434–445. https://doi.org/10.1016/j.tibs.2016.02.005
Cerritelli SM, Iranzo J, Sharma S, Chabes A, Crouch RJ, Tollervey D, El Hage A (2020) High density of unrepaired genomic ribonucleotides leads to Topoisomerase 1-mediated severe growth defects in absence of ribonucleotide reductase. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa103
Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L (2003) Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112:391–401
Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P (2014) Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 10:e1004288. https://doi.org/10.1371/journal.pgen.1004288
Cho JE, Jinks-Robertson S (2017) Ribonucleotides and transcription-associated mutagenesis in yeast. J Mol Biol 429:3156–3167. https://doi.org/10.1016/j.jmb.2016.08.005
Cho JE, Jinks-Robertson S (2018) Topoisomerase I and genome stability: the good and the bad. Methods Mol Biol 1703:21–45. https://doi.org/10.1007/978-1-4939-7459-7_2
Cho JE, Kim N, Jinks-Robertson S (2015) Topoisomerase 1-dependent deletions initiated by incision at ribonucleotides are biased to the non-transcribed strand of a highly activated reporter. Nucleic Acids Res 43:9306–9313. https://doi.org/10.1093/nar/gkv824
Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM (2013) RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res 41:3130–3143. https://doi.org/10.1093/nar/gkt027
Coggins SA, Mahboubi B, Schinazi RF, Kim B (2020) SAMHD1 functions and human diseases. Viruses. https://doi.org/10.3390/v12040382
Coquel F, Silva MJ, Techer H, Zadorozhny K, Sharma S, Nieminuszczy J, Mettling C, Dardillac E, Barthe A, Schmitz AL, Promonet A, Cribier A, Sarrazin A, Niedzwiedz W, Lopez B, Costanzo V, Krejci L, Chabes A, Benkirane M, Lin YL, Pasero P (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557:57–61. https://doi.org/10.1038/s41586-018-0050-1
Corcoles-Saez I, Dong K, Cha RS (2019) Versatility of the Mec1(ATM/ATR) signaling network in mediating resistance to replication, genotoxic, and proteotoxic stresses. Curr Genet 65:657–661. https://doi.org/10.1007/s00294-018-0920-y
Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi–Goutieres syndrome and mimic congenital viral brain infection. Nat Genet 38:910–916. https://doi.org/10.1038/ng1842
Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, Abdel-Hamid MS, Abdel-Salam GM, Ackroyd S, Aeby A, Agosta G, Albin C, Allon-Shalev S, Arellano M, Ariaudo G, Aswani V, Babul-Hirji R, Baildam EM, Bahi-Buisson N, Bailey KM, Barnerias C, Barth M, Battini R, Beresford MW, Bernard G, Bianchi M, Billette de Villemeur T, Blair EM, Bloom M, Burlina AB, Carpanelli ML, Carvalho DR, Castro-Gago M, Cavallini A, Cereda C, Chandler KE, Chitayat DA, Collins AE, Sierra Corcoles C, Cordeiro NJ, Crichiutti G, Dabydeen L, Dale RC, D'Arrigo S, De Goede CG, De Laet C, De Waele LM, Denzler I, Desguerre I, Devriendt K, Di Rocco M, Fahey MC, Fazzi E, Ferrie CD, Figueiredo A, Gener B, Goizet C, Gowrinathan NR, Gowrishankar K, Hanrahan D, Isidor B, Kara B, Khan N, King MD, Kirk EP, Kumar R, Lagae L, Landrieu P, Lauffer H, Laugel V, La Piana R, Lim MJ, Lin JP, Linnankivi T, Mackay MT, Marom DR, Marques Lourenco C, McKee SA, Moroni I, Morton JE, Moutard ML, Murray K, Nabbout R, Nampoothiri S, Nunez-Enamorado N, Oades PJ, Olivieri I, Ostergaard JR, Perez-Duenas B, Prendiville JS, Ramesh V, Rasmussen M, Regal L, Ricci F, Rio M, Rodriguez D, Roubertie A, Salvatici E, Segers KA, Sinha GP, Soler D, Spiegel R, Stodberg TI, Straussberg R, Swoboda KJ, Suri M, Tacke U, Tan TY, te Water Naude J, Wee Teik K, Thomas MM, Till M, Tonduti D, Valente EM, Van Coster RN, van der Knaap MS, Vassallo G, Vijzelaar R, Vogt J, Wallace GB, Wassmer E, Webb HJ, Whitehouse WP, Whitney RN, Zaki MS, Zuberi SM, Livingston JH, Rozenberg F, Lebon P, Vanderver A, Orcesi S, Rice GI (2015) Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A 167A:296–312. https://doi.org/10.1002/ajmg.a.36887
Crow YJ, Shetty J, Livingston JH (2020) Treatments in Aicardi–Goutieres syndrome. Dev Med Child Neurol 62:42–47. https://doi.org/10.1111/dmcn.14268
Devbhandari S, Remus D (2020) Rad53 limits CMG helicase uncoupling from DNA synthesis at replication forks. Nat Struct Mol Biol 27(5):461–471. https://doi.org/10.1038/s41594-020-0407-7
Domkin V, Thelander L, Chabes A (2002) Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J Biol Chem 277:18574–18578. https://doi.org/10.1074/jbc.M201553200
Drolet M (2006) Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59:723–730. https://doi.org/10.1111/j.1365-2958.2005.05006.x
El Hage A, French SL, Beyer AL, Tollervey D (2010) Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24:1546–1558. https://doi.org/10.1101/gad.573310
El Hage A, Webb S, Kerr A, Tollervey D (2014) Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet 10:e1004716. https://doi.org/10.1371/journal.pgen.1004716
Elledge SJ, Davis RW (1990) Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev 4:740–751
Forey R, Poveda A, Sharma S, Barthe A, Padioleau I, Renard C, Lambert R, Skrzypczak M, Ginalski K, Lengronne A, Chabes A, Pardo B, Pasero P (2020) Mec1 Is activated at the onset of normal S phase by low-dNTP pools impeding DNA replication. Mol Cell. https://doi.org/10.1016/j.molcel.2020.02.021
French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, El Hage A, Smith MM, Tollervey D, Smith JS, Beyer AL (2011) Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol 31:482–494. https://doi.org/10.1128/MCB.00589-10
Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8:358–366. https://doi.org/10.1038/ncb1382
Gan H, Yu C, Devbhandari S, Sharma S, Han J, Chabes A, Remus D, Zhang Z (2017) Checkpoint kinase Rad53 couples leading- and lagging-strand DNA synthesis under replication stress. Mol Cell 68(446–455):e443. https://doi.org/10.1016/j.molcel.2017.09.018
Ganai RA, Johansson E (2016) DNA replication—a matter of fidelity. Mol Cell 62:745–755. https://doi.org/10.1016/j.molcel.2016.05.003
Garbacz MA, Lujan SA, Kunkel TA (2020) Opportunities for new studies of nuclear DNA replication enzymology in budding yeast. Curr Genet 66:299–302. https://doi.org/10.1007/s00294-019-01023-4
Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. https://doi.org/10.1038/nature00935
Giannattasio M, Branzei D (2017) S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion. Cell Mol Life Sci 74:2361–2380. https://doi.org/10.1007/s00018-017-2474-4
Gomez-Gonzalez B, Aguilera A (2019) Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev. https://doi.org/10.1101/gad.324517.119
Gunther C, Kind B, Reijns MA, Berndt N, Martinez-Bueno M, Wolf C, Tungler V, Chara O, Lee YA, Hubner N, Bicknell L, Blum S, Krug C, Schmidt F, Kretschmer S, Koss S, Astell KR, Ramantani G, Bauerfeind A, Morris DL, Cunninghame Graham DS, Bubeck D, Leitch A, Ralston SH, Blackburn EA, Gahr M, Witte T, Vyse TJ, Melchers I, Mangold E, Nothen MM, Aringer M, Kuhn A, Luthke K, Unger L, Bley A, Lorenzi A, Isaacs JD, Alexopoulou D, Conrad K, Dahl A, Roers A, Alarcon-Riquelme ME, Jackson AP, Lee-Kirsch MA (2015) Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J Clin Investig 125:413–424. https://doi.org/10.1172/JCI78001
Gupta A, Sharma S, Reichenbach P, Marjavaara L, Nilsson AK, Lingner J, Chabes A, Rothstein R, Chang M (2013) Telomere length homeostasis responds to changes in intracellular dNTP pools. Genetics 193:1095–1105. https://doi.org/10.1534/genetics.112.149120
Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A (2012) Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med 209:1419–1426. https://doi.org/10.1084/jem.20120876
Hiller B, Hoppe A, Haase C, Hiller C, Schubert N, Muller W, Reijns MAM, Jackson AP, Kunkel TA, Wenzel J, Behrendt R, Roers A (2018) Ribonucleotide excision repair is essential to prevent squamous cell carcinoma of the skin. Cancer Res 78:5917–5926. https://doi.org/10.1158/0008-5472.CAN-18-1099
Huang SY, Ghosh S, Pommier Y (2015) Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J Biol Chem 290:14068–14076. https://doi.org/10.1074/jbc.M115.653345
Huang SN, Williams JS, Arana ME, Kunkel TA, Pommier Y (2017) Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J 36:361–373. https://doi.org/10.15252/embj.201592426
Hyjek M, Figiel M, Nowotny M (2019) RNases H: structure and mechanism. DNA Repair (Amst) 84:102672. https://doi.org/10.1016/j.dnarep.2019.102672
Jakobsen KP, Andersen AH, Bjergbaek L (2019a) Abortive activity of Topoisomerase I: a challenge for genome integrity? Curr Genet 65:1141–1144. https://doi.org/10.1007/s00294-019-00984-w
Jakobsen KP, Nielsen KO, Lovschal KV, Rodgaard M, Andersen AH, Bjergbaek L (2019b) Minimal resection takes place during break-induced replication repair of collapsed replication forks and is controlled by strand invasion. Cell Rep 26(836–844):e833. https://doi.org/10.1016/j.celrep.2018.12.108
Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078–1083. https://doi.org/10.1038/nature01900
Kim N, Jinks-Robertson S (2012) Transcription as a source of genome instability. Nat Rev Genet 13:204–214. https://doi.org/10.1038/nrg3152
Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S (2011) Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332:1561–1564. https://doi.org/10.1126/science.1205016
Knighton LE, Delgado LE, Truman AW (2019) Novel insights into molecular chaperone regulation of ribonucleotide reductase. Curr Genet 65:477–482. https://doi.org/10.1007/s00294-018-0916-7
Kojima Y, Tam OH, Tam PP (2014) Timing of developmental events in the early mouse embryo. Semin Cell Dev Biol 34:65–75. https://doi.org/10.1016/j.semcdb.2014.06.010
Kumar D, Viberg J, Nilsson AK, Chabes A (2010) Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res 38:3975–3983. https://doi.org/10.1093/nar/gkq128
Li F, Wang Q, Seol JH, Che J, Lu X, Shim EY, Lee SE, Niu H (2019a) Apn2 resolves blocked 3′ ends and suppresses Top1-induced mutagenesis at genomic rNMP sites. Nat Struct Mol Biol 26:155–163. https://doi.org/10.1038/s41594-019-0186-1
Li X, Jin X, Sharma S, Liu X, Zhang J, Niu Y, Li J, Li Z, Zhang J, Cao Q, Hou W, Du LL, Liu B, Lou H (2019b) Mck1 defines a key S-phase checkpoint effector in response to various degrees of replication threats. PLoS Genet 15:e1008136. https://doi.org/10.1371/journal.pgen.1008136
Lim YW, Sanz LA, Xu X, Hartono SR, Chedin F (2015) Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome. Elife. https://doi.org/10.7554/eLife.08007
Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O’Shea SH, Jinks-Robertson S (2011) Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci USA 108:698–703. https://doi.org/10.1073/pnas.1012363108
Lockhart A, Pires VB, Bento F, Kellner V, Luke-Glaser S, Yakoub G, Ulrich HD, Luke B (2019) RNase H1 and H2 are differentially regulated to process RNA–DNA hybrids. Cell Rep 29(2890–2900):e2895. https://doi.org/10.1016/j.celrep.2019.10.108
Mackenzie KJ, Carroll P, Lettice L, Tarnauskaite Z, Reddy K, Dix F, Revuelta A, Abbondati E, Rigby RE, Rabe B, Kilanowski F, Grimes G, Fluteau A, Devenney PS, Hill RE, Reijns MA, Jackson AP (2016) Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 35:831–844. https://doi.org/10.15252/embj.201593339
Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, Osborn RT, Wheeler AP, Nowotny M, Gilbert N, Chandra T, Reijns MAM, Jackson AP (2017) cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548:461–465. https://doi.org/10.1038/nature23449
Maicher A, Gazy I, Sharma S, Marjavaara L, Grinberg G, Shemesh K, Chabes A, Kupiec M (2017) Rnr1, but not Rnr3, facilitates the sustained telomerase-dependent elongation of telomeres. PLoS Genet 13:e1007082. https://doi.org/10.1371/journal.pgen.1007082
Mathews CK (2015) Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat Rev Cancer 15:528–539. https://doi.org/10.1038/nrc3981
Mikolaskova B, Jurcik M, Cipakova I, Kretova M, Chovanec M, Cipak L (2018) Maintenance of genome stability: the unifying role of interconnections between the DNA damage response and RNA-processing pathways. Curr Genet 64:971–983. https://doi.org/10.1007/s00294-018-0819-7
Morafraile EC, Diffley JF, Tercero JA, Segurado M (2015) Checkpoint-dependent RNR induction promotes fork restart after replicative stress. Sci Rep 5:7886. https://doi.org/10.1038/srep07886
Moriel-Carretero M, Pasero P, Pardo B (2019) DDR Inc., one business, two associates. Curr Genet 65:445–451. https://doi.org/10.1007/s00294-018-0908-7
Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA (2010) Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6:774–781. https://doi.org/10.1038/nchembio.424
Osmundson JS, Kumar J, Yeung R, Smith DJ (2017) Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat Struct Mol Biol 24:162–170. https://doi.org/10.1038/nsmb.3342
Owiti N, Wei S, Bhagwat AS, Kim N (2018) Unscheduled DNA synthesis leads to elevated uracil residues at highly transcribed genomic loci in Saccharomyces cerevisiae. PLoS Genet 14:e1007516. https://doi.org/10.1371/journal.pgen.1007516
Owiti N, Stokdyk K, Kim N (2019) The etiology of uracil residues in the Saccharomyces cerevisiae genomic DNA. Curr Genet 65:393–399. https://doi.org/10.1007/s00294-018-0895-8
Pai CC, Kearsey SE (2017) A critical balance: dNTPs and the maintenance of genome stability. Genes (Basel). https://doi.org/10.3390/genes8020057
Pardo B, Crabbe L, Pasero P (2017) Signaling pathways of replication stress in yeast. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fow101
Pendergraft WF 3rd, Means TK (2015) AGS, SLE, and RNASEH2 mutations: translating insights into therapeutic advances. J Clin Investig 125:102–104. https://doi.org/10.1172/JCI78533
Pizzi S, Sertic S, Orcesi S, Cereda C, Bianchi M, Jackson AP, Lazzaro F, Plevani P, Muzi-Falconi M (2015) Reduction of hRNase H2 activity in Aicardi–Goutieres syndrome cells leads to replication stress and genome instability. Hum Mol Genet 24:649–658. https://doi.org/10.1093/hmg/ddu485
Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P (2012) dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31:883–894. https://doi.org/10.1038/emboj.2011.470
Pommier Y, Sun Y, Huang SN, Nitiss JL (2016) Roles of eukaryotic Topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17:703–721. https://doi.org/10.1038/nrm.2016.111
Potenski CJ, Niu H, Sung P, Klein HL (2014) Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature 511:251–254. https://doi.org/10.1038/nature13292
Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP (2012) Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149:1008–1022. https://doi.org/10.1016/j.cell.2012.04.011
Reijns MAM, Kemp H, Ding J, de Proce SM, Jackson AP, Taylor MS (2015) Lagging-strand replication shapes the mutational landscape of the genome. Nature 518:502–506. https://doi.org/10.1038/nature14183
Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ (2009) Mutations involved in Aicardi–Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41:829–832. https://doi.org/10.1038/ng.373
Rice GI, Meyzer C, Bouazza N, Hully M, Boddaert N, Semeraro M, Zeef LAH, Rozenberg F, Bondet V, Duffy D, Llibre A, Baek J, Sambe MN, Henry E, Jolaine V, Barnerias C, Barth M, Belot A, Cances C, Debray FG, Doummar D, Fremond ML, Kitabayashi N, Lepelley A, Levrat V, Melki I, Meyer P, Nougues MC, Renaldo F, Rodero MP, Rodriguez D, Roubertie A, Seabra L, Uggenti C, Abdoul H, Treluyer JM, Desguerre I, Blanche S, Crow YJ (2018) Reverse-transcriptase inhibitors in the Aicardi–Goutieres syndrome. N Engl J Med 379:2275–2277. https://doi.org/10.1056/NEJMc1810983
Saini N, Roberts SA, Sterling JF, Malc EP, Mieczkowski PA, Gordenin DA (2017) APOBEC3B cytidine deaminase targets the non-transcribed strand of tRNA genes in yeast. DNA Repair (Amst) 53:4–14. https://doi.org/10.1016/j.dnarep.2017.03.003
Sanvisens N, de Llanos R, Puig S (2013) Function and regulation of yeast ribonucleotide reductase: cell cycle, genotoxic stress, and iron bioavailability. Biomed J 36:51–58. https://doi.org/10.4103/2319-4170.110398
Sparks JL, Burgers PM (2015) Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J 34:1259–1269. https://doi.org/10.15252/embj.201490868
Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM (2012) RNase H2-initiated ribonucleotide excision repair. Mol Cell 47:980–986. https://doi.org/10.1016/j.molcel.2012.06.035
Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S (2014) A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 158:327–338. https://doi.org/10.1016/j.cell.2014.04.053
Stuckey R, Garcia-Rodriguez N, Aguilera A, Wellinger RE (2015) Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proc Natl Acad Sci USA 112:5779–5784. https://doi.org/10.1073/pnas.1501769112
Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S (2011) Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 108:692–697. https://doi.org/10.1073/pnas.1012582108
Techer H, Koundrioukoff S, Nicolas A, Debatisse M (2017) The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet 18:535–550. https://doi.org/10.1038/nrg.2017.46
Tercero JA, Longhese MP, Diffley JF (2003) A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11:1323–1336
Tichy ED, Stambrook PJ (2008) DNA repair in murine embryonic stem cells and differentiated cells. Exp Cell Res 314:1929–1936. https://doi.org/10.1016/j.yexcr.2008.02.007
Tran PLT, Pohl TJ, Chen CF, Chan A, Pott S, Zakian VA (2017) PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat Commun 8:15025. https://doi.org/10.1038/ncomms15025
Uehara R, Cerritelli SM, Hasin N, Sakhuja K, London M, Iranzo J, Chon H, Grinberg A, Crouch RJ (2018) Two RNase H2 mutants with differential rNMP processing activity reveal a threshold of ribonucleotide tolerance for embryonic development. Cell Rep 25(1135–1145):e1135. https://doi.org/10.1016/j.celrep.2018.10.019
Volkman HE, Stetson DB (2014) The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol 15:415–422. https://doi.org/10.1038/ni.2872
Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D (2016) S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30:1327–1338. https://doi.org/10.1101/gad.280834.116
Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA (2013) Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell 49:1010–1015. https://doi.org/10.1016/j.molcel.2012.12.021
Williams JS, Clausen AR, Lujan SA, Marjavaara L, Clark AB, Burgers PM, Chabes A, Kunkel TA (2015) Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nat Struct Mol Biol 22:291–297. https://doi.org/10.1038/nsmb.2989
Williams JS, Lujan SA, Kunkel TA (2016) Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat Rev Mol Cell Biol 17:350–363. https://doi.org/10.1038/nrm.2016.37
Yeeles JTP, Janska A, Early A, Diffley JFX (2017) How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell 65:105–116. https://doi.org/10.1016/j.molcel.2016.11.017
Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R (2001) The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20:3544–3553. https://doi.org/10.1093/emboj/20.13.3544
Zhao H, Zhu M, Limbo O, Russell P (2018) RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair. EMBO Rep. 1:1. https://doi.org/10.15252/embr.201745335
Zhou ZX, Lujan SA, Burkholder AB, Garbacz MA, Kunkel TA (2019) Roles for DNA polymerase delta in initiating and terminating leading strand DNA replication. Nat Commun 10:3992. https://doi.org/10.1038/s41467-019-11995-z
Zimmermann M, Murina O, Reijns MAM, Agathanggelou A, Challis R, Tarnauskaite Z, Muir M, Fluteau A, Aregger M, McEwan A, Yuan W, Clarke M, Lambros MB, Paneesha S, Moss P, Chandrashekhar M, Angers S, Moffat J, Brunton VG, Hart T, de Bono J, Stankovic T, Jackson AP, Durocher D (2018) CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559:285–289. https://doi.org/10.1038/s41586-018-0291-z
Acknowledgements
We sincerely thank Robert J. Crouch (National Institutes of Health, USA), Philippe Pasero (Institute of Human Genetics, Montpellier) and David Tollervey (Wellcome Centre for Cell biology, Edinburgh) for critically reading the manuscript. We also thank Shar-Yin N Huang (Center for Cancer Research, National Cancer Institute, NIH) for discussions.
Funding
AEH is a research fellow in the laboratory of Professor David Tollervey, who is funded by the Welcome Trust. SMC is supported by the DIR-Eunice Kennedy Shriver National Institute of Child Health and Human Development National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cerritelli, S.M., El Hage, A. RNases H1 and H2: guardians of the stability of the nuclear genome when supply of dNTPs is limiting for DNA synthesis. Curr Genet 66, 1073–1084 (2020). https://doi.org/10.1007/s00294-020-01086-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-020-01086-8