Abstract
Membrane transport systems active in cellular inorganic phosphate (Pi) acquisition play a key role in maintaining cellular Pi homeostasis, independent of whether the cell is a unicellular microorganism or is contained in the tissue of a higher eukaryotic organism. Since unicellular eukaryotes such as yeast interact directly with the nutritious environment, regulation of Pi transport is maintained solely by transduction of nutrient signals across the plasma membrane. The individual yeast cell thus recognizes nutrients that can act as both signals and sustenance. The present review provides an overview of Pi acquisition via the plasma membrane Pi transporters of Saccharomyces cerevisiae and the regulation of internal Pi stores under the prevailing Pi status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regulation of cellular processes in response to external stimuli is fundamental to all living processes. Independent of whether the cell is a unicellular microorganism or is integrated in the tissue of a higher eukaryote, the nutrient status regulates vital properties, such as cell growth, proliferation, development and functional responses. To achieve this, the cell requires numerous and varied mechanisms by which these stimuli are allowed to communicate across the cell′s surface. The cellular response mediated via an altered gene expression pattern thus results in changes in the activities and functions of cellular proteins. Cellular membranes, composed primarily of lipids and proteins, hence not only serve as a structural backbone separating the interior of the cells from the extracellular milieu and compartmentalizing its interior into specialized functional organelles, but also take part in the regulation of many cellular processes. The functions of many membrane proteins predicted in the genome of Saccharomyces cerevisiae (Goffeau et al. 1997) still remain unknown. However, a large number of specialized integral membrane transport proteins, by which water-soluble solutes can be sensed and moved across the hydrophobic barrier of the phospholipid bilayer have now been identified and characterized. These specific membrane-spanning proteins allow for interaction and exchange between cellular compartments, transmission of signals, transport of essential nutrients, ions and metabolites and communication between the cells and their surrounding environment. Inorganic phosphate, Pi, an essential nutrient for all organisms, is required for structural and metabolic needs, such as the synthesis of nucleic acids, phospholipids and cellular metabolites. Cellular acquisition, storage and release and metabolic integration of Pi rely on the essential participation of numerous enzymes, such as exocellular acid phosphatases (APases), phosphodiesterase(s), phosphate transporters, polyphosphate kinase(s), alkaline phosphatase(s) (ALPase) and endopolyphosphatase(s) (Dawes and Senior 1973; Kaneko et al. 1985; Oshima et al. 1996). The activities of these enzymes needed for the intracellular regulation of Pi and homeostasis are subjected to regulation via the Pi signal transduction pathway (PHO pathway; reviewed by Lenburg and O′Shea 1996; Oshima 1997; Lagerstedt et al. 2000) in response to varying Pi levels (See Table 1 for functions of PHO-related genes and gene products).
Although genetic studies and analyses of the S. cerevisiae genome have produced much information on the components of the PHO regulatory pathway (and to some extent also the mode of interaction of these), little is known about how the cell senses the environmental Pi level and the mechanistic regulation of carrier-mediated Pi acquisition. The systems for Pi transport into the cells have been characterized as a low-affinity and a high-affinity process (see Persson et al. 1999). The low-affinity system satisfies the cellular need for Pi at normal or high external Pi concentrations (Blasco et al. 1976; Roomans et al. 1977), while the high-affinity transport system is mobilized in response to Pi starvation (Martinez and Persson 1998; Martinez et al. 1998; Pattison-Granberg and Persson 2000). Both of these acquisition systems rely on the activity of integral membrane proteins folded into an active conformation within the membrane, where they can harness the free energy stored in the electrochemical ion gradient into the active sequestration of substrate across the plasma membrane. In all organisms and cells, the uptake of Pi is coupled to a downhill movement of H+ or Na+, depending on the ionic current used. This review focuses mainly on recent advances in understanding of the regulation and function of these Pi uptake systems in S. cerevisiae.
Physiological regulation of Pi uptake in S. cerevisiae
The metabolism of many microorganisms such as S. cerevisiae is highly adaptable in order to allow the cells to respond to environmental changes. In these cells, enzyme systems are repressed or derepressed in response to changes, e.g. nutrient status, pH and salinity (Borst-Pauwels and Peters 1977; Hirimburegama et al. 1992; Yale and Bohnert 2001). A rapid response and adaptation to such environmental stress is thus essential for cell survival. Of the kinetically characterized pH-dependent Pi uptake systems in S. cerevisiae, the not yet genetically identified low-affinity system with a K m for external Pi of approximately 1 mM is proposed to be constitutively expressed (Tamai et al. 1985; Borst-Pauwels and Peters 1987). However, a slight increase in Pi uptake is observed under high-Pi conditions (Martinez and Persson 1998). Besides this low-affinity system, a growing family of potentially derepressible Pi transporters is now believed to exist, including Pho84p, Pho87p, Pho88p, Pho89p, Pho90p and Pho91p (Wykoff and O′Shea 2001). In agreement with previous observations, Pho84p is responsible for the majority of Pi uptake into the cells, while Pho89p (Martinez and Persson 1998) and the recently characterized Pho87p, Pho90p and Pho91p seem to play a less significant role (Wykoff and O′Shea 2001; Giots et al. 2003). Of these, the two most characterized derepressible high-affinity Pi transporters are Pho84p and Pho89p, shown to be activated when the cells meet a limitation in external Pi. The derepressible Pho84p transporter, genetically characterized by Bun-ya et al. (1991), catalyzes a H+-coupled Pi uptake (Borst-Pauwels 1993; Berhe et al. 1995; Fristedt et al. 1996, 1999a, 1999b) with a K m for external Pi of 1–15 µM and a proposed stoichiometry of 2–3 H+ per monovalent Pi anion (Cockburn et al. 1975; Borst-Pauwels and Peters 1987). In S. cerevisiae cells grown on both fermentable and non-fermentable carbon sources under Pi starvation, transient expression of the Pho84p transporter occurs (Martinez et al. 1998). Under conditions of non-limited carbon supply, the extracellular Pi concentration serves as a major factor controlling the expression and activity of the transporter (Martinez et al. 1998; Petersson et al. 1999). In contrast, limitations in the carbon source results in rapid degradation of the transporter in the presence of derepressible levels of Pi (Martinez et al. 1998). Hence, the functionality of the Pho84p transporter depends on the availability of both derepressible Pi concentrations and an abundant carbon source, not necessarily glucose (Martinez et al. 1998). Besides the Pi-dependent regulation of the transport process, transport is also dependent on both intra- and extracellular pH conditions (Borst-Pauwels and Peters 1987; Walker 1998). Alterations in metabolic conditions resulting from a shift from anaerobic to aerobic conditions or the replacement of glucose with ethanol have been shown to increase the pH optimum of Pi transport (Borst-Pauwels and Peters 1987). Additionally, the co-transport of Pi with H+ results in a lowered internal pH, a situation favorable for an increased cation uptake (Roomans et al. 1977). Furthermore, strong evidence in support of the fact that Pho84p can by itself participate in a Pi translocation pathway has been obtained by Escherichia coli expression of a histidine-tagged Pho84 protein. After solubilization, purification and unidirectional reconstitution into proteoliposomes, the protein was shown to catalyze a H+-coupled accumulation of Pi (Berhe et al. 1995; Fristedt et al. 1999a). Additional support for H+-coupling by Pho84p was provided by the establishment of a co-reconstituted proteoliposomal system, in which the Pho84-mediated, H+-coupled Pi uptake can be driven by the H+-pumping cytochrome c oxidase from beef heart (Fristedt et al. 1999b). Bivalent cations such as Mn2+ and Co2+ have been shown to stimulate the Pi uptake process in proteoliposomes harboring the purified Pho84 protein, whereas the addition of the chelator EDTA eliminated Pi transport (Fristedt et al. 1999b). Although these findings indicate the possibility that Pi may be transported as a soluble, electroneutral metal–Pi complex, the presence of Mg2+ rather lowered the prevailing electrical gradient (Δp)-driven Pi uptake. Previous studies of intact yeast cells (Bun-ya et al. 1991; Martinez et al. 1998) and inverted plasma membrane vesicles (Fristedt et al. 1996) verified that the Pho84 transporter catalyzes a bi-directional H+-coupled Pi uptake, where the direction of transport is determined by the directionality of the driving force rather than by the orientation of the protein.
The other derepressible, high-affinity Pi transporter, with a K m for external Pi of 0.5 µM, is encoded by the PHO89 gene and mediates a cation-coupled Pi transport with a strong preference for Na+ (Martinez and Persson 1998; Pattison-Granberg and Persson 2000). The rates of Pi uptake catalyzed by these two high-affinity transporters exhibit a strong pH dependence. The Pho84p transporter is maximally active at a pH close to 5.0, a pH at which the Pho89p transporter is largely inactive (Roomans et al. 1977; Martinez and Persson 1998; Pattison-Granberg and Persson 2000). In contrast, the Pho89p transporter is active in the alkaline pH range with a maximal activity at pH 9.5 (Martinez and Persson 1998). Using a yeast strain devoid of the PHO89 gene, it was concluded that Pho84p is functionally independent of Pho89p. Yet when combined, the functional properties of these two Pi transporters provide a unique ability to scavenge Pi over a broad range of different pH environments. The different pH optima and the lack of sequence similarity between the Pho84 and Pho89 proteins probably rule out simple gene duplication. Pho89p-mediated Pi uptake is stimulated by an increased external Na+ concentration (Martinez and Persson 1998; Pattison-Granberg and Persson 2000), with an activity maximum in the presence of 15–25 mM Na+ (Martinez and Persson 1998). This finding led to the suggestion that this transporter is of an electrogenic nature, where transport of a positively charged complex resulting from the combination of one monovalent Pi ion with two Na+ ions may be driven by the Δp across the membrane. However, the contribution from the Na+ chemical gradient, originating from the activity of Na+-coupled ATPases, could not be excluded (Persson et al. 1999). The ability, although to a lesser extent, of the Pho89 transporter to catalyze a Li+-enhanced Pi uptake (Roomans et al. 1977; Martinez and Persson 1998) was suggested to reflect the presence of two sites for Na+ and Li+ involved in the stimulation of Pi uptake (Roomans et al. 1977).
The maximal activity of Pho89p is at least 100-fold lower than that of the Pho84p transporter (assayed at their respective activity optima). When the yeast is grown at acidic conditions, the H+ electrochemical gradient is used for Pho84p-mediated symport of Pi across the membrane. This reaction is coupled to H+ extrusion from the cytoplasm via the essential plasma membrane, H+-ATPase (Pma1p; Van der Rest et al. 1995; Serrano 1996). At alkaline pH conditions, however, the membrane H+ gradient is disrupted and ion-pumping of the plasma membrane Na+-ATPase (Ena1p/Pmr2p) is required for growth (Garciadeblas et al. 1993; Lamb et al. 2001). Ena1p hydrolyzes ATP to pump Na+ out of the cell and thus generates a Na+ gradient across the membrane, which allows the coupled uptake of Na+ and other cations (Garciadeblas et al. 1993; Mendoza et al. 1994). Previously, the activation of Na+-coupled Pi uptake via Pho89p in cells grown in low-Pi media at acidic pH was shown to precede and partially overlap the activity of Pho84p, which is mobilized later during cell growth (Pattison-Granberg and Persson 2000). This supports the idea that up-regulation of both expression and activity of Pho89p is sensitive to small pH fluctuations during the early growth of cells.
Clearly, yeast cells have a developed system for a continued uptake of Pi under poor external Pi conditions. As an additional survival strategy, polyphosphate (polyP) is synthesized and accumulated in the vacuole and other cell compartments and represents a Pi reserve used during times of Pi starvation (Kulaev 1979; Kulaev and Vagabov 1983; Kulaev and Kulakovskaya 2000). Upon transfer of cells to a Pi-deficient medium, a rapid decrease in the polyP content is seen in whole cells and vacuoles, indicating that the polyP pool is used for cellular needs under these growth conditions (Kulaev et al. 1999). This mobilization of stored Pi is accomplished by the concerted action of several different phosphatases (Oshima 1997; Table 1). Furthermore, a lowered capacity to synthesize polyP exerts control on the rate at which Pi is taken up, possibly via a transient increase in the level of Pi in the cytosol. This may in turn lead to a direct negative feedback on Pho84p activity. The polyP thus plays a significant role in increasing the cell′s endurance to unfavorable environmental conditions and in regulating different biochemical processes.
polyP regulation
The central role of polyP on Pi homeostasis has been the subject of several extensive reviews (Kulaev 1979; Wood and Clark 1988; Kornberg et al. 1999; Kulaev and Kulakovskaya 2000). In this review, we wish to address the aspect of the complex relationship of this long-chain polymer with signaling and expression of the genes in the PHO pathway (Jacobson et al. 1982; Oshima 1997; Ogawa et al. 2000).
polyP is a highly ubiquitous molecule common to all organisms, which is formed by a high-energy phosphoanhydrous linkage of orthophosphate molecules (Rao et al. 1998). The occurrence of polyP may even precede DNA and RNA in prebiotic evolution, which may in part explain its widely distributed roles (Kornberg et al. 1999). The exact role depends on cell type, age, subcellular compartment and environmental conditions (Kornberg et al. 1999). In E. coli, polyP has various roles as a means of adapting to changes in the environment (Crooke et al. 1994; Van Dien and Keasling 1999). Indeed, similar roles are also apparent in yeast, where polyP is proposed to play important and diverse roles in cellular metabolism. These include buffering of pH variations of the cytosol (Pick et al. 1990), detoxification and osmoregulation (Castro et al. 1995), acting to sequester metal ions (e.g. Ca2+; Harold 1966; Dunn et al. 1994), facilitating DNA entry into cells (Reusch and Sadoff 1988), regulating stress and survival (Kornberg 1995) and providing storage of Pi (Kornberg et al. 1999). This last attribute suggests a pivotal involvement in Pi homeostasis and interplay with other gene products of the PHO regulatory system (Fig. 1).
A schematic diagram of the potential factors involved in polyphosphate (polyP) metabolism. polyP is located in several organelles, including the nucleus, vacuoles and periplasmic spaces (open rectangles). Enzymes involved in cleavage of the polyP terminal inorganic phosphate (Pi) residue are the exopolyphosphatases Pho5p, Pho11p and Pho12p (located in the periplasmic space), Ppx1p (cytosolic) and Pho8p (vacuolar). Pho84p and Pho89p lead to the enhanced uptake of extracellular Pi. Endopolyphosphatase, Ppn1p (containing one putative transmembrane domain and located in the vacuolar membrane, cleaves long-chain polyP into shorter chain-lengths of 3 Pi (P3) and 60 Pi (P60) residues. The vacuolar transporter chaperons, Vtc1p–Vtc4p, are most likely to be involved in trafficking proteins to the vacuole and in docking and membrane fusion, rather than in a direct involvement with polyP metabolism. Of these, only Vtc4p does not possess any putative transmembrane domains. Pho9p (Pep4p) is a protein peptidase required for the maturation of vacuolar proteins, including Pho8p. Phm6p–Phm8p, located in the cytosol, have an as yet undefined biological function. The elusive Pi transporter of the vacuole membrane (black oval) has only been semi-characterized by biochemical methods and no gene for this protein is yet known
polyPs in yeast can be divided into two types: the acid-soluble polyPs (with an average chain-length of four orthophosphate residues) and the acid-insoluble polyPs (which range in length from four Pi residues up to thousands of Pi residues; Kulaev 1979; Schuddemat et al. 1989). A large discrepancy exists as to where most polyP is stored, whether in the vacuolar compartment, or in other organelles (Trilisenko et al. 2002). Some reports document that polyP can account for up to 10% of the cellular dry weight, 40% of the total cellular Pi content and a vacuolar accumulation of up to 95% of the total cellular polyP (Urech et al. 1978; Kulaev 1979; Castro et al. 1995). A counter belief is that polyP is present in lower amounts in the vacuole and that variations in the growth stage, strain and detection methods affect estimated polyP levels (Trilisenko et al. 2002). A smaller amount of polyP is also present at the cell surface (Tijssen and Van Steveninck 1984), in the cytoplasm (Kulaev and Kulakovskaya 2000), in mitochondria (Beauvoit et al. 1989) and in the nucleus (Schuddemat et al. 1989), although the average chain-length is shorter in these compartments. The nucleus of S. cerevisiae was found to contain polyphosphatase activity (Lichko et al. 1996). However, the role played by polyP in the nucleus and its potential role in the regulation of PHO genes has so far not been investigated. Over-expression of yeast exopolyphosphatase (PPX1) in E. coli was recently shown to confer a decrease in polyP, resulting in enhanced UV and mytomycin C sensitivity and suggesting a possible role in the regulation of SOS genes (Tsutsumi et al. 2000). The presence of polyP in all these other compartments suggests either that enzymes involved in synthesis/breakdown are also present in these organelles or, alternatively, that specific mechanisms mediate uptake/extrusion of polyP (Lichko et al. 1998; Kulaev and Kulakovskaya 2000). More general APases are located in multiple organelles and may play a role in polyP turnover, e.g. Pho5p, Pho11p and Pho12p are located in the extracellular periplasmic space (Haguenauer-Tsapis et al. 1986), while the alkaline phosphatase, Pho8p, is localized in the vacuole (Plankert et al. 1991), glycerol phosphatases such as Hor2p are located in the cytosol (Norbeck et al. 1996) and polyphosphatase activity was shown to be localized in the mitochondria (Lichko et al. 2000). These enzymes are capable of hydrolyzing a variety of substrates, including nucleic acids, phosphosugars, phospholipids and phosphoproteins (Oshima 1997).
One of the enzymes implicated in the breakdown of polyP in yeast is encoded by the PPX1 gene, originally referred to as ScPPX1 to distinguish it from the PPX gene previously identified in E. coli (Wurst and Kornberg 1994). The 396-amino-acid protein encoded by the yeast PPX1 gene possesses an exo-nuclease activity that elicits polyP degradation by a stepwise removal of the terminal Pi residue (polyPn → polyPn−1 + Pi; Wurst and Kornberg 1994; Wurst et al. 1995). The rate of hydrolysis for Ppx1p is about 40-fold greater than for the Ppx of E. coli and releases 30,000 Pi residues min−1 enzyme molecule−1 at 37 °C (Wurst and Kornberg 1994). However, a yeast strain lacking the PPX1 gene did not show a significant increase in the level of polyP (Wurst et al. 1995). Ppx1p from the wild-type strain lacks a signal sequence, is primarily located in the cytosol and is not enriched in a purified vacuolar fraction (Wurst et al. 1995). Indeed, the PPX1 gene shows no variation with respect to Pi availability or the regulation of other genes in the PHO pathway.
Further investigations led to the characterization of an endopolyphosphatase, Ppn1p (Phm5p), a homodimer of 35 kDa with a single putative N-terminal transmembrane (TM) domain (Kumble and Kornberg 1996, Ogawa et al. 2000). This enzyme catalyzes the non-processive cleavage of long chains of polyP (P750), to produce shorter chains of 60 Pi units (P60) and 3 Pi units (P3; Kumble and Kornberg 1996). The endopolyphosphatase activity is greater in yeast, in contrast to the barely detectable activity in E. coli, suggesting the involvement of the compensatory effect of high exopolyphosphatase activity in bacteria. The presumption that the Ppn1p enzyme is located in the vacuole is supported by the finding that mutant strains, where proteolytic enzymes involved in the processing of vacuolar proteins are absent, results in a 20-fold lower expression level of Ppn1p, as compared with wild-type strains (Kumble and Kornberg 1996; Sethuraman et al. 2001). Interestingly, it was also reported that the enzyme activity was inhibited by 50% and by 100% in the presence of 20 mM Pi and 10 mM pyrophosphate (PPi), respectively (Kumble and Kornberg 1996). Thus, a feedback mechanism appears to exist to prevent the wasteful degradation of polyP during times when the end products, i.e. Pi and PPi, are abundant in the cell.
In contrast to the considerable knowledge gained about both the exo- and endopolyphosphatase enzymes, very little is known about enzymes involved in polyP synthesis (kinases). It is anticipated that enzymes involved in the synthesis of this polymer are under an equally stringent regulation, in order to maintain a correct balance of polyP, perhaps even more so considering that the activity of Ppx1p is about 20-fold greater than the reported activity of a kinase (Wurst et al. 1995). However, no gene homologous to the bacterial polyP kinase has yet been identified in yeast (Ogawa et al. 2000). The feeble enzyme activity reported for the kinase which catalyses the reversible reaction ADP + polyPn ⇌ ATP + polyPn–1 and results in the transfer of Pi residues from polyP to ADP at a rate of 4 nmol of Pi residues min−1 mg−1 protein, is even less in the reverse direction, i.e. for the synthesis of polyP by condensation of the terminal Pi of ATP (Felter and Stahl 1973; Wurst et al. 1995). Perhaps other kinases are involved to a greater or lesser extent in the synthesis of polyP from different substrates (e.g. 1,3-diphosphoglycerate + polyPn ⇌ 3-phosphoglycerate + polyPn+1), which is catalyzed by the enzyme 1,3-diphosphoglycerate:polyP phosphotransferase (Schuddemat et al. 1989).
A double mutation of the PPN1 and PPX1 genes resulted in a rapid loss of viability in the stationary phase (Sethuraman et al. 2001). The loss of viability may be due to the increased chelating capacity of polyP for the divalent ions (Ca2+, Mg2+, Mn2+) required for cellular processes, which in this double mutant reached up to 20%, by dry weight of the cell. Alternatively, the shorter P60 and P3 chains might be involved in cellular functions (Sethuraman et al. 2001).
DNA microarray studies indicate a whole battery of known and uncharacterized genes implicated in the regulation of polyP and Pi (Ogawa et al. 2000). This set of genes includes: those encoding (1) the high-affinity Pi transporters (i.e. PHO84, PHO89), (2) a protein involved in trafficking of at least one Pho protein (PHO86), (3) phosphatases, of which the APases encoded by PHO5, PHO11 and PHO12 are localized in the periplasmic space and the ALPase encoded by PHO8 is localized in the vacuole, (4) proteins involved in polyP synthesis, such as Phm1-4p, and (5) genes known (PHO81) and suggested (YPL110c, SPL2) to be regulators of PHO genes. This set also includes some genes of different known and unknown functions [e.g. HOR2/GPP2 (a dl-glycerol phosphatase), HIS1 (an ATP phosphoribosyl transferase), PHM5–PHM8].
PHM1, PHM2, PHM3 and PHM4 display varying degrees of perturbation in their capacity to accumulate polyP in the vacuole of S. cerevisiae. These genes are identical to those previously identified by Cohen and colleagues: VTC1 (PHM4), VTC2 (PHM1), VTC3 (PHM2) and VTC4 (PHM3; Cohen et al. 1999; Ogawa et al. 2000). Vtc1p (vacuolar transporter chaperon) corresponds to a hydrophobic protein of 129 amino acid residues (14.4 kDa), structurally organized into three putative TM domains shown to co-fractionate by immunoprecipitation with a subunit of the vacuolar ATPase, Vma10p. A vtc1 deletion mutant was shown to confer a reduction in the amount of v-ATPase in the vacuolar membrane and a reduction in the amount of the plasma membrane H+-ATPase, Pma1p (Cohen et al. 1999). Vtc2p and Vtc3p share a 58% similarity at the amino acid level and are of approximately equal size, being 828 and 835 amino acids (95.4 kDa, 96.6 kDa), respectively. Both proteins contain a large, globular N-terminal domain and two or three predicted TM regions in the C-terminal parts. Vtc4p is similar to Vtc2p and Vtc3p, except that it lacks the hydrophobic domain (Cohen et al. 1999) and is only contained in the membrane fraction when Vtc1p and either Vtc2p or Vtc3p are present (Ogawa et al. 2000). The Vtc4 protein has a predicted sequence of 648 amino acid residues. The N-terminal globular domain of several members of the Vtc protein family shares a significant homology with Pho81p and Pho87p (Cohen et al. 1999). All single-gene deletion mutants of vtc1–vtc4 are viable in high-Pi media (Ogawa et al. 2000). For cells grown at low-Pi conditions, it was observed that, after the initial uptake of Pi via the high-affinity plasma membrane transporters with a concomitant increase in the intracellular Pi level, no further Pi influx occurred in the vtc4, vtc1 mutant strains and the double mutant, vtc2–vtc3. The inability of these mutants to produce polyP molecules serving as a Pi sink prevented further Pi being loaded into these cells. Thus, the homeostasis of the intracellular Pi level appears to be governed by the synthesis of polyP, which exerts a negative-feedback loop controlling the rate of Pi uptake by the high-affinity Pho84p transporter across the plasma membrane (Ogawa et al. 2000). The single vtc2 mutation displayed a Pi uptake activity similar to that of the wild type, with only a minor decrease in the polyP level. The Vtc1–Vtc4 proteins are thought to be involved in the transport of Pi into the vacuole, the metabolism of polyP (Ogawa et al. 2000), or the fusion of vacuolar membranes (Müller et al. 2002).
Other up-regulated genes identified in the study of Ogawa et al. (2000) were PHM5–PHM8. Although no specific role has yet been assigned to the gene products of PHM6–PHM8, the levels of mRNA for PHM6 and PHM8 were induced almost 5-fold by treatment with the drug 2-(2-hydroxyethylamino)-6-(3-chloroanilino)-9-isopropylpurine, which decreased the expression of both CDC28 and PHO85 mRNA levels (Gray et al. 1998). To ensure a continued supply of the Pi present within the cell, the activation of genes for the expression both of proteins involved in Pi acquisition from the medium and also of proteins involved in the liberation of Pi from stored polyP in the vacuole requires a coordinated control. This necessitates a high degree of interplay between the gene products involved in polyP metabolism and Pi acquisition from the external media, although these may not necessarily overlap in time or may be subjected to differing degrees of control, as was reported for the expression of PHO5 and PHO8 (Münsterkötter et al. 2000).
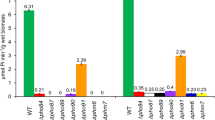
A clear degradation and mobilization of polyP, as visualized by 31P-NMR analysis during growth of S. cerevisiae at Pi-limiting conditions, precedes the maximal expression of the high-affinity Pho84 Pi transporter. It is apparent from the 31P-NMR spectra shown in Fig. 2A that the level of Pi was approximately 10-fold reduced and that the peak corresponding to polyP was already absent during the initial lag phase. At later stages of growth, the internal Pi pool was replenished because the induction of the high-affinity Pho84p results in sequestering available Pi from the medium. This is in agreement with the idea that the degradation of polyP has already taken place prior to the maximal expression of the Pho84p transporter (Fig. 2B). Further addition of Pi to the growth media at the point of maximal expression of the Pho84p Pi transporter was shown to result in a rapid internalization and targeting of the protein to the vacuole for degradation, a process completed within 45 min (Lagerstedt et al. 2002). The absence of replenished polyP during this period possibly reflects that the activity of the kinase is extremely feeble, as compared with the phosphatases (Wurst et al. 1995). This weak kinase activity may explain the delay in polyP synthesis and the observed polyP overload scenario upon the further addition of Pi, originally described by Harold (1966) and more recently described by Andreeva et al. (2001).
Analysis of Pi and polyP in Saccharomyces cerevisiae cells. A 31P NMR showing the intracellular levels of Pi and polyP in S. cerevisiae cells expressing Pho84WT and grown on high-Pi and low-Pi media. Cells were harvested after growth to an absorbance at 590 nm (A 590) of 5.0 in high-Pi medium (spectrum I) or to A 590 of 1.0, 2.5, 3.5, 5.0 and 7.0 in low-Pi medium (spectra II, III, IV, V, VI, respectively). At each point, 4 g of cells were harvested, treated with 10% perchloric acid, neutralized with a saturated solution of potassium hydrogen carbonate and used for 31P NMR studies. Peaks were assigned from the literature and by the addition of known compounds during perchloric acid extraction. a Sugar phosphates, including glucose-6-phosphate and fructose-6-phosphate, b glycerylphosphorylethanolamine, c glycerylphosphorylcholine, d γ-phosphate of NTPs, e α-phosphate of NTPs and f - β-phosphate of NTPs and the terminal phosphate signals from polyphosphate and triphosphate, polyP i middle phosphate of polyphosphate molecules. B Western analysis for Pho84-MYC expression from cells isolated during sample collection described for A. Samples were isolated from cells grown in high-Pi medium (lane I) or to A 590 of 1.0, 2.5, 3.5, 5.0 and 7.0 in low-Pi medium (lanes II, III, IV, V, VI, respectively)
In addition, the formation of the phosphoanhydride bond and the synthesis of polyP is an energy-consuming process and this may also be true for the uptake of Pi across the vacuole membrane. This is supported by the findings of Castro and colleagues (1995), who reported that ATP might act as either a precursor or regulator of polyP synthesis. In mutants deficient in the vacuolar ATPase, vma4, only 1% of the polyP level is retained, as compared with wild-type cells (Beauvoit et al. 1991). However, by lowering the intravacuolar pH, a low but significant level of polyP accumulates in vma4 mutants, suggesting that the v-ATPase is not essential for polyP synthesis itself (Nelson and Nelson 1990), but is rather the maintenance of a pH gradient across the vacuolar membrane. Clearly, more in-depth studies are required to elucidate the precise function of the VTC1–VTC4 genes and their participation in Pi uptake across the vacuole membrane, polyP metabolism and/or chaperoning vacuolar transport. Although a vacuolar Pi exchange mechanism has been proposed (Bourne 1990, 1991; Booth and Guidotti 1997), neither the gene nor the protein responsible for a concentration-dependent and H+ gradient-dependent Pi transport have been identified.
The PHO regulatory pathway
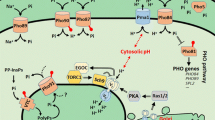
As a response of Pi limitation, at least 22 PHO-related genes in S. cerevisiae are derepressed, leading to elevated transcript levels, as judged by DNA microarray analysis (Ogawa et al. 2000), together with genetic and biochemical studies (reviewed by Oshima 1997; Persson et al. 1999; see Table 1). The expression of these PHO genes is under the tight regulation of the PHO regulatory pathway. A complex of the cyclin-dependent kinase (CDK), Pho85p, one of its cyclins, Pho80p, and the CDK inhibitor (CKI), Pho81p, together with the transcription factors Pho4p and Pho2p constitute the core components of this transcriptional regulation of PHO genes (Fig. 3). Access of the transcriptional regulators Pho2p and Pho4p to specific DNA sites is aided or hindered by the chromatin structure; and hence the nature of DNA packaging is important for the regulation of gene expression. It is documented that PHO5 transcription requires Arg82, a nuclear-located inositol polyphosphate kinase, for remodeling the chromatin structure of the promoter region (Steger et al. 2003). Acetylation, methylation or phosphorylation of histones can all modify the chromatin-packing arrangement. Additional factors implicated in chromatin remodeling include the formation of SWI/SNF complexes that alter histone/DNA interactions in the promoter regions of certain genes e.g. PHO8 and PHO84. PHO8 transcription, under Pi-starvation conditions, occurs after the targeting of SWI/SNF and acetylation of H3 in a Gcn5p-dependent manner, whereas acetylation of H3 by Gcn5p for derepressed PHO5 transcription is SWI/SNF-independent (Krebs et al. 2000). Transcriptional activation of other genes in the PHO regulon which are also shown to be highly dependent on SWI/SNF and Gcn5p acetylation include PHO11, PHO12 and PHO84 (Sudarsanam et al. 2000).
Regulation of PHO genes at low and high external Pi conditions. At low external Pi concentrations (left panel) PHO genes such as PHO84 are up-regulated. This occurs by binding the active transcription factors (ovals) Pho4p and Pho2p to the promoter regions of the PHO genes. The synthesized Pho84p is transported to the plasma membrane in a Pho86p-dependent manner, where it exerts its function as a competent high-affinity Pi transporter. When the external Pi is fully exhausted, or when a repressible concentration of Pi is added to the culture, Pho84p is endocytosed and thereby inactivated (boxes), with the vacuole as the final destination. The potential of Pho4p in transcriptional activation is governed by its cellular location, which in turn is dependent on its phosphorylated state. At low Pi conditions, Pho4p is transported into the nucleus via Pse1p. The cyclin-dependent kinase (CDK) Pho85p and its cyclin Pho80p are, at these conditions, inhibited by the CDK inhibitor Pho81p and are thus unable to phosphorylate Pho4p. In contrast, at high Pi conditions (right panel) Pho4p is phosphorylated, complexed with Msn5p and Gsp1p and transported into the cytoplasm. In the cytoplasm, the GTPase activation protein Rna1 mediates a dissociation of the complex
To monitor the status of the PHO pathway, the activity of a secreted acid phosphatase (the gene product of PHO5, which is transcriptionally induced as a response to Pi starvation) is commonly used (Toh-e et al. 1973; Oshima 1997). Deletion analysis of the PHO5 promoter reveals two regulatory elements (or upstream activator sequences; UAS), UASp1 and UASp2 (Rudolph and Hinnen 1987), to which Pho4p is shownto bind in vivo upon Pi starvation, but not under high-Pi conditions (Venter et al. 1994). When the chromatin region surrounding the PHO5 promoter cannot be remodeled, Pho4p can bind to the accessible site of UASp1 but not UAS2p, which is integrated into part of the nucleosome structure (Venter et al. 1994; Steger et al. 2003). The binding sequences for the Pho4 protein, CACGTG or CACGTT, have been described (Oshima 1997) and at least one copy of these sequences can be found in the immediate promoter region (500 bp upstream of the coding region) of all but one of the other described PHO-regulated genes (Ogawa et al. 2000), suggesting that Pho4p indeed is a transcriptional activator for these genes. The Pho4p protein consists of 312 amino acid residues (Yoshida et al. 1989) and contains four functional domains (Ogawa and Oshima 1990). In vitro glutathione S-transferase pull-down experiments have shown a direct interaction between Pho4p and yeast TFIIB (Wu and Hampsey 1999). Based on a Pho4p-dependent V8 protease accessibility, the authors also suggest a Pho4p-mediated conformational change of TFIIB during the process of transcriptional initiation. The structure of the DNA-binding domain of Pho4p, resolved to 2.8 Å, shows that this particular domain folds into a C-terminal (amino acids 251–312) basic helix-loop-helix (bHLH) motif and that Pho4p binds to the DNA sequence as a homodimer (Shimizu et al. 1997). The bundle topology of the two Pho4p monomers (folded into a parallel, left-handed four-helix bundle stabilized by van der Waals interactions) is identical to the structures of other bHLH/Zip proteins (Shimizu et al. 1997), such as Max and USF (Ferré-D′Amaré et al. 1993, 1994, respectively). Using native gel electrophoresis technology and an in vivo Pho5p phosphatase activity assay, a cysteine to alanine (C300A) mutation of Pho4p shows this particular cysteine is crucial for Pho4p dimerization, for binding the homodimer to its UAS in the promoter region of PHO5 and for the transcriptional activation of the PHO regulon (Shao et al. 1998). The backbone dynamics of the bHLH domain in the absence of DNA, nonspecifically bound to DNA and bound to cognate DNA, has been probed by NMR techniques (Cave et al. 2000). This secondary structure study shows that, relative to the unbound bHLH domain of Pho4p, a similar and large change in conformation and backbone dynamics in the basic domain occurs as the bHLH domain binds to either the nonspecific or cognate DNA.
By itself, the homeodomain transcription factor Pho2p (also known as Bas2p or Grf10p) binds DNA [consensus sequence (T/C)TAA(T/A)T(T/G)AAT; Barbaric et al. 1996] with low affinity (Brazas and Stillman 1993) and therefore, for the mediation of its function, Pho2p interacts with several other proteins. The pleiotropic effector Pho2p is involved in the regulation of a diverse array of other genes. Together with Swi5p, it binds cooperatively to a regulatory element in the HO promoter (Brazas et al. 1995). Pho2p is also involved in the regulation of HIS4 (Arndt et al. 1987), TRP4 (Braus et al. 1989) and certain ADE genes (Daignan-Fornier and Fink 1992). For its function in PHO regulation, Pho2p acts through multiple DNA-binding sites. At the PHO5 promoter region, one of these sites significantly overlaps with the Pho4p-binding site at UASp1, while another two sites flank the second Pho4p-binding site, UASp2. Binding to these sites occurs in a cooperative manner with Pho4p (Barbaric et al. 1996). In this context, two critical functions for Pho2p have been described, the first being to recruit Pho4p to the DNA and the second being to enhance its activation potential once bound to the DNA. For UASp1, recruitment of Pho4p seems to be the crucial role of Pho2p, while at UASp2 it is mainly the second function of Pho2p that is required (Barbaric et al. 1998). Based on mutational screening combined with activity assays of reporter genes, the region from positions 343 and 390 in Pho2p have been suggested to be important for the protein–protein interaction between Pho2p and Pho4p (Bhoite et al. 2002).
Pho85p is one of five CDKs found in S. cerevisiae (discussed by Andrews and Measday 1998). In general, CDKs are inactive as monomers and require binding of a cyclin for their activity (Morgan 1995); and at least ten different cyclins are known to bind Pho85p (Measday et al. 1997). These Pho85p cyclins (Pcls) are involved in cell cycle and/or metabolic regulation. For example, Pcl5p is required for the function of Pho85p in phosphorylating Gcn4p (Shemer et al. 2002), a yeast transcriptional activator of amino acid biosynthetic genes (Hope and Struhl 1985) that also activates genes involved in glycogen homeostasis (Natarajan et al. 2001). Furthermore, for the targeting of the glycogen synthase, Gsy2p, Pho85p is dependent on Pcl8p and Pcl10p (Huang et al. 1998). In the regulation of the cell cycle, Pho85p interacts with the G1 cyclins, Pcl1p and Pcl2p (Espinoza et al. 1994; Measday et al. 1994). For the phosphorylation of Glc8p (the yeast orthologue of the mammalian phosphatase inhibitor-2, which in turn regulates the yeast Glc7 serine/threonine protein phosphatase-1; Nigavekar et al. 2002), the predominant Pho85 cyclins appear to be Pcl6p and Pcl7p (Tan et al. 2003).
In the Pi assimilation process, the kinase function of Pho85p is activated by interaction with Pho80p and the kinase activity of this Pho80p–Pho85p cyclin–CDK complex regulates the status of the PHO regulon by phosphorylating Pho4p and possibly other regulatory elements. The phosphorylation of Pho4p occurs when yeasts are grown in Pi-rich medium (about 10 mM Pi; Kaffman et al. 1994); and five serines with a consensus phosphorylation site sequence SPXI/L are modified, as shown by in vitro and in vivo experiments (O′Neill et al. 1996). Pho4p is concentrated to the nucleus when yeast cells are starved for Pi and it is predominantly cytoplasmic when yeast cells are grown in Pi-rich medium (O′Neill et al. 1996). Furthermore, in a pho85 or pho80 mutant strain when Pho4p is hypophosphorylated, Pho4p is concentrated in the nucleus at both high and low external Pi concentrations (O′Neill et al. 1996). In contrast, Pho4p is predominantly cytoplasmic in a pho81 mutant strain grown at high- or low-Pi conditions. Clearly, the phosphorylated state of Pho4p is important for its localization in the cell and hence for its activity in transcriptional regulation of the PHO genes. The transport of Pho4p out of the nucleus is mediated by the receptor Msn5p (Kaffman et al. 1998a). In this process, the hyperphosphorylated Pho4p is accessible for complex formation with Msn5p and Gsp1p, a yeast Ran homologue, after which the complex is transported into the cytoplasm. For the formation of the complex, the Gsp1p has to contain bound GTP (Kaffman et al. 1998a); and then hydrolysis to GDP, an event possibly stimulated by the GTPase-activating protein, Rna1p, dissociates the complex in the cytoplasm.
At low Pi concentrations, the hyperphosphorylation of Pho4p is abolished by inactivation of the Pho85p–Pho80p kinase complex by the CKI, Pho81p (Schneider et al. 1994); and activation of the PHO genes, including PHO81, is enabled. Pho4p, in its unphosphorylated form, is presumably transported into the nucleus through the nonclassic import pathway that utilizes the importin beta family member, Pse1/Kap121p (Kaffman et al. 1998b). In contrast, in its phosphorylated state, Pho4p is unable to bind to Pse1p (Kaffman et al. 1998b), thus preventing transport of Pho4p into the nucleus at repressible conditions. The individual roles of the phosphorylation sites of Pho4p (named SP1, SP2, SP3, SP4, SP6; Komeili and O′Shea 1999) and their kinetic parameters, together with site preference (Jeffery et al. 2001), have been determined. Inactivation of nuclear Pho4p occurs first by phosphorylation of SP6. This phosphorylation prevents the interaction of Pho4p with its transcriptional co-activator Pho2p, resulting in an immediate repression of the genes dependent on Pho4p–Pho2p. Phosphorylation on SP2 and SP3 inactivates Pho4p by promoting its rapid export from the nucleus. Import of Pho4p into the nucleus is blocked by phosphorylation of SP4. As predicted by computer modeling, 87–92% of the Pho4p molecules that are phosphorylated on SP2 and SP3 are also phosphorylated on SP4 and SP6 (Jeffery et al. 2001). A futile cycle, where exported Pho4p is directly imported back into the nucleus again, via interaction with Pse1, is thus prevented.
In vitro experiments show that Pho2p can also be phosphorylated by a kinase (Liu et al. 2000), possibly indicating a similar mechanism.
The positive feedback-regulated inhibition of Pho85p–Pho80p exerted by Pho81p, makes Pho81p a key player in the PHO regulatory pathway. Pho81p contains a tandem repeat of six ankyrin consensus regions. The ankyrin motif is recognized in several hundred proteins (Bork 1993), including CKIs; and it is regarded to be important for protein–protein interaction (discussed by Sedgwick and Smerdon 1999). Ogawa et al. (1995) reported that the fifth and sixth repeats, plus some additional amino acids (in total 141 amino acids), are sufficient for proper in vitro inhibition of the Pho85p–Pho80p complex, suggesting that the ankyrin repeat indeed plays an essential role in the CKI/CDK complex interaction. Recently, it was shown that a minimum domain of 80 amino acids residing C-terminally of the six ankyrin repeats is necessary and sufficient for Pho81p inhibition of Pho85p–Pho80p in response to low Pi conditions (Huang et al. 2001). How the signal for Pi limitation is transduced to the Pho81p and where the signal originates are not yet fully understood. We observed that, upon incubation of cells in a Pi-limiting environment, the first source of Pi to be mobilized is the internal polyP pool stored in the vacuole (See Fig. 2). Although there is no evidence to link the levels of polyP and other Pi compounds, e.g. inositol polyphosphates with regulation of the PHO regulatory pathway, polyP biosynthesis can occur simultaneously with RNA synthesis as a means to sequester liberated Pi (Crooke et al. 1994; Kulaev and Kulakovskaya 2000).
Structural and functional basis of the Pho84 and Pho89 Pi transporters
Pho84p belongs to the family of Pi:H+ symporters (TC number 2.A.1.9; Saier 2000) and is a member of the major facilitator superfamily (MFS; Pao et al. 1998). This protein has been shown to be highly homologous to several hexose sugar transporters (HXT) of mammalian, yeast and bacterial origin (Bun-ya et al. 1991; Bisson et al. 1993; Hendersson 1993; André 1995; Nelissen et al. 1995; Kruckeberg 1996) and to identified Pi transporters in yeast, fungi and plants. Based on hydropathic analysis of the primary amino acid sequence (587 amino acids), a secondary structure model of this hydrophobic integral membrane protein has been proposed (Fig. 4A; Persson et al. 1999), in which 12 TM domains are connected by hydrophilic loops and connected with extended N- and C-termini located at the same membrane phase. As in the case for several HXT (Kruckeberg 1996; Boles and Hollenberg 1997), the Pho84 transporter exhibits a significant number of conserved residues between each of the two halves of the protein (Persson et al. 1999), separated by a central, large hydrophilic loop harboring 75 amino acid residues between TM VI and VII. The duplication is proposed to have arisen by an internal gene duplication event prior to the divergence of the MFS families (Rubin et al. 1990; Pao et al. 1998). Several conserved sequence motifs are found in the Pho84 protein. A Walker nucleotide-binding consensus motif, GX4GKT (as yet uncharacterized; Walker et al. 1982), is found in the C-terminal connection of the large central loop to TM segment VII, as the sequence GgwkyGKi (not fully conserved residues are indicated by lowercase letters). As a consequence of the internal duplication within the two 6+6 TM units, the consensus sequence Div/iGRK (Kruckeberg 1996) is found in the hydrophilic loops connecting TM segments II and III and TM segments VIII and IX. In another MFS protein (the human Glut1 glucose transporter) the positive charges in the corresponding sequence of that protein are suggested to be important for proper membrane topology (Sato and Mueckler 1999). The fact that the structural organization of these two proteins shares common features, such as 12 TM segments and a centrally positioned hydrophilic loop sequence, together with the common localization of the conserved motifs, suggest a similar importance of these positively charged amino acids in the folding process of the Pho84p. Also, the consensus sequence IPEs/tp/kRK is found in the hydrophilic domains connecting putative TM segments VI and VII and in the C-terminal extension of TM XII. The function of this duplicated motif has not yet been established. It can, however, not be ruled out that the PEST-like sequence (Rechsteiner and Rogers 1996) specifies a motif recognized in regulated degradation of the protein. The Pho84 protein contains 12 native cysteine residues, of which five are predicted to be located in putative TM regions III, VI, VIII, IX and X, with the remaining seven in the hydrophilic domains of the protein. A cysteine-less variant of Pho84p, in which all 12 cysteine residues are replaced with serine residues, was demonstrated to be stably expressed in vivo under derepressing conditions and active in H+-coupled Pi transport across the yeast plasma membrane (Berhe et al. 2001). In the absence of a protein crystal structure, identification of individual amino acid residues that are in close physical contact with each other in the functional structure may be achieved by low-resolution techniques, such as site-directed crosslinking. For this, cysteine-scanning mutagenesis and crosslinking of selectively introduced pairs of cysteine residues, followed by subsequent analysis of disulfide bond formation prove to be valuable in the elucidation of structural and dynamic aspects of the membrane protein structure and function (Frillingos et al. 1998; Grünewald et al. 2002; Kohler et al. 2002). It can be anticipated that the cysteine-scanning technique will be important in depicting the structural organization of the Pho84p transporter, a prerequisite for the elucidation of the Pi translocation mechanism.
Secondary structure model of the Pho84p (A) and Pho89p (B) transporters derived using the Top-Pred algorithm (Claros and von Heijne 1994) The one-letter amino acid code is used and the 12 putative transmembrane segments are shown as cylinders. Bold-faced amino acid residues represent residues duplicated within the proteins. The positions of the 12 and five cysteine residues of Pho84p and Pho89p, respectively, are boxed. The SINNDAKSS-like sequence of Pho84p is underlined
Like Pho84p, the 574-amino-acid sequence of the Pho89 transporter (TC number 2.A.20.2), based on hydropathic analysis, is predicted (Claros and von Heijne 1994) to be organized into 12 discrete hydrophobic TM domains (Persson et al. 1999; Fig. 4B). The sequence identity between these two proteins is as low as 15%. In spite of this, a similar internal duplication of amino acid residues, as seen with Pho84p, can be seen in Pho89p. Here, rather than the symmetric duplication seen for Pho84p, the large hydrophilic segment containing 110 amino acid residues is predicted to connect TM VII and VIII and is located on the opposite membrane face from that of the N- and C-termini (Persson et al. 1999). The 41 duplicated residues of Pho89p are located in putative TM segments I–V, in IX–XII and in the hydrophilic domains connecting these segments (Persson et al. 1999). Pho89p is a homologue of the mammalian type III Na+-Pi transporters. Several of these type III transporters have a dual function and, apart from their Pi transport capability, they also serve as the cell receptor for several retroviruses. An experimentally based topological model for one of these proteins, namely the Pit2p Pi transporter/retrovirus receptor sharing 28% identity with Pho89p (Salaün et al. 2001), is in agreement with the proposed model of Pho89p, with 12 TM regions and a large hydrophilic loop connecting TM VII and TM VIII. Furthermore, the N- and C-terminal extensions of Pit2p are shown to be extracellular. Recently, Böttger and Pedersen (2002) described two glutamates in Pit2p that are critical to Na+-dependent Pi transport function in this protein. These amino acid residues are conserved within the type III Na-Pi transporters, in PHO-4 from Neurospora crassa and in Pho89 (Böttger and Pedersen 2002), which would suggest a similar importance for Na+-coupled Pi uptake in these proteins. The corresponding glutamates in Pho89p are localized to the central region of TM II and in the suggested extracellular face of TM XI.
Regulation and trafficking
In S. cerevisiae, transcription of the PHO84 gene is initiated when cells are grown in a low-Pi medium containing approximately 200–300 μM Pi and is repressed under conditions of Pi adequacy (reviewed by Oshima 1997; Lagerstedt et al. 2000). Enhanced capacity by cells for Pi uptake correlates with elevated transcript levels and with the amount of synthesized protein detected, by Northern and Western blot analyses, respectively (Martinez et al. 1998; Lagerstedt et al. 2002). In a similar fashion, the repressible APase, PHO5, is also believed to be transcriptionally regulated (Tamai et al. 1985). However, other studies based on Northern analyses show there is only a subtle increase (1.2- to 2.0-fold) in the amount of PHO84 transcript in a quadruple Pi transporter-deletion strain lacking the genes PHO87, PHO89, PHO90 and PHO91 (Wykoff and O′Shea 2001). These authors raise the possibility that the compensatory effect seen for the non-Pho84p-mediated Pi uptake pathway occurs via an enhanced Pho84p activity, due to its accumulation in the plasma membrane, which is probably the result of a post-transcriptional modification. Further exploration of such a post-translational regulatory mechanism is anticipated to be informative and provide an insight into this proposed additional control level, allowing for a rapid adaptation of the cell in response to changes in the Pi status.
Recent reports on the transcriptional response of S. cerevisiae cells when shifted to alkaline conditions identify an increased expression of between 71 transcripts (Causton et al. 2001) and 500 transcripts (Lamb et al. 2001), depending on the pH range investigated and the technologies used. From these studies, it can also be concluded that, in addition to low Pi levels, mild alkaline stress can also give rise to a transcriptional induction of several genes related to Pi metabolism, i.e. PHO84, PHO89, PHO12, PHM2, PHM3 and PHM4. Of these, PHO89 is proposed to be induced to a higher level by mild alkalization treatment, rather than by Pi starvation alone. The induced expression of PHO89 by alkalization is, in part, maintained even in pho4 and pho2 mutant cells, but is totally abolished in a crz1 mutant (a transcription factor activated by calcium/calcineurin; Serrano et al. 2002). Both PHO89 and ENA1 are shown to give a very rapid and transient response, whereas PHO84 induction is slower. Upon mild alkalization, the PHO84 mRNA transcript is about 12-fold enhanced after a 25–45 min incubation period (Serrano et al. 2002). After activation of the gene and de novo synthesis, the Pho84p must be correctly targeted to its final destination within the cell. The correct sorting of Pho84p, lacking a signal sequence, is mediated through the participation of Pho86p (Lau et al. 2000). Pho86p is a 34-kDa protein consisting of 311 amino acid residues with two strongly hydrophobic segments in its N-terminal half, suggested to be membrane-associated (Yompakdee et al. 1996a). In the absence of Pho86p, Pho84p is localized to the endoplasmic reticulum (ER) and fails to be targeted for its plasma membrane destination. Pho86p has been found to be an ER resident protein required for the correct packaging of Pho84p into COPII vesicles (Lau et al. 2000). Pho86p lacks any recognizable retrieval signal and is not itself packaged into the COPII vesicles, but remains associated with the ER, therefore belonging to a class of outfitters: resident ER proteins that facilitate the loading of cargo into transport vesicles (Herrmann et al. 1999). The function of Pho86p is likely to be specific to Pho84p, since it has no effect on the galactose transporter that shares sequence similarity with Pho84p. However, the exact mechanism whereby Pho86p ensures correct packaging of Pho84p is not known. It could possibly be involved in folding, maturation and/or oligomerization of Pho84p in the ER, allowing it to assume a conformation competent for incorporation into COPII vesicles. Another possibility is that Pho86p is involved in recruiting COPII proteins to the ER. It is not known whether Pho86p interacts directly with Pho84p. If it does, this could explain the specificity of Pho86p for Pho84p. In contrast to PHO84 and PHO89, PHO86 is transcribed in both high- and low-Pi media. However, in low-Pi media, the levels of PHO86 transcript are slightly more elevated. PHO86 thus appears to be under a less stringent control by variations in Pi. Disruption of PHO86 is not lethal to cells, although a consequence of a lack of Pho86p manifests itself as a decrease in Pi uptake activity, as is the case for a pho84 deletion strain.
The rate of Pi uptake by Pho84p increases in parallel with the exponential growth phase in low-Pi media and reaches its maximum rate at the mid- to late-exponential growth phase, before rapidly declining. The highest transport activity is achieved when the extracellular Pi concentration is in the range 50–70 μM. The onset of the decline in transport activity coincides with a situation when the extracellular Pi is very low (10 μM), which is close to the reported K m value for the transporter (Martinez et al. 1998). This would support the hypothesis that the derepression of PHO84 is under the control of the extracellular Pi level and that its inactivation is also subject to the same control (Bun-ya et al. 1991; Martinez et al. 1998). The suggestion that the signal for derepression is external Pi and not internal Pi is also supported by experiments by Bostian and coworkers (1983). Two alternatives may be distinguished explaining why the disruption of PHO84 causes constitutive expression of the Pho5p APase. Either the Pho84p plays a direct role in sensing high Pi, thus controlling the gene activation via the PHO regulon or, alternatively, insufficient levels of Pi are taken up by these cells, signaling Pi starvation even under high-Pi conditions (Wykoff and O′Shea 2001). Interestingly, pho84 mutant cells release a substantial amount of Pi into the media early in the growth phase (Pattison-Granberg and Persson 2000). At later stages of growth, the cells are in part able to compensate for this liberation and to accumulate Pi, although at a slower rate. Pho84p shares 39% similarity with the transporter-like glucose sensors, Snf3p and Rgt2p (Özcan et al. 1996). PHO84 is located upstream of all other known pathway components and is thus a candidate for a dual role, with a capability not only for Pi uptake but also for sensing Pi levels. For a recent review on transporter-like sensors, see Forsberg and Ljungdahl (2001). Recently, Giots et al. (2003) showed that either deletion of the Pho84p transporter or destruction of the H+ gradient across the plasma membrane leads to an abolished phosphate signaling. Pho87p, a protein proposed to interact with Pho84p (Bun-ya et al. 1996) is also shown to sustain rapid phosphate signaling. However, Pho84p does not display any sequence homology to Pho87p, nor does it contain a typical N-terminal extension (Bun-ya et al. 1996). However, Wykoff and O′Shea (2001) suggest that the sensor is more likely an internal sensor, although no direct link to Pi or the polyP status of the vacuole is implicated.
Internal Pi pools satisfying a demand for rapid internal Pi signaling may possibly act in balance with activation of scavenging proteins in response to external stimuli. Under conditions when the cells meet no Pi limitations, free Pi is predominantly stored in the form of polyP, whereas only low amounts of these Pi reserves are maintained during Pi starvation (Vagabov et al. 2000; Fig. 2A). It appears that intracellular polyP pools are responsible for sustaining cell growth for a limited period when the cell first encounters a Pi limitation and that the maximal expression of Pho84p occurs after polyP has been depleted (Fig. 2). Indeed, mutants defective in aspects of polyP metabolism are not seen to have any affect on the PHO signaling pathway. After replenishment of Pi in the media, cells no longer require a high-affinity scavenging mechanism. The construction of a chimeric Pho84-green fluorescent protein (Pho84-GFP) and its expression from the chromosomal location of PHO84 under the control of its native upstream promoter has allowed the detailed intracellular localization study of the trafficking of the Pho84p during Pi starvation and replenishment studies (Petersson et al. 1999). These studies indicate the existence of a down-regulatory pathway, with the vacuole being the terminal destination of the Pho84 protein under restrictive Pi conditions, as has been shown for several other plasma membrane receptors and nutrient transporters destined for degradation in this acidic compartment (Riballo et al. 1995; Hicke and Rietzman 1996; Horak and Wolf 1997; Krampe et al. 1998; Kruckeberg et al. 1999; Graschopf et al. 2001). The best characterized of these is the mating pheromone α-factor and its plasma membrane receptor, Ste2p, which require both End3 and End4 proteins for internalization (Raths et al. 1993).
Although Pho84p has been shown to be endocytosed in an End4p-dependent manner (Lau et al. 2000), the proteolytic pathway involved in the degradation of Pho84p still remains to be fully elucidated. Whether the Pho84p, like many other plasma membrane proteins in yeast, is phosphorylated and/or ubiquitinated prior to endocytosis and vacuolar processing (reviewed by Hicke 1999; Rotin et al. 2000) is so far unknown. The targets for ubiquitination in substrate proteins are lysine residues with which isopeptide bonds are formed between the C-terminal glycine of the ubiquitin peptide and the ε amino group of the lysine. Of the 25 lysine residues contained in the primary structure of Pho84p, 16 are cytoplasmically located, according to the existing secondary model (Persson et al. 1999). It remains to be seen whether these residues are functionally important for ubiquitination/endocytosis. The amino acid sequence SINNDAKSS, located within the C-terminal tail of the membrane-bound Ste2 α-factor pheromone receptor, constitutes the target site for signaling endocytosis, whereby the lysine residue contained within this sequence is ubiquitinated, an event preceded by phosphorylation of the serine residues (Hicke and Riezman 1996; Hicke et al. 1998). However, substitution and deletion mutagenesis of a similar, partly conserved C-terminal sequence, nkNNDieSS (residues not fully conserved are indicated by lowercase letters), of both GFP- and MYC-epitope-tagged Pho84 proteins, followed by in vivo analysis of the localization and function of the transporter, do not reveal any alteration in the degradation pattern, as compared with the wild-type protein (Lagerstedt et al. 2002). Degradation of the Pho84 protein thus seems to be regulated by factors other than this sequence and the last 18 amino acid residues of the C-terminal tail. A detailed analysis of the trafficking of the Pho89p is still lacking. In vivo studies of both GFP- and MYC-tagged Pho89p are presently hampered by the fact that this protein is synthesized at low levels in low-Pi-grown cells, as compared with the synthesis of Pho84p (Pattison-Granberg, Lagerstedt and Persson, unpublished data).
Although Pho84p has been shown to be solely responsible for H+-coupled Pi uptake in a reconstituted model system (Berhe et al. 1995; Fristedt et al. 1999a, 1999b), several other proteins are proposed to be involved in Pi transport and/or its regulation (Oshima 1997). These auxillary proteins, i.e. Pho87p (Bun-ya et al. 1996), Pho88p (Yompakdee et al. 1996b) and Gtr1p (Bun-ya et al. 1992), are proposed to be associated with the Pi transport system, possibly serving as receptors for Pi signals from the environment or altering the intrinsic stability of Pho84p or otherwise acting as regulatory units in the adaptation to changing metabolic conditions in the cell.
PHO87 encodes a protein of 923 amino acid residues with a highly charged N-terminal half connected to a C-terminal half consisting of 12 putative membrane-spanning segments, as in Pho84p. The pho87 deletion mutant does not show any significant defects in growth or Pi uptake when compared with wild-type cells (Bun-ya et al. 1996). However, a double disruption of PHO87 and PHO86 shows a higher acid phosphatase synthesis in high-Pi medium than either of the single disruptants. The involvement of Pho86p in a correctly localized function of Pho87p can so far not be excluded. It is unclear as to the precise role Pho87p plays in Pi acquisition, although it is suggested to play a role in relaying a signal on the availability of external Pi (Giots et al. 2003). Pho87p shows some sequence similarity to Pho81p and the Vtc proteins, indicating a possible role in the transcriptional regulation of the PHO genes.
Pho88p is a 21-kDa protein consisting of 188 amino acid residues. Pho88p is a membrane protein, with a hydropathic plot similar to Pho86p, containing two hydrophobic segments (Yompakdee et al. 1996a). The upstream region of the PHO88 gene does not contain any recognizable Pho4p-binding sites and the gene is transcribed in both high- and low-Pi media, in contrast to other PHO genes. Disruption of PHO88 shows that it is not essential, but is important for cell growth. The pho88 deletion mutant exhibits slower growth than wild-type cells and a diminished Pi uptake capacity when grown in low-Pi medium. It is suggested that Pho88p might be involved in modulating Pho81p activity in response to Pi signals (Yompakdee et al. 1996b).
GTR1 (GTP-binding protein resemblance) encodes a protein consisting of 310 amino acid residues containing, in its N-terminal region, the characteristic tripartite consensus element for binding GTP, conserved in GTP-binding proteins, except for a histidine replacing the generally conserved asparagine residue in element III (Bun-ya et al. 1992; Nakashima et al. 1999). A conformational change of the protein caused by transition from the GDP-bound to the GTP-bound form results in a change in the regulatory function of the protein, allowing it to interact with other proteins. The asparagine-to-histidine substitution may confer a somewhat reduced GTP-binding activity on Gtr1p. The calculated molecular size of the predicted Gtr1 protein is 35.8 kDa. The fact that disruption of gtr1 leads to a lowered Pi uptake activity in cells grown in low-Pi media suggests that the Gtr1 protein, together with Pho84p, is involved in the mechanism of Pi uptake or its regulation. Gtr1p is also important for cell growth, since cells with a disrupted gtr1 reveal a reduced growth rate at 30 °C and no growth at all at 15 °C (Hirose et al. 1998). Expression of GTR1 is very low, regardless of the external Pi concentration, suggesting the gene is subjected to relaxed control or possibly no control via Pi signaling, in contrast to Pho84p. Since G proteins are recognized to be obligatory in opening ionic channels or in conferring co-modulating effects, together with other stimuli, such as the membrane potential needed for channel opening (Brown and Birnbaumer 1990), it cannot be excluded that such a function is true also for Gtr1p. Alternatively, Gtr1p may be directly active in the intracellular localization of Pho84p, a role in agreement with the function of other GTP-binding proteins involved in the translocation of proteins (Balch 1990). Since the gtr1 deletion strain grows slowly on both high- and low-Pi media, the Gtr1p function might apply to both the high- and low-affinity Pi-uptake systems.
Since cells with the pho84 pho89 double mutation are still viable, even in low-Pi media, the low-affinity transporter could be essential for cellular Pi acquisition. Recently, two new PHO genes, PHO90 and PHO91, were identified as possibly constituting the expression of the low-affinity transporter (Wykoff and O′Shea 2001). Pho90p and Pho91p both have about 30% sequence identity with Pho87p; and they are all suggested to be involved in Pi transport (Wykoff and O′Shea 2001). However, experiments show they have a lower affinity for Pi than Pho84p. Deletion of all of the proposed Pi transporters (PHO84, PHO87, PHO89, PHO90, PHO91) is lethal to cells. Interestingly, over-expression of Git1p, an organic Pi transporter involved in the transport of glycerophosphoinositol (Patton-Vogt and Henry 1998), restores viability to these cells. Approximately 25 predicted proteins in the yeast genome share at least 20% identity with Pho84p; and it is possible that some of these may also act as transporters for Pi or Pi-containing compounds (Costanzo et al. 2001).
In conclusion, the genes associated with Pi acquisition and mutants thereof open the way to detailed studies of their properties, regulation and physiological role. Yeast is indeed an invaluable tool and its use as a model permits valuable information to be extrapolated into other species. An understanding of the cellular strategies for sensing and uptake of this essential nutrient should provide new insight into the mechanistic regulation of substrate transport and utilization, not only in yeast, but also in other organisms. It is clear that Pi metabolism encroaches on many different areas of extremely active research.
References
André B (1995) An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11:1575–1611
Andreeva NA, Kulakovskaya TV, Kulaev IS (2001) Two exopolyphosphatases of the cytosol of the yeast S. cerevisiae: comparative characteristics. Biochemistry (Mosc) 66:147–153
Andrews B, Measday V (1998) The cyclin family of budding yeast: abundant use of a good idea. Trends Genet 14:66–72
Arima K, Oshima T, Kubota I, Nakamura N, Mizunaga T, Toh-e A (1983) The nucleotide sequence of the yeast PHO5 gene: a putative precursor of repressible acid phosphatase contains a signal peptide. Nucleic Acids Res 11:1657–1672
Arndt KT, Styles C, Fink GR (1987) Multiple global regulators control HIS4 transcription in yeast. Science 237:874–880
Balch WE (1990) Small GTP-binding proteins in vesicular transport. Trends Biochem Sci 15:473–477
Barbaric S, Münsterkötter M, Svaren J, Horz W (1996) The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res 24:4479–4486
Barbaric S, Münsterkötter M, Goding C, Horz W (1998) Cooperative Pho2–Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol Cell Biol 18:2629–2639
Beauvoit B, Rigoulet M, Guerin B, Canioni P (1989) Polyphosphates as a source of high energy phosphates in yeast mitochondria: a 31P-NMR study. FEBS Lett 252:17–21
Beauvoit B, Rigoulet M, Raffard G, Canioni P, Guerin B (1991) Differential sensitivity of the cellular compartments of Saccharomyces cerevisiae to protonophoric uncoupler under fermentative and respiratory energy supply. Biochemistry 30:11212–11220
Berhe A, Fristedt U, Persson BL (1995) Expression and purification of the high-affinity phosphate transporter of Saccharomyces cerevisiae. Eur J Biochem 227:566–572
Berhe A, Zvyagilskaya R, Lagerstedt JO, Pratt JR, Persson BL (2001) Properties of the cysteine-less Pho84 phosphate transporter of Saccharomyces cerevisiae. Biochem Biophys Res Commun 287:837–842
Bhoite LT, Allen JM, Garcia E, Thomas LR, Gregory ID, Voth WP, Whelihan K, Rolfes RJ, Stillman DJ (2002) Mutations in the Pho2 (Bas2) transcription factor that differentially affect activation with its partner proteins Bas1, Pho4, and Swi5. J Biol Chem 277:37612–37618
Bisson LF, Coons DM, Kruckeberg AL, Lewis DA (1993) Yeast sugar transporters. Crit Rev Biochem Mol Biol 28:259–308
Blasco F, Ducet G, Azoulay E (1976) Demonstration of 2 phosphate transport systems in Candida tropicalis. Biochimie 58:351–357
Boles E, Hollenberg CP (1997) The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev 21:85–111
Booth JW, Guidotti G (1997) Phosphate transport in yeast vacuoles. J Biol Chem 272:20408–20413
Bork P (1993) Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins 17:363–374
Borst-Pauwels GWFH (1981) Ion transport in yeast. Biochim Biophys Acta 650:88–127
Borst-Pauwels GWFH (1993) Mutual interaction of ion uptake and membrane potential. Biochim Biophys Acta 1145:15–24
Borst-Pauwels GW, Peters PH (1977) Effect of the medium pH and the cell pH upon the kinetical parameters of phosphate uptake by yeast. Biochim Biophys Acta 466:488–495
Borst-Pauwels GWFH, Peters PHJ (1987) Phosphate uptake in Saccharomyces cerevisiae. In: Torriani-Gorini A, Rothman FG, Silver S, Wright A, Yagil E (eds) Phosphate metabolism and cellular regulation of microorganisms. ASM Press, Washington, D.C., pp 205–209
Bostian KA, Lemire JM, Cannon LE, Halvorson HO (1980) In vitro synthesis of repressible yeast acid phosphatase: identification of multiple mRNAs and products. Proc Natl Acad Sci USA 77:4504–4508
Bostian KA, Lemire JM, Halvorson HO (1983) Physiological control of repressible acid phosphatase gene transcripts in Saccharomyces cerevisiae. Mol Cell Biol 3:839–853
Böttger P, Pedersen L (2002) Two highly conserved glutamate residues critical for type III sodium-dependent phosphate transport revealed by uncoupling transport function from retroviral receptor function. J Biol Chem 277:42741–42747
Bourne RM (1990) A 31P-NMR study of phosphate transport and compartmentation in Candida utilis. Biochim Biophys Acta 1055:1–9
Bourne RM (1991) Net phosphate transport in phosphate-starved Candida utilis: relationships with pH and K+. Biochim Biophys Acta 1067:81–88
Braus G, Mösch HU, Vogel K, Hinnen A, Hütter R (1989) Interpathway regulation of the TRP4 gene of yeast. EMBO J 8:939–945
Brazas RM, Stillman DJ (1993) The Swi5 zinc-finger and Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter. Proc Natl Acad Sci USA 90:11237–11241
Brazas RM, Bhoite LT, Murphy MD, Yu YX, Chen YY, Neklason DW, Stillman DJ (1995) Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J Biol Chem 270:29151–29161
Brown AM, Birnbaumer L (1990) Ionic channels and their regulation by G protein subunits. Annu Rev Physiol 52:197–213
Bun-ya M, Nishimura M, Harashima S, Oshima Y (1991) The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11:3229–3238
Bun-ya M, Harashima S, Oshima Y (1992) Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae. Mol Cell Biol 12:2958–2966
Bun-ya M, Shikata K, Nakade S, Yompakdee C, Harashima S, Oshima Y (1996) Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr Genet 29:344–351
Castro CD, Meehan AJ, Koretsky AP, Domach MM (1995) In situ 31P nuclear magnetic resonance for observation of polyphosphate and catabolite responses of chemostat-cultivated Saccharomyces cerevisiae after alkalinization. Appl Environ Microbiol 61:4448–4453
Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, et al (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12:333–337
Cave JW, Kremer W, Wemmer DE (2000) Backbone dynamics of sequence specific recognition and binding by the yeast Pho4 bHLH domain probed by NMR. Protein Sci 9:2354–2365
Claros MG, Heijne G von (1994) TopPredII: an improved software for membrane protein structure prediction. Comput Appl Biosci 10:685–686
Cockburn M, Earnshaw P, Eddy AA (1975) The stoichiometry of the absorption of protons with phosphate and l-glutamate by yeasts of the genus Saccharomyces. Biochem J 146:705–712
Cohen A, Perzov N, Nelson H, Nelson N (1999) A novel family of yeast chaperons involved in the distribution of V-ATPase and other membrane proteins. J Biol Chem 274:26885–26893
Costanzo MC, Crawford ME, Hirschman JE, Kranz JE, Olsen P, Robertson LS, Skrzypek MS, Braun BR, Hopkins KL, Kondu P, Lengieza C, Lew-Smith JE, Tillberg M, Garrels JI (2001) YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res 29:75–79
Crooke E, Akiyama M, Rao NN, Kornberg A (1994) Genetically altered levels of inorganic polyphosphate in Escherichia coli. J Biol Chem 269:6290–6295
Daignan-Fornier B, Fink GR (1992) Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA 89:6746–6750
Dawes EA, Senior PJ (1973) The role and regulation of energy reserve polymers in microorganisms. Adv Microb Physiol 10:135–266
Devenish RJ, Prescott M, Roucou X, Nagley P (2000) Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim Biophys Acta 1458:428–442
Dunn T, Gable K, Beeler T (1994) Regulation of cellular Ca2+ by yeast vacuoles. J Biol Chem 269:7273–7278
Espinoza FH, Ogas J, Herskowitz I, Morgan DO (1994) Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science 266:1388–1391
Felter S, Stahl AJ (1973) Enzymes for metabolism of polyphosphates in yeast. 3. Purification and properties of polyphosphate-ADP-phosphotransferase. Biochimie 55:245–251
Ferré-D'Amaré AR, Prendergast GC, Ziff EB, Burley SK (1993) Recognition by Max of its cognate DANN trough a dimeric b/HLH/Z domain. Nature 363:38–45
Ferré-D'Amaré AR, Pognonec P, Roeder RG, Burley SK (1994) Structure and function of the b/HLH/Z domain of USF. EMBO J 13:180–189
Forsberg H, Ljungdahl PO (2001) Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr Genet 40:91–109
Frillingos S, Sahin-Toth M, Wu J, Kaback HR (1998) Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J 12:1281–1299
Fristedt U, Berhe A, Ensler K, Norling B, Persson BL (1996) Isolation and characterization of membrane vesicles of Saccharomyces cerevisiae harboring the high-affinity phosphate transporter. Arch Biochem Biophys 330:133–141
Fristedt U, Weinander R, Martinsson HS, Persson BL (1999a) Characterization of purified and unidirectionally reconstituted Pho84 phosphate permease of Saccharomyces cerevisiae. FEBS Lett 458:1–5
Fristedt U, Van Der Rest M, Poolman B, Konings WN, Persson BL (1999b) Studies of cytochrome c oxidase-driven H+_coupled phosphate transport catalyzed by the Saccharomyces cerevisiae Pho84 permease in coreconstituted vesicles. Biochemistry 38:16010–16015
Garciadeblas B, Rubino F, Quintero FJ, Banuelos MA, Haro R, Rodrigues-Navarro A (1993) Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol Gen Genet 236:363–368
Giots F, Donaton MCV, Thevelein JM (2003) Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 47:1163–1181
Gouffeau A, Aert R, Agostini-Carbone ML, et al (1997) The yeast genome directory. Nature 387 [Suppl]:1–105
Graschopf A, Stadler JA, Hoellerer MK, Eder S, Sieghardt M, Kohlwein SD, Schweyen RJ (2001). The yeast plasma membrane protein Alr1 controls Mg2+ homeostasis and is subject to Mg2+-dependent control of its synthesis and degradation. J Biol Chem 276:16216–16222
Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG (1998) Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science 281:533–538
Grünewald M, Menaker D, Kanner BI (2002) Cysteine-scanning mutagenesis reveals a conformationally sensitive reentrant pore-loop in the glutamate transporter GLT-1. J Biol Chem 277:26074–26080
Haguenauer-Tsapis R, Nagy M, Ryter A (1986) A deletion that includes the segment coding for the signal peptidase cleavage site delays release of Saccharomyces cerevisiae acid phosphatase from the endoplasmic reticulum. Mol Cell Biol 6:723–729
Harold FM (1966) Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol Rev 30:772–794
Henderson PJF (1993) The 12-transmembrane helix transporters. Curr Opin Cell Biol 5:708–721
Herrmann JM, Malkus P, Schekman R (1999) Out of the ER–outfitters, escorts and guides. Trends Cell Biol 9:5–7
Hicke L (1999) Gettin′ down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol 9:107–112
Hicke L, Riezman H (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277–287
Hicke L, Zanolari B, Riezman HJ (1998) Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol 141:349–358
Hirimburegama K, Durnez P, Keleman J, Oris E, Vergauwen R, Mergelsberg H, Thevelein JM (1992) Nutrient-induced activation of trehalase in nutrient-starved cells of the yeast Saccharomyces cerevisiae: cAMP is not involved as second messenger. J Gen Microbiol 138:2035–2043
Hirose E, Nakashima N, Sekiguchi T, Nishimoto T (1998) RagA is a functional homologue of the S. cerevisiae Gtr1p in the Ran/Gsp1-GTPase pathway. J Cell Sci 111:11–21
Hohmann S, Meacock PA (1998) Thiamin metabolism and thiamin diphosphate-dependent enzymes in the yeast Saccharomyces cerevisiae: genetic regulation. Biochim Biophys Acta 1385:201–219
Hope IA, Struhl K (1985) GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell 43:177–188
Horak J, Wolf DH (1997) Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol 179:1541–1549
Huang D, Moffat J, Wilson WA, Moore L, Cheng C, Roach PJ, Andrews B (1998) Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol Cell Biol 18:3289–3299
Huang S, Jeffery DA, Anthony MD, O′Shea EK (2001) Functional analysis of the cyclin-dependent kinase inhibitor Pho81 identifies a novel inhibitory domain. Mol Cell Biol 21:6695–6705
Jacobson L, Halmann M, Yariv J (1982) The molecular composition of the volutin granule of yeast. Biochem J 201:473–479
Jeffery DA, Springer M, King DS, O′Shea EK (2001) Multi-site phosphorylation of Pho4 by the cyclin-CDK Pho80–Pho85 is semi-processive with site preference. J Mol Biol 306:997–1010
Kaffman A, Herskowitz I, Tjian R, O′Shea EK (1994) Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80–PHO85. Science 263:1153–1156
Kaffman A, Rank NM, O′Neill EM, Huang LS, O′Shea EK (1998a) The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396:482–486
Kaffman A, Rank NM, O′Shea EK (1998b) Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev 12:2673–2683
Kaneko Y, Toh-e A, Oshima Y (1982) Identification of the genetic locus for the structural gene and a new regulatory gene for the synthesis of repressible alkaline phosphatase in Saccharomyces cerevisiae. Mol Cell Biol 2:127–137
Kaneko Y, Tamai Y, Toh-e A, Oshima Y (1985) Transcriptional and post-translational control of PHO84 expression by PHO regulatory genes in Saccharomyces cerevisiae. Mol Cell Biol 5:248–252
Kaneko Y, Hayashi N, Toh-e A, Banno I, Oshima Y (1987) Structural characteristics of the PHO8 gene encoding repressible alkaline phosphatase in Saccharomyces cerevisiae. Gene 58:137–148
Kaneko Y, Toh-e A, Banno I, Oshima Y (1989) Molecular characterization of a specific p-nitrophenylphosphatase gene, PHO13, and its mapping by chromosome fragmentation in Saccharomyces cerevisiae. Mol Gen Genet 220:133–139
Karpichev IV, Cornivelli L, Small GM (2002) Multiple regulatory roles of a novel Saccharomyces cerevisiae protein, encoded by YOL002c, in lipid and phosphate metabolism. J Biol Chem 277:19609–19617
Kohler K, Forster IC, Stange G, Biber J, Murer H (2002) Identification of functionally important sites in the first intracellular loop of the NaPi-IIa cotransporter. Am J Physiol Ren Physiol 282:687–696
Komeili A, O′Shea EK (1999) Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284:977–980
Komeili A, O′Shea EK (2000) Nuclear transport and transcription. Curr Opin Cell Biol 12:355–360
Kornberg A (1995) Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J Bacteriol 177:491–496
Kornberg A, Rao NN, Ault-Riche D (1999) Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 68:89–125
Krampe S, Stamm O, Hollenberg CP, Boles E (1998) Catabolite inactivation of the high-affinity hexose transporters Hxt6 and Hxt7 of Saccharomyces cerevisiae occurs in the vacuole after internalization by endocytosis. FEBS Lett 441:343–347
Krebs JE, Fry CJ, Samuels ML, Peterson CL (2000) Global role for chomatin remodeling enzymes in mitotic gene expression. Cell 102:587–598
Kruckeberg AL (1996) The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol 166:283–292
Kruckeberg AL, Ye L, Berden JA, Dam K van (1999) Functional expression, quantification and cellular localization of the Hxt2 hexose transporter of Saccharomyces cerevisiae tagged with the green fluorescent protein. Biochem J 339:299–307
Kulaev IS (1979) The biochemistry of inorganic polyphosphates. Wiley, New York
Kulaev I, Kulakovskaya T (2000) Polyphosphate and phosphate pump. Annu Rev Microbiol 54:709–734
Kulaev IS, Vagabov VM (1983) Polyphosphate metabolism in microorganisms. Adv Microb Physiol 24:83–171
Kulaev I, Vagabov V, Kulakovskaya T (1999) New aspects of polyphosphate metabolism and function. J Biosci Bioeng 88:111–129
Kumble KD, Kornberg A (1996) Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J Biol Chem 271:27146–27151
Lagerstedt JO, Kruckeberg AL, Berden JA, Persson BL (2000) The yeast phosphate transporting system: regulated trafficking of the Pho84 phosphate transporter. In: Hohmann S, Nielsen S (eds) Molecular biology and physiology of water and solute transport: fundamental research and applied aspects. Kluwer/Plenum, New York, pp 405–414
Lagerstedt JO, Zvyagilskaya R, Pratt JR, Pattison-Granberg J, Kruckeberg AL, Berden JA, Persson BL (2002) Mutagenic and functional analysis of the C-terminus of Saccharomyces cerevisiae Pho84 phosphate transporter. FEBS Lett 526:31–37
Lamb TM, Xu W, Diamond A, Mitchell AP (2001) Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276:1850–1856
Lau WW, Schneider KR, O′Shea EK (1998) A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics 150:1349–1359
Lau WT, Howson RW, Malkus P, Schekman R, O′Shea EK (2000) Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter Pho84p. Proc Natl Acad Sci USA 97:1107–1112
Lemire JM, Willcocks T, Halvorson HO, Bostian KA (1985) Regulation of repressible acid phosphatase gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 5:2131–2141
Lenburg ME, O′Shea EK (1996) Signaling phosphate starvation. Trends Biochem Sci 21:383–387
Lichko LP, Kulakovskaia TV, Kulaev IS (1996) Characteristics of polyphosphatase activity of isolated mitochondria from Saccharomyces cerevisiae. Biokhimiya 61:1664–1671
Lichko L, Kulakovskaya T, Kulaev I (1998) Membrane-bound and soluble polyphosphatases of mitochondria of Saccharomyces cerevisiae: identification and comparative characterization. Biochim Biophys Acta 1372:153–162
Lichko LP, Kulakovskaya TV, Kulaev IS (2000) Purification and characterization of a soluble polyphosphatase from mitochondria of Saccharomyces cerevisiae. Biochemistry (Mosc) 65:355–360
Liu C, Yang Z, Yang J, Xia Z, Ao S (2000) Regulation of the yeast transcriptional factor PHO2 activity by phosphorylation. J Biol Chem 275:31972–31978
Martinez P, Persson BL (1998) Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol Gen Genet 258:628–638
Martinez P, Zvyagilskaya R, Allard P, Persson BL (1998) Physiological regulation of the derepressible phosphate transport in Saccharomyces cerevisiae. J Bacteriol 180:2253–2256
Measday V, Moore L, Ogas J, Tyers M, Andrews B (1994) The PCL2 (ORFD)–PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science 266:1391–1395
Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman AM, Andrews B (1997) A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol 17:1212–1223
Mendoza I, Rubio F, Rodrigues-Navarro A, Pardo JM (1994) The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem 269:8792–8796
Morgan DO (1995) Principles of CDK regulation. Nature 374:131–134
Müller O, Bayer MJ, Peters C, Andersen JS, Mann M, Mayer A (2002) The Vtc proteins in vacuole fusion: coupling NSF activity to V(0) trans-complex formation. EMBO J 21:259–269
Münsterkötter M, Barbaric S, Horz W (2000) Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J Biol Chem 275:22678–22685
Nakashima N, Noguchi E, Nishimoto T (1999) Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 152:853–867
Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21:4347–4368
Nelissen B, Mordant P, Jonniaux J-L, De Wachter R, Gouffeau A (1995) Phylogenetic classification of the major superfamily of membrane transport facilitator, as deduced from yeast genome sequencing. FEBS Lett 377:232–236
Nelson H, Nelson N (1990) Disruption of genes encoding subunits of the yeast vacuolar H(+)-ATPase causes conditional lethality. Proc Natl Acad Sci USA 87:3503–3507
Nigavekar SS, Tan YS, Cannon JF (2002) Glc8 is a glucose-repressible activator of Glc7 protein phosphatase-1. Arch Biochem Biophys 404:71–79
Nishimura H, Kawasaki Y, Kaneko Y, Nosaka K, Iwashima A (1992a) A positive regulatory gene, THI3, is required for thiamine metabolism in Saccharomyces cerevisiae. J Bacteriol 174:4701–4706
Nishimura H, Kawasaki Y, Kaneko Y, Nosaka K, Iwashima A (1992b) Cloning and characteristics of a positive regulatory gene, THI2 (PHO6), of thiamin biosynthesis in Saccharomyces cerevisiae. FEBS Lett 297:155–158
Norbeck J, Pahlman AK, Akhtar N, Blomberg A, Adler L (1996) Purification and characterization of two isoenzymes of dl-glycerol-3-phosphatase from Saccharomyces cerevisiae. Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J Biol Chem 271:13875–13881
Nosaka K (1990) High affinity of acid phosphatase encoded by PHO3 gene in Saccharomyces cerevisiae for thiamin phosphates. Biochim Biophys Acta 1037:147–154
Ogawa N, Oshima Y (1990) Functional domains of a positive regulatory protein, pho4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol Cell Biol 10:2224–2236
Ogawa N, Noguchi K, Yamashita Y, Yasuhara T, Hayashi N, Youshida K, Oshima Y (1993) Promoter analysis of the PHO81 gene encoding a 134 kDa protein bearing ankyrin repeats in the phosphatase regulon of Saccharomyces cerevisiae. Mol Gen Genet 238:444–454
Ogawa N, Noguchi K, Sawai H, Yamashita Y, Yompakdee C, Oshima Y (1995) Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol Cell Biol 15:997–1004
Ogawa N, DeRisi J, Brown PO (2000) New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell 11:4309–4321
O′Neill EM, Kaffman A, Jolly ER, O′Shea EK (1996) Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin-CDK complex. Science 271:209–212
Oshima Y (1997) The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst 72:323–334
Oshima Y, Ogawa N, Harashima S (1996) Regulation of phosphatase synthesis in Saccharomyces cerevisiae. Genetics 149:1707–1715
Özcan S, Dover J, Rosenwald AG, Wölfl S, Johnston M (1996) Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA 93:12428–12432
Pao SS, Paulsen IT, Saier MH Jr (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34
Pattison-Granberg J, Persson BL (2000) Regulation of cation-coupled high-affinity phosphate uptake in the yeast Saccharomyces cerevisiae. J Bacteriol 182:5017–5019
Patton-Vogt JL, Henry SA (1998) GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149:1707–1715
Persson BL, Petersson J, Fristedt U, Weinander R, Berhe A, Pattison J (1999) Phosphate permeases of Saccharomyses cerevisiae: structure, function and regulation. Biochim Biophys Acta 1422:255–272
Petersson J, Pattison J, Kruckeberg AL, Berden JA, Persson BL (1999) Intracellular localization of an active green fluorescent protein-tagged Pho84 phosphate permease in Saccharomyces cerevisiae. FEBS Lett 462:37–42
Pick U, Bental M, Chitlaru E, Weiss M (1990) Polyphosphate hydrolysis—a protective mechanism against alkaline stress? FEBS Lett 274:15–18
Plankert U, Purwin C, Holzer H (1991) Yeast fructose-2,6-bisphosphate 6-phosphatase is encoded by PHO8, the gene for nonspecific repressible alkaline phosphatase. Eur J Biochem 196:191–196
Rao NN, Liu S, Kornberg A (1998) Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol 180:2186–2193
Raths S, Rohrer J, Crausaz F, Riezman H (1993) end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol 120:55–65
Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21:267–271
Reusch RN, Sadoff HL (1988) Putative structure and function of a poly-beta-hydroxybutyrate/calcium phosphate channel in bacterial plasma membranes. Proc Natl Acad Sci USA 85:4176–4180
Riballo E, Herweijer M, Wolf DH, Lagunas R (1995) Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J Bacteriol 177:5622–5627
Roomans GM, Blasco F, Borst-Pauwels GW (1977) Cotransport of phosphate and sodium by yeast. Biochim Biophys Acta 467:65–71
Rotin D, Staub O, Haguenauer-Tsapis R (2000) Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin–protein ligases. J Membr Biol 176:1–17
Rubin RA, Levy SB, Heinrikson RL, Kézdy FJ (1990) Gene duplication in the evolution of the two complementing domains of gram-negative bacterial tetracycline efflux protein. Gene 87:7–13
Rudolph H, Hinnen A (1987) The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proc Natl Acad Sci USA 84:1340–1344
Saier MH Jr (2000) A functional–phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev 64:354–411
Salaün C, Rodrigues P, Heard JM (2001) Transmembrane topology of PiT-2 phosphate transporter-retrovirus receptor. J Virol 75:5584–5592
Sato M, Mueckler M (1999) A conserved amino acid motif (R-X-G-R-R) in the Glut1 glucose transporter is an important determinant of membrane topology. J Biol Chem 274:24721–24725
Schneider KR, Smith RL, O′Shea EK (1994) Phosphate-regulated inactivation of the kinase PHO80–PHO85 by the CDK inhibitor PHO81. Science 266:122–126
Schuddemat J, Boo R de, Leeuwen CC van, Broek PJ van den, Steveninck J van (1989) Polyphosphate synthesis in yeast. Biochim Biophys Acta 1010:191–198
Schurr A, Yagil E (1971) Regulation and characterization of acid and alkaline phosphatase in yeast. J Gen Microbiol 65:291–303
Sedgwick SG, Smerdon SJ (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci 24:311–316
Seedorf M, Silver PA (1997) Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA 94:8590–8595
Sengstag C, Hinnen A (1987) The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual amino acid composition. Nucleic Acids Res 15:233–246
Serrano R (1996) Salt tolerance in plants and microorganisms: toxicity targets and defence responses. Int Rev Cytol 165:1–52
Serrano R, Ruiz A, Bernal D, Chambers JR, Arino J (2002) The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol Microbiol 46:1319–1333
Sethuraman A, Rao NN, Kornberg A (2001) The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98:8542–8547
Shao D, Creasy CL, Bergman LW (1998) A cysteine residue in helix II of the bHLH domain is essential for homodimerization of the yeast transcription factor Pho4p. Nucleic Acids Res 26:710–714
Shemer R, Meimoun A, Holtzman T, Kornitzer D (2002) Regulation of the transcription factor gcn4 by pho85 cyclin pcl5. Mol Cell Biol 22:5395–5404
Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T (1997) Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J 16:4689–4697
Steensma HY de, Jonge P de, Kaptein A, Kaback DB (1989) Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: localization of a repeated sequence containing an acid phosphatase gene near a telomere of chromosome I and chromosome VIII. Curr Genet 16:131–137
Steger DJ, Haswell ES, Miller AL, Wente SR, O′Shea EK (2003) Regulation of chromatin remodeling by inositol polyphosphatases. Science 299:114–116
Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD (1998) INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem 273:11852–11861
Sudarsanam P, Iyer VR, Brown PO, Winston F (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97:3364–3369
Tamai Y, Toh-e A, Oshima Y (1985) Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. J Bacteriol 164:964–968
Tan YS, Morcos PA, Cannon JF (2003) Pho85 phosphorylates the Glc7 protein phosphatase regulator Glc8 in vivo. J Biol Chem 278:278147–278153
Tijssen JP, Van Steveninck J (1984) Detection of a yeast polyphosphate fraction localized outside the plasma membrane by the method of phosphorus-31 nuclear magnetic resonance. Biochem Biophys Res Commun 119:447–451
Toh-e A, Kakimoto S (1975) Genes coding for the structure of the acid phosphatases in Saccharomyces cerevisiae. Mol Gen Genet 143:65–70
Toh-e A, Shimauchi T (1986) Cloning and sequencing of the PHO80 gene and CEN15 of Saccharomyces cerevisiae. Yeast 2:129–139
Toh-e A, Ueda Y, Kakimoto S-I, Oshima Y (1973) Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol 113:727–738
Toh-e A, Tanaka K, Uesono Y, Wickner RB (1988) PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol Gen Genet 214:162–164
Trilisenko LV, Vagabov VM, Kulaev IS (2002) The content and chain length of polyphosphates from vacuoles of Saccharomyces cerevisiae VKM Y-1173. Biochemistry (Mosc) 67:592–596
Tsutsumi K, Munekata M, Shiba T (2000) Involvement of inorganic polyphosphate in expression of SOS genes. Biochim Biophys Acta 1493:73–81
Uesono Y, Tanaka K, Toh-e A (1987) Negative regulators of the PHO system in Saccharomyces cerevisiae: isolation and structural characterization of PHO85. Nucleic Acids Res 15:10299–10309
Uesono Y, Tokai M, Tanaka K, Tohe A (1992) Negative regulators of the PHO system of Saccharomyces cerevisiae: characterization of PHO80 and PHO85. Mol Gen Genet 231:426–432
Urech K, Durr M, Boller T, Wiemken A, Schwencke J (1978) Localization of polyphosphate in vacuoles of Saccharomyces cerevisiae. Arch Microbiol 116:275–278
Vagabov VM, Trilisenko LV, Kulaev IS (2000) Dependence of inorganic polyphosphate chain length on the orthophosphate content in the culture medium of the yeast Saccharomyces cerevisiae. Biochemistry (Mosc) 65:349–54
Van der Rest ME, Kamminga AH, Nakano A, Anraku Y, Poolman B, Konings WN (1995) The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev 59:304–322
Van Dien SJ, Keasling JD (1999) Effect of polyphosphate metabolism on the Escherichia coli phosphate-starvation response. Biotechnol Prog 15:587–593
Venter U, Hörz W (1989) The acid phosphatase genes PHO10 and PHO11 in S. cerevisiae are located at the telomeres of chromosomes VIII and I. Nucleic Acids Res 17:1353–1369
Venter U, Svaren J, Schmitz J, Schmid A, Hörz W (1994) A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J 13:4848–4855
Walker GM (1998). Yeast physiology and biotechnology. Wiley, New York, pp 81–82
Walker JE, Saraste M, Runswick MH, Gay NJ (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951
Wang Z, Wilson WA, Fujino MA, Roach PJ (2001) The yeast cyclins Pc16p and Pc17p are involved in the control of glycogen storage by the cyclin-dependent protein kinase Pho85p. FEBS Lett 506:277–280
Wood HG, Clark JE (1988) Biological aspects of inorganic polyphosphates. Annu Rev Biochem 57:235–260
Wu WH, Hampsey M (1999) An activation-specific role for transcription factor TFIIB in vivo. Proc Natl Acad Sci USA 96:2764–2769
Wurst H, Kornberg A (1994) A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purification and characterization. J Biol Chem 269:10996–11001
Wurst H, Shiba T, Kornberg A (1995) The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J Bacteriol 177:898–906
Wykoff DD, O′Shea EK (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159:1491–1499
Yale J, Bohnert HJ (2001) Transcript expression in Saccharomyces cerevisiae at high salinity. J Biol Chem 276:15996–16007
Yompakdee C, Bun-ya M, Shikata K, Ogawa N, Harashima S, Oshima Y (1996a) A putative new membrane protein, Pho86p, in the inorganic phosphate uptake system of Saccharomyces cerevisiae. Gene 171:41–47
Yompakdee C, Ogawa N, Harashima S, Oshima Y (1996b) A putative membrane protein, Pho88p, involved in inorganic phosphate transport in Saccharomyces cerevisiae. Mol Gen Genet 251:580–590
Yoshida K, Kuromitsu Z, Ogawa N, Oshima Y (1989) Mode of expression of the positive regulatory genes PHO2 and PHO4 of the phosphatase regulon in Saccharomyces cerevisiae. Mol Gen Genet 217:31–39
Zhu H, Riggs AF (1992) Yeast flavohemoglobin is an ancient protein related to globins and a reductase family. Proc Natl Acad Sci USA 89:5015–5019
Acknowledgements
Work in the authors' laboratories was supported by research grants from the Human Frontier Science Organization and the Swedish Royal Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann
Rights and permissions
About this article
Cite this article
Persson, B.L., Lagerstedt, J.O., Pratt, J.R. et al. Regulation of phosphate acquisition in Saccharomyces cerevisiae . Curr Genet 43, 225–244 (2003). https://doi.org/10.1007/s00294-003-0400-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-003-0400-9