Abstract
The main objective of this investigation was to characterize a collection of actinomycetes strains isolated from unexplored polluted ecosystems and to evaluate their antimicrobial potential in order to discover interesting bioactive compounds. Based on morphological and culture characters, 32 different strains were isolated: 20 strains from compost heap, seven strains from manure, and five strains from waste water. As expected, the genus Streptomyces was the most prevalent followed by the genus Micromonospora. Analysis of the antimicrobial activities of the isolated strains showed that those from compost heap were more efficient against the tested microorganisms (Candida albicans, Methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherichia coli). Several bioactive compounds were identified by liquid chromatography (LC) combined with mass spectrometry (MS) and then analyzed by both MEDINA’s database, which contains the most common secondary metabolites, and Dictionary of Natural Products Chapman & Hall. Many interesting well-known and unknown biomolecules were identified. Quinomycin A and Daidzein were the most fascinating compounds isolated, respectively, by Streptomyces sp. WW2 and Streptomyces sp. WW4. The most active strain was identified based on 16S rDNA’s sequences and it seems to be a new strain. The crude extract of the strain CH12 was analyzed and the UV absorption spectra and mass spectra (MS) of the main active compound were reported. It’s an interesting compound (possible purpuromycin) with the molecular formula C26H18O13.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Actinomycetes are filamentous Gram-positive bacteria having a high coefficient of Chargaff (60–70%) [1]. In general, they are strict aerobic bacteria but some are facultative or obligatory anaerobic, heterotrophic, and/or chemosynthetic, but mostly chemo-heterotrophic able to use a wide variety of nutritional sources, including various complex polysaccharides [2]. Actinomycetes are able to produce several bioactive secondary metabolites including antibacterial, antifungal, and antiparasitic agents, and so on. [3]. Actinobacteria are ubiquitous colonizing different ecological niches such as soil, aquatic environments, and even extreme ecosystems with high temperature, pressure, salt content, and/or hostile pH [4].
The existence and role of actinomycetes in the polluted sites become increasingly studied.
The biological treatment by composting is a big business based on the potential of some microorganisms. It involves converting organic waste material into a stable and valuable product which is called compost [5]. Actinomycetes are part of these microorganisms widely involved in the process of degradation of organic matter [6].
In addition, the wastewater treatment by soil infiltration and percolation has long been used as a simple and inexpensive way of wastewater management worldwide [7]. It has recently been demonstrated that the antagonistic activities of actinomycetes could contribute significantly to the mechanisms of elimination of microbes in wastewater treatment plants [8].
The majority of the strains isolated from such ecosystems are actinomycetes belonged to the genus Streptomyces (ex. Streptomyces violaceorubidus). These isolates showed a broad spectrum of activity against a variety of pathogenic microorganisms (yeasts (ex. Candida albicans), Gram-negative bacteria (ex. Salmonella spp.),and Gram-positive (ex. Staphylococcus aureus)). These results indicate the potential involvement of antagonistic actinomycetes in the elimination of pathogens [3].
With the aim of discovering promising bioactive compounds from actinomycetes strains, we examined such rarely unexplored ecosystem. Furthermore, these actives actinomycetes can be used for a variety of bioremediation purposes including biotransformation, biodegradation, and so on.
Material and Methods
Sample Collection
Three different polluted sites, by organics and inorganics waste, are studied. In fact, strains are isolated from the following:
-
Compost heap (CH): located near the sebkha of Monastir, 35°45'N 010°46'E.
-
Manure (MN): Sheep manure used for plant fertilization.
-
Waste water (WW): from the National Sanitation Utility (ONAS) in Monastir, Tunisia
A weight of one gram of each solid sample was air-dried and incubated at 28 °C for seven days in an atmosphere saturated with moisture [9].
Wastewater sampling was done according to the Rodier's method, using a ballasted plunger at which is fixed a sterile polyethylene flask [10]. Water collected is introduced into sterile glass flasks of 250 ml and transported in an icebox at the laboratory [11]. The analyses are carried out on fresh samples.
Actinomycetes were isolated by serial dilution method [12]. Stock solution was prepared by diluting 1 g of sediment in 9 ml of sterile saline water and shaken well by using vortex mixer. From the stock solution, 1 ml was used to prepare the final volume of ten serial dilutions (10–1–10–5) by serial dilution method. Finally, 0.1 ml of suspension from each dilution was used to spread in three replicates over the surface of Glucose-Yeast Extract-Agar (GYEA) medium [13]. The media supplemented with cycloheximide (50 µg/ml) to prevent fungal contamination.
The plates were and incubated at 28 °C for 7–14 days and observed periodically for the growth of actinomycetes [12]. Isolated colonies were picked and restreaked several times in order to obtain pure cultures. The obtained isolates were stored at – 80 °C in agar blocks submerged in 20% of glycerol, or liquid cultures supplemented with 10% glycerol.
A total number of 32 actinomycetes were isolated: 20 strains from compost, seven from manure, and five strains from waste water. These strains were examined for colony and micromorphological criteria. Pigmentation, colonial elevation, consistency, and opacity were studied. The morphological features of spores, sporangia, and aerial and substrate mycelium were observed and recorded. Actinomycetes were recognized on the basis of these morphological criteria [14].
Antimicrobial Screening
Several culture media were applied in order to test antimicrobial activities of the isolated actinomycetes [15]. The designed media and their composition are as follows (per one liter of distilled water): GCM (glucose 5 g, peptone of soya 20 g, MOPS 10.5 g, yeast extract 1.5 g, CaCl2 0.34 mM, pH 6.8). GHSA (MgSO4*7H2O 0.6 g, glucose 10 g, soya flour 10 g, yeast extract 0.5 g, MOPS (morpholinopropanesulfonic acid) 21 g, CaCl2 20 mM, trace element solution 2 ml, pH 6.8) (trace element solution as described in Kieser et al.) [16]. FPY-12 culture medium contains 20 g fructose, 10 g glucose, 10 g maltose, 5 g bacto peptone, 5 g amicase, 1 ml trace elements (For 100 ml: 100 mg Cl2Mn.4H2O, 100 mg Cl2Zn,100 mg Cl2Fe.4H2O, 50 mg NaI, pH 7.0). DEF-15: 40 g sucrose, 2 g ClNH4, 2 g SO4Na2, 1 g K2HPO4, 1 g Cl2Mg.6H2O, 1 g ClNa, 1 ml trace elements, pH 7.0 and add 2 g Co3Ca after autoclave. DEF-15S has the same composition as DEF-15 medium with some modifications which are the amount of sucrose (5 g) and the addition of soluble potato starch (20 g), which are used to promote slow-growing actinomycetes. KHC: 20 g dextrin from corn type I, 10 g beta cyclo dextrin, 20 g tomato paste, 10 g primary yeast, 5 mg CaCl2.6H2O, pH 7.2. FR23: 5 g glucose, 30 g soluble starch from potato, 20 g cane molasses, 20 g pharmamedia (cottonseed flour), pH 7.0. GPA medium contains 45 g glucose, 10 g peptonized milk, 1.5 g ardamina, pH 7. R358: 10 g starch from potato, 4 g bacto yeast extract, 2 g bacto peptone, 5 ml FeSO4.7, 5 ml KBr, pH 7.0. APM-9: 50 g glucose, 12 g soluble starch from potato, 30 g soy flour, 2 mg COCl2.6H2O, 7 g CO3Ca (added after adjusting pH), pH 7.0, used for the slow-growing actinomycetes strains.
Microfermentation and Organic Extraction
Isolated actinomycetes were pre-cultured in ATCC-2 liquid media (g l-1: 20 soluble starch from potato, 10.0 dextrose, 5.0 NZ amine type E, 3.0 difco beef extract, 5.0 bacto peptone, 5.0 bacto yeast extract, pH adjusted to 7.0 with NaOH prior to the addition of 1 g CaCO3). An aliquot of 0.8 µl from each culture was put in the wells of a master plate (MP). MPs were used to carry out microfermentations in 96-deep well plates designated by Duetz system assay) following the approach described by Duetz et al. The ten previously described media were inoculated with spores/mycelia, and incubated at 28ºC (120 rpm), during 7 days for Streptomyces and Nocardia, and during 15 days for Micromonospora, Pseudonocardia, and the non-Streptomyces strains. Bacterial broth was then subjected to an organic extraction with 800 ml acetone and 80 µl of dimethylsulfoxyde (DMSO) per well. The extract was concentrated to dryness and used for the analysis of the antimicrobial activities.
Analysis of the Antimicrobial Activity
The antimicrobial activity was analyzed using the Kirby Bauer diffusion method [17].
Antimicrobial activity of the isolated actinomycetes was assessed, after organic extraction, using Tecan Aquarius Robot Spotlight. Clinically relevant pathogenic microorganisms were targeted: Candida albicans, Methicillin-resistant Staphylococcus aureus (MRSA), Staphylococcus aureus ATCC6538P, Acinetobacter baumanniiATCC5973, Pseudomonas aeruginosa ATCC5919, and Escherichia coli ESS (Gram-negative bacteria are originally from A.L. Demain, Department of Biology, Massachusetts Institute of Technology, Cambridge). Bacteria were grown on Luria–Bertani (LB) and brain–heart infusion (BHI) for Gram-negative and Gram-positive bacteria culture, respectively. Candida albicans was grown on Sabouraud agar medium. Bacteria were incubated at 37 °C for 24 h and Candida albicans at 28 °C for 48 h.
Purification and Partial Characterization of the Antimicrobials Compounds
Cellular extracts showing the greatest antimicrobial activity in solid culture were dereplicated by LC–MS using an Agilent 1200 HPLC coupled with a Rapid Resolution Mass Spectrometer. The column used was charged with 1 µl of the vacuum-dried sample resuspended in 100 µl 50% DMSO. Mobile phase A contained 10% of acetonitrile, 90% of water and 1.3 mM of trifluoroacetic acid/ammonium formate, and B contained 90% of acetonitrile, 10% of water and 1.3 mM of trifluoroacetic acid/ammonium formate). Gradient 10% B increasing linearly to 100% B over 6 min, holding at 100% B for 25 min and then decreasing linearly to 10% B in 2 min. The flow rate was 0.3 ml/min and electrospray ionization (ESI) was adjusted to positive mode. Retention time and the mass of each component were compared to the high-resolution database at the MEDINA Foundation and the “Chapman & Hall Dictionary of Natural Products” [18]. The compound was considered identified if a match was found between the exact mass, molecular formula, and producing organism. Molecules were considered potential new compounds if they could not be identified using the Chapman & Hall database.
Identification of the Most Active Strain CH12
The strain was firstly identified on the basis of colony and microbiological criteria. Cultural characteristics of the isolated strain were determined after 2 weeks of incubation at 28 °C on all tested media. The morphological features of surface ornamentation, spores, sporangia, and aerial and substrate mycelium were observed by light microscopy.
The molecular identification of the strain was also realized. Genomic DNA was extracted as previously described [16]. The 16S rDNA was amplified on the basis of the PCR method using primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) [19].
Phylogenetic and molecular evolutionary analyses were conducted using software included in MEGA version 6.0 package. Tree topologies were evaluated by bootstrap analysis based on 1000 resamplings of the neighbor-joining dataset [19].
Statistical Analysis
Statistical analysis was performed using the Statview 4.57 software (Abacus Concepts Inc., Berkeley, CA, USA). Data were subjected to three-way analysis of variance (ANOVA) in order to evaluate the effect of actinomycetes strains, culture media, and marker microorganisms on antimicrobial activities. Data correspond to the average and the standard deviation (SD) from three independent measures and values were considered significant when P ≤ 0.05 (For revue, Trabelsi I, 2016).
The Accession Number
Sequence has been deposited in the DNA Data Bank of Japan (DDBJ) (https://www.ddbj.nig.ac.jp/index-e.html) with accession number LC720405.
Results and Discussion
Microbiological Analysis
As indicated above, a total of 32 actinomycetes strains were isolated from the three different samples: 20 strains from compost heap, seven strains from manure, and five strains from waste water.
Based on phenotypic characteristics macro and microscopic analysis, 4 different genera were described: Streptomyces (22 isolates), Micromonospora (6 isolates), Pseudonocardia (2 isolates), and Nocardia (1 isolates). Only one strain, named non-Streptomyces, could not be classified on the basis of the phenotypic characters.
These results prove the abundance of the actinomycetes in the composting sites as reported previously by Tiquia et al. [20]. In fact, the actinomycetes became numerous during the cooling stage of composting and represent the more important fraction with respect to fungi and other bacteria [5]. Moreover, the genus Streptomyces showed a notable dominance in all the sampling sites and especially in compost. It seems to be the most important actinobacteria in ecological function [21]
Pre-analysis of the Antimicrobial Activity
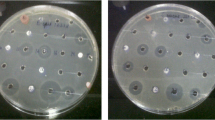
Only five strains from the 20 isolated from the compost heap are inoffensive against all tested microorganisms. An interesting strain called CH12 showed a high antimicrobial potential against all tested pathogens especially against Gram-negative bacteria. However, actinomycetes strains isolated from manure have limited antibacterial activities and absence of antifungal ones. Likewise, most strains isolated from waste water showed only antibacterial potential against Gram-positive and Gram-negative tested bacteria.
Growth of actinomycetes in liquid media is very important for the production of secondary metabolites. GHSA media seems to be the most adequate to the growth of actinomycetes strains. Several authors showed the influence of the medium composition on the production of antimicrobial molecules [15, 22].
ANOVA reported in Tables 1 revealed that antimicrobial activity varied significantly (at P ≤ 0.01) depending on the producing actinomycetes strains, the culture media , and the microorganisms test. A significant interaction (at P ≤ 0.01) between these factors was also detected.
Purification and Partial Characterization of Bioactive Molecules
As detailed in methods, cellular extracts from liquid culture were subjected to chemical dereplication orderly. Several bioactive molecules were identified, on the basis of MEDINA’s database and the dictionary of natural products Chapman and Hall, such as quinomycin A produced by Streptomyces sp WW2 and daidzein produced by Streptomyces sp. WW4, both isolated from waste water.
Quinomycin A is a cyclic octadepsipeptide of the quinoxaline family, produced by various strains of Streptomycetes. It is a potent inhibitor of DNA transcription showing great antitumour and antibiotic activities, especially against methicillin-resistant Staphylococus aureus. Liu et al. presented, in 2008, the first report on the insecticidal properties of antibiotic quinomycin A [23].
Daidzein is an isoflavone which has interesting biological activities (antioxidants, antimicrobials, free radical scavengers, metal chelators, and antibacterial agents). Thus, the isolation and synthesis of isoflavones have become frequent research issues [24].
Many detected compounds do not match with any known bioactive molecules present in MEDINA’s database, which contains the most common secondary metabolites. These compounds can be subject of others assays to search for new antibiotics.
The strain CH12 showed the most remarkable activity among all studied strains. The analysis of the crude extract showed an interesting compound with the molecular formula C26H18O13 (Fig. 1). It has been identified as possible purpuromycin.
Purpuromycin was isolated for the first time from Actinoplanes [25]. It’s an hydroxy derivative of r-rubromycin, an antibiotic isolated from Streptomyces. Even though the two antibiotics are chemically similar, they present a different biological activity. Rubromycins in fact are recognized to be effective only on Gram-positive bacteria while purpuromycin shows good activity also on Gram-negative bacteria and fungi. This can explain the intensive antimicrobial activity of the CH12 strain.
Molecular Identification of the CH12 Strain
Phylogenetic analysis using the 16S rDNA sequence shows that isolate CH12 seems to be a new actinomycetes strain. The similarity level has reached 98% with many Streptomyces strains as Streptomyces djakartensis strain TTIO5.3, Streptomyces plicatus strain B4-7, and Streptomyces rochei strain MML2604. In fact, much higher 16S rDNA similarities have been found between Streptomyces sp. DS9 and Streptomyces strains grouped in a cluster supported by significant bootstrap values (100%) (Fig. 2).
Neighbor-joining phylogenetic tree derived from the 16S rRNA gene sequences of the most active actinomycetes isolated in this work and the most similar sequences of the databases. Percentages at the nodes represent the levels of bootstrap support from 1,000 re-sampled datasets; only values of 70% or above are shown. The scale bar indicates 0.1 substitutions per nucleotide position. Accession numbers are given in parentheses
Conclusion
Environmental pollution is becoming a major problem. Bioremediation, which involves the use of living organisms or their products, is becoming increasingly applied. Actinobacteria have proven to be an effective tool to perform this procedure. Indeed, actinomycetes, which are affected by extreme pollution conditions, are capable of producing a wide range of active biomolecules that can be used in different fields.
During this study, strains of actinomycetes were isolated from various polluted sites. The results obtained prove the high potential of this relatively unexplored environment in the screening of new secondary metabolites produced by putative novel actinomycetes strains. Further studies are required to exploit these strains in bioremediation processes.
Data Availability
The datasets used or analyzed during the study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
Stackebrandt E, Rainey FA, Ward-Rainey NL (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Evol Microbiol 47(2):479–491
Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clément C, Ouhdouch Y, van Wezel GP (2015) Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev 80(1):1–43. https://doi.org/10.1128/MMBR.00019-15
Bensultana A, Yedir O, Lahcen H, Mezrioui NE, Leila R (2010) Isolation and characterization of wastewater sand filter actinomycetes. World J Microbiol Biotechnol 26:481–487. https://doi.org/10.1007/s11274-009-0194-0
Bull AT (2011). Actinobacteria of the extremobiosphere. Reference Work entry. Extremophiles Handbook. 1203–1240
Mokni-Tlili S, Jaoua L, Murano F, Jedidi N, Hassen A (2009) Study of the effects of urban organic residues on the distribution of culturable actinomycetes in a Tunisian agricultural soil. Waste Manag Res 27(3):224–232. https://doi.org/10.1177/0734242X08090405
Simujide H, Chen A, Chun-Jie W, Ma L, Ma B (2013) Microbial activities during mesophilic composting of manure and effect of calcium cyanamide addition. Int Biodeterior Biodegradation 83:139–144. https://doi.org/10.1016/j.ibiod.2013.05.003
Wotton RS (2002) Water purification using sand. Hydrobiologia 469:193–201
Salah El-Din Mohamed W, Zaki DFA (2019) Evaluation of antagonistic actinomycetes isolates as biocontrol agents against wastewater-associated bacteria. Water Sci Technol 79(12):2310–2317. https://doi.org/10.2166/wst.2019.231
Cavalla M, Eberlin T (1994) Isolement des streptomycètes du sol. L’opéron 19:13–17
Rodier J. (1984). L’analyse de l’eau : eaux naturelles, eaux résiduaires, eau de mer, 7e édition, DUNOD, BORDAS (Éditeurs), Paris, France, 1365 p.
Kitouni M, Boudemagh A, Oulmi L, Reghioua S, Boughachiche F, Zerizer H et al (2005) Isolation of actinomycetes producing bioactive substances from water, soil and tree bark samples of the north–east of Algeria. J Myco Méd 15:45–51. https://doi.org/10.1016/j.mycmed.2004.12.005
Gebreyohannes G, Moges F, Sahile S, Raja N, Reetha D (2013) Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of LakeTana Ethiopi. Asian Pac J Trop Biomed 3(6):426–435. https://doi.org/10.1016/S2221-1691(13)60092-1
Boudjella H, Bouti K, Zitouni A, Mathieu F, Lebrihi A, Sabaou N (2006) Taxonomy and chemical characterization of antibiotics of Streptosporangium Sg 10 isolated from a Saharan soil. Microbiol Res 161(4):288–298. https://doi.org/10.1016/j.micres.2005.10.004
Murray PR, Brenner DJ, Bryant MP, Holt HG, Krieg NR, Moulder JW, Pfenning N, Sneath PHA, Staley JT (1984) Streptomycetes and Related Genera. Bergeys manual of systematic bacteriology. Williams & Wilkins, Philadelphia, pp 2451–2508
Trabelsi I, Oves D, Manteca A, Genilloud O, Altalhi A, Nour M (2016) Antimicrobial activities of some actinomycetes isolated from different rhizospheric soils in Tunisia. Curr Microbiol 73(2):220–227. https://doi.org/10.1007/s00284-016-1053-5
Kieser, T., M.J. Bibb, M.J. Buttner, K.F. Chater and D.A. Hopwood (2000). Practical Streptomyces Genetics. John Innes Foundation, Norwich
Biemer JJ (1973) Antimicrobial susceptibility testing by the Kirby-Bauer disc diffusion method. Ann Clin Lab Sci 3:135–140
Buckingham J (2013) Dictionary of Natural Products on CD-ROM. Chapman & Hall, London
Badji B, Zitouni A, Mathieu F, Lebrihi A, Sabaou N (2006) Antimicrobial compounds produced by Actinomadura sp. AC104 isolated from an Algerian Saharan soil. Can J Microbiol 52(4):373–382
Tiquia SM (2002) Evolution of extracellular enzyme activities during manure composting. J Appl Microbiol 92(4):764–775
Xu LH, Li QR, Jiang CL (1996) Diversity of soil actinomycetes in Yunnan. China Appl Environ Microbiol 62:244–248
Boudemagh A, Kitouni M, Boughachiche F, Hamdiken H, Oulmi L et al (2005) Isolation and molecular identification of actinomycete microflora, of some saharian soils of south east Algeria (Biskra, EL-Oued and Ourgla) study of antifungal activity of isolated strains. J Myco Med 15:39–44. https://doi.org/10.1016/j.mycmed.2004.12.004
Liu H, Sheng Q, Yongxia W, Wenjun L (2008) Insecticidal action of Quinomycin A from Streptomyces sp. KN-0647, isolated from a forest soil. World J Microbiol Biotechnol 24(10):2243–2248. https://doi.org/10.1007/s11274-008-9736-0
Roh C, Seo SH, Choi KY, Cha M, Pandey BP, Kim JH, Park JS, Kim DH, Chang IS, Kim BG (2009) Regioselective hydroxylation of isoflavones by Streptomyces avermitilis MA-4680. J Biosci Bioeng 108(1):41–46. https://doi.org/10.1016/j.jbiosc.2009.02.021
Coronelli C, Pagani H, Bardone MR, Lancini GC (1974) Purpuromycin, a new antibiotic isolated from Actinoplanes ianthinogenes N sp. J Antibiot 27:161–8
Acknowledgements
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code: (22UQU4331312DSR08).
Funding
Not applicable
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
Not applicable.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable
Consent for Publication
Not applicable
Microbial Deposition in Repository
The GenBank accession number for the 16S rRNA sequence of strain CH12 is LC720405.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Trabelsi, I., Soltane, R., Hassine-Zaafrane, M. et al. Study of the Antimicrobial Potential of Actinomycetes Isolated from Organic and Inorganic Waste. Curr Microbiol 79, 372 (2022). https://doi.org/10.1007/s00284-022-03024-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03024-y