Abstract
Verticillium wilt is a major disease of cotton that considerably decreases yield and crop quality. Soil microbial communities play an important role in plant health. Therefore, biocontrol bacteria that regulate microbial communities in rhizosphere soil can improve plant resistance to pathogens. Previously, the antagonistic strain Bacillus axarquiensis TUBP1 was screened and found to act against Verticillium dahliae with 43% biocontrol effect in cotton fields. We studied the effect of Bacillus axarquiensis TUBP1 with a green fluorescent protein (GFP) gene marker on the microbial community structure of cotton rhizosphere soil and cotton yield and quality. Cotton Verticillium wilt incidence, soil biochemical properties, and soil bacterial and fungal communities were analyzed. Results showed that bacterial and fungal abundance in cotton rhizosphere soil was temporarily changed after applying B. axarquiensis TUBP-315GFP. However, Bacillus significantly increased, whereas V. dahliae significantly decreased. The incidence of cotton Verticillium wilt after treatment with B. axarquiensis TUBP-315GFP was significantly lower and cotton production increased by 40.6%. Our findings indicated that the application of B. axarquiensis TUBP-315GFP can change microbial community structure of cotton rhizosphere soil, leading to a reduction in the incidence of cotton Verticillium wilt and increasing cotton yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cotton Verticillium wilt, caused by Verticillium dahliae Kleb, is a major soil-borne disease, which seriously reduces cotton yield and quality in cotton-producing areas globally [1]. The disease is difficult to control because the pathogen forms microsclerotia in the soil that can survive for long periods, and fungicides fail to eliminate the fungus once it has entered the xylem of host plants [2]. Some control measures have been attempted, including improved agricultural management practices and the integrated use of resistant cultivars and biocontrol agents. The use of biological agents to control Verticillium wilt in cotton has attracted much attention [3, 4]. Biocontrol strains play a beneficial role in plant growth either directly or indirectly under disease stress, which includes antibiosis, nutrient competition, and systemic resistance induction. Bacillus strains, as spore-forming bacteria that are more easily stored, are the best candidates for controlling many plant diseases, and some important biocontrol agents, including B. subtilis strain GB03, B. pumilus GB34, B. licheniformis SB3086, B. subtilis GB122, B. amyloliquefaciens GB99, B. amyloliquefaciens ZM9, B. subtilis N11 [5], and B. amyloliquefaciens RWL-1 [6] have been reported.
The application of biocontrol agents can also alter microbial community structure in rhizosphere soil, which can improve the disease resistance of plants, and this has become a focal point in the use of biocontrol agents. The structure of microbial communities in the rhizosphere soil of ginseng was altered after applying B. subtilis 50-1 [7]. The relative abundance of Fusarium oxysporum and the mortality of ginseng replantation significantly decreased. After the application of biocontrol bacteria, the mortality of ginseng after different seedling years was significantly reduced.
In our previous work, we isolated a strain of TUBP1 that exhibited antagonistic activity against V. dahliae resulting in a 43% biocontrol effect in cotton fields [8]. The experimental results showed perturbation of the plasma membrane of the spores and the hyphae of V. dahliae when they were treated with the TUBP1 protein, which induced mitochondrial damage in V. dahliae leading to apoptosis [9]. Then, we successfully constructed B. axarquiensis TUBP1 with a green fluorescent protein (GFP) signature, which didn’t change the bioactivity in vitro. B. axarquiensis TUBP1 was able to colonize cotton plants and the rhizosphere soil [10]. However, little is known about how microbial communities, soil biochemical properties, and cotton Verticillium wilt incidence respond to the inoculation of rhizosphere soil with B. axarquiensis TUBP-315GFP. In the present study, we aimed to determine whether, (i) microbial community composition shifted in cotton rhizosphere soil, (ii) Verticillium wilt and B. axarquiensis TUBP-315GFP were correlated, (iii) soil biochemical properties and cotton quality index responded B. axarquiensis TUBP-315GFP inoculation in cotton rhizosphere soil.
Materials and Methods
Test Strain
Bacillus axarquiensis TUBP-315GFP were provided by the Key Laboratory of Biological Conservation and Utilization, Tarim University. And the fermentation supernatant of B. axarquiensis TUBP-315GFP was obtained after it was fermented at 37 °C and 180 rpm/min for 48 h in Luria–Bertani medium (LB, Invitrogen, Carlsbad, USA).
Overview of Test Site and Material
The cotton variety tested in the present study was Xinluzhong 70, which was also provided by the Key Laboratory of Biological Conservation and Utilization, Tarim University. The experiment was carried out in the experimental field of the 12th regiment farm (40°32′ N, 81°17′ E; altitude 1011 m) in Alar City, Xinjiang Province, China from March to October of 2018–2019, sandy soil in the experimental field, growing cotton all the year round. The basic chemical properties of the 0–20-cm layer of arable soil in this area is pH 7.4, organic matter 8.09 g/kg, available nitrogen (AN) 39.02 mg/kg, available phosphorus (AP) 27.83 mg/kg, and available potassium (AK) 92.63 mg/kg. The average temperature is 20 °C during the day and 4 °C at night, annual sunshine hours are 2900 h, frost-free period is 205–219 days, and annual average rainfall is 50 mm.
Experimental Design and Sampling
Three treatments and one control were set up at the test site. Three different concentrations of B. axarquiensis TUBP-315GFP fermentation broth (106, 108, or 1010 CFU/mL) were applied during the four growth periods of cotton (referred to as T1.1, T1.2, T1.3, T2.1, T2.2, T2.3, T3.1, T3.2, T3.3, T4.1, T4.2, and T4.3, respectively), and the control group was given the same volume of Luria–Bertani medium (referred to as CK1, CK2, CK3, and CK4).
Each treatment was 60 m2, and each treatment was repeated three times (contained ten lines with pianting spacing of 60 cm). 0–20-cm root irrigation inoculated with B. axarquiensis TUBP-315GFP. At each treatment stage, 500 g rhizosphere soil was extracted from the same depth and divided into two samples. One 250 g sample was put into an aseptic bag and quickly put into a dry ice box and stored at − 80 °C. A total of 48 soil samples were obtained for the four treatments at four growth stages and subjected to high-throughput sequencing. The other 250 g sample was stored at 4 °C for analysis of the physical and chemical properties of the rhizosphere soil.
Effects of B. axarquiensis TUBP-315GFP on Enzyme Activities and Chemical Properties of Cotton Rhizosphere Soil
The physical and chemical properties of the soil were determined by the following methods. Total nitrogen (TN) was estimated using the Kjeldahl method [11]. Organic matter (OM) was determined using the potassium bichromate titrimetric method [12]. AN was determined using the alkali diffusion method [13]. AP was extracted with 0.5 M NaHCO3, and then the content in solution was calculated using molybdenum antimony colorimetric determination [14]. Soil AK was determined with the fame photometric method [15]. Following which for each soil sample the 1:5 soil–water extract was prepared [16, 17], determined using a DDS-11A digital conductivity instrument electrical conductivity (TS).
Soil enzymatic activity reagents were bought from the Suzhou Keming Biotechnology Co., Ltd, Su Zhou, China [14]. Alcalase protease activity (ALPT) was determined using casein as the substrate [18]. Alkaline phosphatase activity (AKP) was determined using phenolphthalein phosphate as the substrate [19]. Urease activity (UE) was determined using the urea-phenol red method [20]. Polyphenol oxidase (PPO) can oxidize monophenol and diphenol to produce quinone under aerobic conditions to determine the activity of PPO [21]. Sucrase activity (SC) was determined using the colorimetric method [20]. Catalase (CAT) can decompose hydrogen peroxide, so that the absorbance (A240) of the reaction solution decreases with the reaction time. CAT activity can be measured by measuring the rate of change in absorbance [22].
Soil DNA Extraction and High-Throughput Sequencing
Total microbial DNA in the rhizosphere soil was extracted by the combined SDS-CTAB method [23] with a soil DNA kit (Shanghai Shenggong Co.). Briefly, total microbial DNA of 5 g rhizosphere soil was crude extracted by the SDS-CTAB method, and then, 0.25 g of the crude extract was used for DNA extraction, according to the manufacturer's protocol from the Ezup column soil DNA extraction kit of Shanghai Shenggong. Finally, the quality of the total DNA of the extracted soil microorganisms was detected by gel electrophoresis. The qualified genomic DNA was sent to Beijing Nuohe Zhiyuan Bioinformation Technology Co., Ltd. for amplification of the V3–V4 and ITS regions of 16S rDNA and high-throughput sequencing using the Illumina MiSeq 2500 sequencing platform.
Data Analysis and Processing
UPARSE (v7.0.1001, http://www.drive5.com/uparse/) [24] software was used to cluster all effective tags of all samples. By default, 97% identity was used to cluster the sequences into operational taxonomic units (OTUs). Then, representative sequences of OTUs were selected. According to the algorithm principle, the sequences with the highest frequency in OTUs were selected as representative sequences of OTUs. Species annotation was carried out for OTUs representative sequences, and species annotation analysis was carried out with mothur software and SSUrRNA [25] database of SilVA [26] (threshold value was 0.8–1). To obtain the taxonomic information, we used the mothur software package to divide the 16S rDNA sequences into OTUs with a threshold of 97% and constructed a dilution curve [27]. The dilution curve and species accumulation box chart were used to analyze species richness and determine the rationality of sampling. The alpha diversity was estimated by calculating the Simpson and Shannon indexes in the QIIME (v1.7.0) software [28]. The chao index is often used to quantitatively estimate the abundance of microbial flora. The Shannon index reflects the diversity of the samples, and the higher the Shannon index, the higher the diversity of the community. Beta diversity was estimated as the microbial community composition of different samples. The similarity clustering tree was constructed by QIIME software. Principal component analysis (PCA) and non-metric multi-dimensional scaling (NMDS) were conducted in R (Version 2.15.3), and can be used to assess the species composition differences among communities in different habitats, and to determine the differences among samples to reveal the microbial diversity of the rhizosphere soil.
Influence of B. axarquiensis TUBP-315GFP on Cotton Yield and Quality
Cotton yield after B. axarquiensis TUBP-315GFP application was determined by hand harvesting the four central rows in each treatment on October, 2018. Then, we calculated the yield per acre of cotton, and determined the quality of cotton based on the following indicators: length, strength, and micronaire value of cotton fiber [29].
Statistical Analysis
All results of the experiments are presented as the mean ± standard deviation of three independent experiments. The single factor analysis of variance (ANOVA) was performed to test the mean value of multiple groups of data, based on Tukey’s multiple range tests use SPSS statistics (version 18.0) software. P < 0.05 was considered to be significant, and P < 0.01 was considered to be very significant. Metastats and distance-based redundancy analysis (db-RDA) were conducted in R (Version 2.15.3). Metastats reflects significant differences in the communities between groups. RDA reflects the influence of environmental factors on the composition of different sample communities.
Results
Enhancement of Yield and Quality of Cotton Through Inoculation with B. axarquiensis TUBP-315GFP
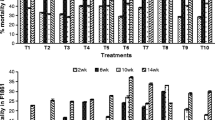
The incidence of cotton Verticillium wilt was detected, and the results are shown in Fig. 1. Compared with CK, the incidence of cotton Verticillium wilt was significantly lower after cotton root was inoculated with B. axarquiensis TUBP-315GFP at the budding, flowering, and bolling stages (P < 0.05). However, there were no significant differences in the incidence of cotton Verticillium wilt between T2 and T3.
Effects of Bacillus axarquiensis TUBP-315GFP on the incidence of Verticillium wilt in cotton. TUBP1 changed the incidence of cotton Verticillium wilt (the symptoms of cotton Verticillium wilt were mainly reflected after the flower bud stage). GFP, green fluorescent protein. The inoculation concentration of T1 was 106 CFU/mL. The inoculation concentration of T2 was 108 CFU/mL. The inoculation concentration of T3 was 1010 CFU/mL. Different letters (a, b, c) indicate statistically significant differences (P < 0.05) between inoculated and control plants according to t-test. Error bars represent SD (n = 3)
The yield and quality of cotton were evidently changed after cotton rhizosphere soil was inoculated with B. axarquiensis TUBP-315GFP (Table 1). The cotton yield in the B. axarquiensis TUBP-315GFP-treated group significantly increased compared with that of CK, of which the yield of T2 increased by 40.6% (P < 0.05). However, cotton yield did not significantly change in the rhizosphere soil in T3, which indicated that cotton yield was closely related to the application concentration of B. axarquiensis TUBP-315GFP. Furthermore, biocontrol agents have been reported to improve crop quality [30]. Thus, eight cotton quality indexes FL (fiber length), UI (uniformity index), STR (fiber strength), ELO (fiber elongation), MIC (micronaire), REF (reflectivity), FY (fiber yellowness), and SHI (short fiber index) were evaluated after cotton rhizosphere soil was inoculated with different concentrations of B. axarquiensis TUBP-315GFP (Table 1). Among them, LE, UI, STR, and SHI significantly changed in the rhizosphere soil in T2 compared with that in CK (P < 0.05). However, ELO, MIC, REY, and FY were not evidently changed compared with CK. Therefore, cotton yield and some quality indexes improved when cotton rhizosphere soil was inoculated with B. axarquiensis TUBP-315GFP. All these results indicate that cotton Verticillium wilt decreased, while cotton yield was increased, and some cotton quality index were also significantly changed after cotton rhizosphere soil was inoculated with B. axarquiensis TUBP-315GFP.
Soil Enzyme Activity Response to B. axarquiensis TUBP-315GFP
In addition to the qualitative and quantitative changes induced by the microbial inoculants on the rhizosphere microbial community, there were also changes in the functioning of the system, as evaluated by soil enzyme activity (Table 2). The activities of PPO, ALPT, SC, UE, and AKP all increased to some extent compared with those in CK after the application of B. axarquiensis TUBP-315GFP. Both UE and SC activity significantly increased with the advancement of cotton growth stage and peaked during the bolling stage (The change of CAT is not significant). The enzyme activities of UE and SC increased significantly with the advance of cotton growth period, among them, the activity of UE in budding stage increased significantly by 18.08%, bolling stage SC enzyme increased by 36.01%. The soil enzyme activity changed in rhizosphere soil inoculated with B. axarquiensis TUBP-315GFP, the activities of PPO, ALPT, SC, UE, and AKP showed a clear upward trend, and the activity of UE and SC was also closely related to the growth period of cotton. After the application of bacterial agent, the possible mechanism is that the diversity of microbial community has changed after the action of bacterial agent, and the increase of the number of microorganisms and the increase of growth rate have promoted the enzyme activity to the greatest extent.
Soil Property Response to B. axarquiensis TUBP-315GFP
Bacillus axarquiensis TUBP-315GFP treatment affected the soil properties (Table 3). In the presence of B. axarquiensis TUBP-315GFP, soil TN, AP, and AK were significantly higher than those in CK during the period of T1–T4 (P < 0.05). Besides, NN and TS content did not differ between the treatment groups and CK. The soil with the same concentration of B. axarquiensis TUBP-315GFP, flowering stage has the highest content of TN and AK, the content of AP is the highest in bolling stage. Therefore B. axarquiensis TUBP-315GFP increased the contents of TN, AK, and AP in soil, and improved the original soil environment.
Microbial Diversity Response to B. axarquiensis TUBP-315GFP
All collected samples were analyzed using IonS5TMXL platform Novogene Bioinformatics Technology Co., Ltd (Beijing, China), and the sequencing results are shown in Table 4. After cutting and filtering the reads, 79,097 bacterial reads were obtained on average for each sample, and 74,351 effective reads were obtained on average after quality control; the effective rate of quality control was 94.02%. At 97% identity, the sequences were clustered into 5877 OTUs. Then, the OTU sequences were annotated against the Silva132 database (27 January 2019). A total of 2058 (35.02%) OTUs were annotated at generic level.
The average number of fungal reads per sample was 82,560, and the average number of effective reads was 78,975. The effective rate of quality control was 95.72%. With 97% identity, the sequences were clustered into 4582 OTUs. Then, the OTU sequences were annotated against the UNITE database. A total of 1146 (25.01%) OTUs were annotated at generic level.
The Chao values represent richness, and the Shannon diversity index indicates both richness and evenness. For both of these indices, higher values indicate higher diversity. The α-diversity of soil microbial communities slightly increased in the rhizosphere at the flowering and bolling stages for all samples (Fig. 2A–D). However, there was no significant difference between the treatment groups and CK at the flowering and bolling stages. In addition, the diversity index of the treatment groups was higher than that in CK based on the ANOVA at early stages, implying that diversity was higher in the treatment groups. The Chao 1 and Shannon values in the treatment groups with rhizosphere soil inoculated with B. axarquiensis TUBP-315GFP were apparently higher than those in CK at the budding stage, which indicated that B. axarquiensis TUBP-315GFP inoculants affected the diversity of microbial communities at early stages after inoculation.
Diversity indices in different growth stages and treatments in the rhizosphere soil of cotton. A, B Chao 1 index of cotton rhizosphere soil for bacteria and fungi, respectively; C, D Shannon index of cotton rhizosphere soil for bacteria and fungi, respectively. Differences are P < 0.05 as measured by T-test
Multi-Sample Comparative Analysis
PCA and NMDS were applied to evaluate microbial community structure in rhizosphere soil treated with B. axarquiensis TUBP-315GFP (Fig. 3). Bacterial community structure is shown in Fig. 3A; the contribution rate of the first (PC1) and second (PC2) principal components was 11.52% and 7.46%, respectively. Fungal community structure is shown in Fig. 3B; the contribution rate of the PC1 and PC2 based on PCA was 7.54% and 5.18%, respectively. The application of B. axarquiensis TUBP-315GFP inoculants affected the composition of indigenous rhizosphere microbial communities at the seedling stage, whereas no major effect was evident at the flowering and bolling stages. However, the bacterial and fungal community showed a clear temporal shift in the different groups. The NMDS results were similar to those of PCoA, as shown in Fig. 3C–D, indicating that bacterial and fungal community structure differed between the seedling stage and the other three growing stages. Besides, at the budding stage, bacterial and fungal community structure showed significant differences between CK and the treatment groups. However, microbial community structure was not evidently changed between CK and the treatment groups at the flowering and bolling stages after applying B. axarquiensis TUBP-315GFP.
Effect of Bacillus axarquiensis TUBP-315GFP application on the rhizosphere bacterial and fungal community in cotton rhizosphere soil. A, B Bacterial and fungal PCA analysis of cotton rhizosphere soil, respectively. C, D Bacterial and fungal NMDS analysis of cotton rhizosphere soil, respectively. The differences of diversity index between groups were analyzed by R software, and Tukey test and wilcox test were used. GFP green fluorescent protein, PCA principle components analysis, NMDS non-metric multi-dimensional scaling
Bacterial Composition Response to B. axarquiensis TUBP-315GFP
Metastats analysis was performed to evaluate the rhizosphere bacterial community that influenced the composition of microbial communities, and the results indicated that Proteobacteria, Acidobacteria, Latescibacteria, and Gemmatimonadetes, were the major taxa shaping the rhizosphere community (Supplementary Fig. S1, A).
However, bacterial diversity significantly shifted after inoculation with B. axarquiensis TUBP1-315GFP at the seedling and budding stages. The most abundant bacterial genera were Arthrobacter, Streptococcus, Lysobacter, Sphingomonas, Pseudomonas, Bacillus, Bifidobacterium, and Pontibacter. Among them, Pontibacter and Streptococcus were significantly decreased, whereas Bauldia, Acidobacteria, Haliangium, Pseudarthrobacter, Arthrobacter, Actinomarinales, Sphingomonas, Lysobacter, and Bacillus significantly increased (P < 0.01) (Fig. 4A). The content of Bacillus, as the main biocontrol bacterium, in the soil indicates the incidence of cotton Verticillium wilt. In the budding stage, Bacillus was the dominant genus in the treatment groups. Figure 4B shows that the content of B. axarquiensis TUBP-315GFP in the rhizosphere soil increased with the change in cotton growth period. In particular, the content of B. axarquiensis TUBP-315GFP in the treatment groups was significantly higher than that in CK at the budding, flowering, and bolling stages. In addition to the seedling stage, however, there was almost no significant difference in the content of B. axarquiensis TUBP-315GFP in the soil between the seedling and CK groups.
Changes in main species of bacteria and fungi in cotton rhizosphere soil. A Top-10 species of cotton rhizosphere soil bacteria, B Bacillus changes in cotton rhizosphere soil, A1 top-10 species of cotton rhizosphere soil fungi, B1 change in Verticillium dahliae in cotton rhizosphere soil. A, A1 The bacterial and fungal richness and diversity indices (Abundance-based) were estimated using Mothur (version v.1.30.1). B, B1 Different letters (a, b, c) indicate statistically significant differences (P < 0.05) between inoculated and control plants according to t-test. Error bars represent SD (n = 3)
Fungal Composition Response to B. axarquiensis TUBP-315GFP
Meta Stat analysis was performed to evaluate the variation in rhizosphere fungal community influenced by B. axarquiensis TUBP1 inoculants. Ascomycota, Mortierellomycota, and Basidiomycota were the major taxa shaping the rhizosphere community between the CK and treatment groups, and there were no evident differences between them (Supplementary Fig. S1, A1). However, from the top-12 phyla, in contrast with those in CK, Mortierellomycota, Rozellomycota, and Basidiomycota were higher in the budding, flowering, and bolling stages of the treatment groups. The most abundant fungal genera in the rhizosphere soil were Chaetomium, Preussia, Stemphylium, Trichosporon, and Pseudeurotium (P < 0.05) (Fig. 4A1). The dominant species in the treatment groups were Papiliotrema flavescens, Acremonium, and Cladosporium chasmanthicola at the seedling, budding, and flowering stages, respectively.
Verticillium dahliae was the main fungal pathogen, and its content in the rhizosphere soil was detected by the dilution-plate method. Figure 4B1 shows that the content of V. dahliae decreased after inoculation with B. axarquiensis TUBP-315GFP at the budding, flowering, and bolling stages compared with CK.
Correlation Between Soil and Environmental Factors
It can be seen from Fig. 5A that, based on the analysis of dbRDA, the contribution rate of PC1 (dbrda1) and PC2 (dbrda2) was 29.99% and 25.27%, respectively, and ALPT was negatively correlated with other environmental factors. For the first axis, the more important environmental factors were AP, AN, and PPO, whereas on the second axis, the important environmental factors were AKP, CAT, AK, and ALPT. In the three growth stages after the seedling stage, soil bacterial diversity in the T2.2, T3.2, and T4.2 treatment groups was most affected by environmental factors.
Effects of TUBP-315GFP application on cotton rhizosphere soil and environmental factors. RDA, the influence of environmental factors on the composition of different sample communities. A dbRDA analysis of bacteria and environmental factors, B dbRDA analysis of fungi and environmental factors. Redundancy analysis (RDA) was employed to examine effects of TUBP-315GFP application on cotton rhizosphere soil and environmental factors using the vegan package of R software (Version 3.0.2). GFP green fluorescent protein
As can be seen from Fig. 5B, based on dbRDA analysis, the contribution rate of PC1 (dbRDA1) and PC2 (dbRDA2) was 30.08% and 19.2%, respectively. Similar to bacteria, ALPT was negatively correlated with other environmental factors for fungi. For the first axis, the more important environmental factors were TS, CAT, UE, and AP, whereas for the second axis, the important environmental factors were TN and AN. Soil fungal diversity was most closely related to ALPT in the seedling stage. The fungal diversity in the budding and flowering stages was greatly affected by environmental factors.
For bacteria, based on dbRDA analysis, the contribution rate of PC1 (dbRDA1) and PC2 (dbRDA2) was 49.26% and 35.79%, respectively (Fig. 6A). T3 and T4 were more closely related to the relative abundance of Bacillus (BRA), whereas CK2 and CK3 in CK were more closely related to the relative abundance of V. dahliae (VARA). Clostridium butyricum, Rhodovulum sp., Pontibacter populi, and Bifidobacterium pseudocatenulatum were more closely related to VARA, and Bacillus halotolerans and Rhizobium helanshanense were more closely related to BRA. The application of biocontrol bacteria B. axarquiensis TUBP1 not only changed the abundance of V. dahliae and Bacillus in the soil but also affected the abundance of bacterial populations of other species in the soil (Fig. 6A). For fungi, based on dbRDA analysis, the contribution rate of PC1 (dbRDA1) and PC2 (dbRDA2) was 62.88% and 25.11%, respectively (Fig. 6B). Similar to bacteria, CK2 and CK3 were more closely related to VARA, and T2, T3, and T4 were more closely related to BRA. Alternaria alternata and Thielaviopsis basicola were more related to BRA, while Cladosporium limoniforme and C. chasmanthicola were more related to VARA. It showed that the application of the biocontrol bacteria TUBP1 also changed the population abundance of other fungi in the soil (Fig. 6B).
Effects of biological factors on bacteria and fungi in cotton rhizosphere soil. RDA, the influence of environmental factors on the composition of different sample communities. A dbRDA analysis of bacteria and biological factors, B dbRDA analysis of fungi and biological factors. Redundancy analysis (RDA) was employed to examine effects of biological factors on bacteria and fungi in cotton rhizosphere soil using the vegan package of R software (Version 3.0.2)
Discussion
Understanding the biocontrol mechanisms of B. axarquiensis TUBP1 against cotton Verticillium wilt under field conditions will improve its biological control effect. Previous studies reported this bacterial control agent as a potential candidate against V. dahliae via peptide-T-inducing mitochondrial damage and mitochondria-mediated apoptotic cell death [9]. In the cotton field, B. axarquiensis TUBP1 exhibited efficient biocontrol effects against the Verticillium wilt pathogen. The GFP-tagged B. axarquiensis TUBP1 was obtained by electrotransformation, and it colonized different parts of the cotton plants from the rhizosphere soil (data unpublished). In the present study, we evaluated its efficiency in control of Verticillium wilt and its effects on the cotton rhizosphere microbial community in a field trial. The application of B. axarquiensis TUBP-315GFP biocontrol agents can significantly reduce the incidence of cotton Verticillium wilt and increase cotton yield. FL, UI, STR, and SHI were also significantly changed when cotton plants were treated with B. axarquiensis TUBP-315GFP at 108 CFU/mL (Table 1). Bacillus strains are potent biological control agents against diseases of plants and crops, including tomatoes [31], peppers [32], and tobacco [33].
Bacillus spp. are also considered as bioorganic fertilizers because they promote the growth and quality of crops. Besides, crop yield and quality have a positive relationship between some soil properties, including alkaloid content, pH, nitrate K, and total soluble salt [34]. Our results are largely consistent with those of these previous studies in that TN, AP, and AK in the treatment groups were significantly higher than in CK (Table 3). These factors could have contributed to the increased yield and quality of cotton.
Based on Spearman’s rank correlation coefficient, Bacillus were strongly and positively associated with TS, AP, UE, and CAT. In contrast, negative associations between cotton Verticillium wilt and the relative abundance of Sporichthya, Achromobacter, Burkholderia, Comamonas, Ramlibacter, and Pontibacter were observed.
The soil microbial community structure is closely related to plant health status and is shaped by, e.g., biocontrol bacteria inoculants, crop rotation, fertilization, and tillage. Some studies reported that Paenibacillus polymyxa CP-S316 evidently changed the rhizosphere microbial community [35].
It is important to understand whether applications of B. axarquiensis TUBP-315GFP can change the composition of the indigenous populations of soil microorganisms. In the present study, we discovered that the cotton soil rhizosphere microbial community was temporarily changed at the seeding and budding stages after B. axarquiensis TUBP-315GFP inoculation. These results are in agreement with those of previous studies that reported some biocontrol agents affecting and sometimes destroying the original microflora but only temporarily [36,37,38,39,40].
Notably, Bacillus significantly increased, whereas V. dahliae significantly decreased over the whole growing stage. Besides, the diversity of bacterial species evidently changed at the seeding and budding stages in B. axarquiensis TUBP-315GFP-treated groups. Some beneficial microbes, including Bauldia, Acidobacteria, Haliangium, Pseudarthrobacter, Arthrobacter, Actinomarinales, Sphingomonas, Lysobacter, and Bacillus, significantly increased in the treatment groups, indicating that the biocontrol inoculant B. axarquiensis TUBP-315GFP may promote the growth of cotton and improve the quality.
Plants growing in soil develop close associations with soil microorganisms, which inhabit the areas around, on, and inside their roots [41]. In the study of gramineae plant and soybean, the rhizosphere microbial community changes for a whole life cycle, and the trend is that the soil-derived microbial communities gradually differentiates from some specific microbial communities [42, 43]. Chaparro et al. [44] showed that rhizosphere bacterial communities at the seedling stage of Arabidopsis thaliana were distinct from vegetative, bolting, and flowering stages. Our experimental results showed that in the four growth periods of cotton, the species diversity of rhizosphere soil microorganisms was significantly different due to the difference of growth period, the species richness of bacteria in seeding stage soil is the most uniform, and the species diversity of bacteria increases gradually with the passage of growth cycle. Bacillus strain is an effective biocontrol agent for plant diseases [6, 45], and the degree of colonization and proliferation on the host plant affects the biocontrol effect [46].
Conclusion
The application of B. axarquiensis TUBP-315GFP inoculants affected the composition of cotton rhizosphere microbial communities at the seedling and budding stages, but no major effects on the microbial communities were observed at the flowering and bolling stages. Especially, Bacillus significantly increased, while V. dahliae significantly decreased. The incidence of cotton Verticillium wilt after treatment with B. axarquiensis TUBP-315GFP was significantly lower, and cotton production increased by 40.6%. With 108 CFU/mL of B. axarquiensis TUBP-315GFP in the soil, the influence on microbial diversity was the most significant, and the application of B. axarquiensis TUBP-315GFP effectively reduced the incidence of cotton Verticillium wilt and increased cotton yield. These results provide a reference for future research direction on biofertilizers. The present study indicated that the application of B. axarquiensis TUBP-315GFP can change the microbial community composition of the cotton rhizosphere soil, thereby reducing the incidence of cotton Verticillium wilt and increasing cotton yield. In the future, compost fermented with B. axarquiensis TUBP-315GFP and the detection of beneficial bacteria may be employed to improve the biocontrol of cotton Verticillium wilt disease (Fig. 7).
References
Li S, Zhang N, Zhang Z, Luo J, Shen B, Zhang R, Shen Q (2013) Antagonist Bacillus subtilis HJ5 controls Verticillium wilt of cotton by root colonization and biofilm formation. Biol Fertil Soils 49:295–303. https://doi.org/10.1007/s00374-012-0718-x
Zhang J, Hu HL, Wang XN, Yang YH, Zhang CJ, Zhu HQ, Shi L, Tang CM, Zhao MW (2020) Dynamic infection of Verticillium dahliae in upland cotton. Plant Biol (Stuttg) 22:90–105. https://doi.org/10.1111/plb.13037
Yao ZS, Chen ZY, Zheng XB, Zhang J, Huang DF (2003) Genetically marking of natural biocontrol bacterium Bacillus subtilis strains with green fluorescent protein gene. Sheng Wu Gong Cheng Xue Bao 19:551–555. https://doi.org/10.1016/j.apsoil.2011.03.013
Shi YW, Li C, Yang HM, Zhang T, Gao Y, Chu M, Zeng J, Lin Q, OuTiKuEr LYG, Huo XD, Lou K (2017) Colonization study of gfp- tagged Achromobacter marplatensis strain in sugar beet. J Microbiol 55:267–272. https://doi.org/10.1007/s12275-017-6371-1
Zhang N, Wu K, He X, Li SQ, Zhang ZH, Shen B, Yang XM, Zhang RF, Huang QW, Shen QR (2011) A new bioorganic fertilizer can effectively control banana wilt by strong colonization with Bacillus subtilis n11. Plant Soil 344:87–97. https://doi.org/10.1007/s11104-011-0729-7
Shahzad R, Khan AL, Bilal S, Asaf S, Lee IJ (2017) Plant growth-promoting endophytic bacteria versus pathogenic infections: an example of Bacillus amyloliquefaciens RWL-1 and Fusarium oxysporum f. sp. lycopersici in tomato. PeerJ 5:e3107. https://doi.org/10.7717/peerj.3107
Dong LL, Xu J, Zhang LJ, Cheng RY, Wei GF, Wei GF, Su H, Yang J, Qian J, Xu R, Chen SL (2018) Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta pharmaceutica sinica B 8:272–282. https://doi.org/10.1016/j.apsb.2017.12.011
Zeng H, Ding HP, Tian J, Zhang LL (2018) Pore- forming mechanism of TUBP1 protein act on verticillium dahliae. Process Biochem 73:6–14. https://doi.org/10.1016/j.procbio.2018.07.024
Zeng H, Li T, Tian J, Zhang LL (2018) TUBP1 protein lead to mitochondria-mediated apoptotic cell death in Verticillium dahliae. Int J Biochem Cell Biol 103:35–44. https://doi.org/10.1016/j.biocel.2018.08.001
Wang B, Wan CX, Zeng H (2020) Colonization on cotton plants with a GFP labeled strain of Bacillus axarquiensis. Curr Microbiol 77:3085–3094. https://doi.org/10.1007/s00284-020-02071-7
Yanu P, Jakmunee J (2015) Flow injection with in- line reduction column and conductometric detection for determination of total inorganic nitrogen in soil. Talanta 144:263–267. https://doi.org/10.1016/j.talanta.2015.06.002
Chen ZH, Zhang YL, Jia YH, Chen LJ, Liu XB, Wu ZJ (2011) Degradation characteristics of transgenic cotton residues in soil by Fourier transform infrared spectroscopy. Spectrosc Spectr Anal 31:77–81. https://doi.org/10.3964/j.issn.1000-0593(2011)01-0077-05
Jiao SY, Li JR, Li YQ, Jia JW, Xu ZY (2019) Soil C, N, and P distribution as affected by plant communities in the Yellow River Delta, China. PLoS ONE 14:e0226887. https://doi.org/10.1371/journal.pone.0226887
Liu YB, Zhao LN, Wang ZR (2018) Changes in functional gene structure and metabolic potential of the microbial community in biological soil crusts along a revegetation chronosequence in the Tengger desert. Soil Biol Biochem 126:40–48. https://doi.org/10.1007/s00438-017-1347-8
Li Y, Fang F, Wei J, Wu X, Tan D (2019) Humic acid fertilizer improved soil properties and soil microbial diversity of continuous cropping peanut: a three-year experiment. Sci Rep 9:12014. https://doi.org/10.1038/s41598-019-48620-4
Kim JM, Roh AS, Choi SC, Choi EJ, Ahn MT, Kim BK, Lee SK (2016) Soil pH and electrical conductivity are key edaphic factors shaping bacterial communities of greenhouse soils in Korea. J Microbiol 54:838–845. https://doi.org/10.1007/s12275-016-6526-5
Liu GM, Yang JS, Yao RJ (2006) Electrical conductivity in soil extracts: chemical factors and their intensity. Pedosphere 16:100–107. https://doi.org/10.1016/s1002-0160(06)60031-3
Higahara H, Matsubara M, Nakai M, Okunuki K (1986) Crytalline bacterial proteinase from Bacillus subtilis. Biochem 45:189–194
Tabatabai MA (1982) Soil enzymes. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analyses, part 2, chemical and microbiological properties, 2nd edn. American Society of Agronomy, Madison, pp 903–947
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem 3:14–26. https://doi.org/10.1021/ac60147a030
Sinsabaugh RL, Lauber MN, Ahmed B, Zeglin LH (2008) Stoichiometry of soil enzyme activity at global scale. Ecol Lett 11:1252–1264. https://doi.org/10.1111/j.1461-0248.2008.01245
Beers RF, Sizer TW (1952) A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem 195:133–140. https://doi.org/10.1016/S0074-7696(08)60016-9
Zhang YG, Li DQ, Wang HM, Xiao QM (2005) Extraction method of soil microbial DNA for molecular ecology study. Ying Yong Sheng Tai Xue Bao 16:956–960
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. https://doi.org/10.1101/gr.112730.110
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/aem.00062-07
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web- based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van HD, Weber CF (2009) Introducing mothur: open-source, platform- independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Li H, Xu H, Song L, Bai CX, Sun YY, Tian XQ, Bai CX, Li YH, Jiang Y, Ge J, Wang XL, Wen HY (2020) Alterations of gut microbiota contribute to the progression of unruptured intracranial aneurysms. Nat Commun 11:3218. https://doi.org/10.1038/s41467-020-16990-3
Liu XY, Teng ZH, Wang JX, Wu TT, Zhang ZQ, Deng XP, Fang XM, Tan ZY, Ali I, Liu DX, Zhang J, Liu DJ, Liu F, Zhang ZS (2017) Enriching an intraspecific genetic map and identifying QTL for fiber quality and yield component traits across multiple environments in Upland cotton (Gossypium hirsutum L.). Mol Genet Genomics 292:1281–1306. https://doi.org/10.1007/s00438-017-1347-8
Alori ET, Babalola OO (2018) Microbial inoculants for improving crop quality and human health in Africa. Front Microbiol 9:2213. https://doi.org/10.3389/fmicb.2018.02213
Wei Z, Yang XM, Yin SX, Shen Q, Ran W, Xu YC (2011) Efficacy of Bacillus- fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Appl Soil Ecol 48:152–159. https://doi.org/10.1016/j.apsoil.2011.03.013
Joo GJ, Kim YM, Lee IJ, Song KS, Rhee IK (2004) Growth promotion of red pepper plug seedlings and the production of gibberellins by Bacillus cereus, Bacillus macroides and Bacillus pumilus. Biotech Lett 26:487–491. https://doi.org/10.1023/B:BILE.0000019555.87121.34
Yuan S, Wang L, Wu K, Shi J, Wang M, Yang X (2014) Evaluation of Bacillus- fortified organic fertilizer for controlling tobacco bacterial wilt in greenhouse and field experiments. Appl Soil Ecol 75:86–94. https://doi.org/10.1016/j.apsoil.2013.11.004
D’Hose T, Cougnon M, De Vliegher A, Vandecasteele B, Viaene N, Cornelis W, Reheul D (2014) The positive relationship between soil quality and crop production: a case study on the effect of farm compost application. Appl Soil Ecol 75:189–198. https://doi.org/10.1016/j.apsoil.2013.11.013
Sui J, Ji C, Wang X, Liu Z, Sa R, Hu Y, Wang C, Li Q, Liu X (2019) A plant growth-promoting bacterium alters the microbial community of continuous cropping poplar trees’ rhizosphere. J Appl Microbiol 126:1209–1220. https://doi.org/10.1111/jam.14194
Winding A, Binnerup SJ, Pritchard H (2004) Non- target effects of bacterial biological control agents suppressing root pathogenic fungi. FEMS Microbiol Ecol 47:129–141. https://doi.org/10.1016/S0168-6496(03)00261-7
Prévost K, Couture G, Shipley B, Brzezinski R, Beaulieu C (2006) Effect of chitosan and a biocontrol streptomycete on field and potato tuber bacterial communities. Biocontrol 51:533–546. https://doi.org/10.1007/s10526-005-4240-z
Savazzini F, Longa CMO, Pertot I, Gessler C (2008) Real- time PCR for detection and quantification of the biocontrol agent Trichoderma atroviride strain SC1 in soil. J Microbiol Methods 73:185–194. https://doi.org/10.1016/j.mimet.2008.02.004
Chowdhury SP, Dietel K, Rändler M, Schmid M, Junge H, Borriss R, Grosch R (2013) Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS ONE 8:e68818. https://doi.org/10.1371/journal.pone.0068818
Kröber M, Wibberg D, Grosch R, Eikmeyer F, Verwaaijen B, Chowdhury SP, Schlüter A (2014) Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front Microbiol 5:252. https://doi.org/10.3389/fmicb.2014.00252
Hartman K, Tringe SG (2019) Interactions between plants and soil shaping the root microbiome under abiotic stress. Portland Press Open Acces 476:2705–2724. https://doi.org/10.1042/BCJ20180615
Daniel BM, Vogel C, Bai Y, Julia AV (2016) The plant microbiota: systems-level insights and perspectives. Annu Rev Genet 50:211–234. https://doi.org/10.1146/annurev-genet-120215-034952
Akifumi S, Yoshikatsu U, Takahiro Z, Hisabumi T, Kazufumi Y (2014) Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS ONE 9:e100709. https://doi.org/10.1371/journal.pone.0100709
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8:790–803. https://doi.org/10.1038/ismej.2013.196
Zouari I, Jlaiel L, Tounsi S, Trigui M (2015) Biocontrol activity of the endophytic Bacillus amyloliquefaciens strain CEIZ-11 against Pythium aphanidermatum and purification of its bioactive compounds. Biol Control 2016:54–62. https://doi.org/10.1016/j.biocontrol.2016.05.012
Huang HC, Qiu JP (2005) Research advance in controlling plant diseases by Bacillus subtilis. J Zhejing Agric Sci. https://doi.org/10.3969/j.issn.0528-9017.2005.03.022
Funding
Our research was supported by the National Science Foundation (31260013), Regional project of Xinjiang Production and Construction Corps (2018BB044), and Microbial Resources Utilization Innovation Team in Key Field of Tarim University (TDZKCQ202001).
Author information
Authors and Affiliations
Contributions
CG and BW contributed to performing the experiments and writing the initial draft; HZ and G-cM contributed to the guidance of experimental operations; HZ contributed to financial support for this work.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conficts of interest.
Research Involving Human and Animal Participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, C., Wang, B., Ma, Gc. et al. Green Fluorescent Protein-Tagged Bacillus axarquiensis TUBP1 Reduced Cotton Verticillium Wilt Incidence by Altering Soil Rhizosphere Microbial Communities. Curr Microbiol 78, 3562–3576 (2021). https://doi.org/10.1007/s00284-021-02618-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02618-2