Abstract
Verticillium dahliae was one of the most important diseases caused Verticillium wilt of cotton. In our previous study, Bacillus axarquiensis TUBP1 was screened and found to be an antagonistic strain against V. dahliae with 43% biocontrol effect in the cotton field. In order to uncover the functional mechanism of B. axarquiensis against Verticillium wilt in cotton, the colonization of B. axarquiensis labeled with a green fluorescent protein (GFP) was investigated in cotton plants and the rhizosphere soil. Firstly, a plasmid (pHT-315) containing gfp gene was successfully transformed into wild B. axarquiensis TUBP1 and labeled a green fluorescence by electroporation, which didn’t change the bioactivity in vitro. In gnotobiotic conditions, cotton seeds were then inoculated with the gfp-labeled strain and grown in green house. Observation with a confocal laser scanning microscope and a scanning electron microscope showed that GFP-labeled B. axarquiensis TUBP1 infected cotton roots and widely distributed in epidermis, cortical parenchyma, intercellular spaces, the xylem vessels, and pith cells as well as root hair cells through cracks formed at the lateral root junctions, followed by a slow migration from roots to stems and leaves. Quantitative fluorescence and flow cytometry (FACS) approaches showed a gradual decrease in the number of TUBP1-315gfp with increasing inoculation time. However, TUBP1-315gfp levels were detectable till 45 days after planting. In contrast, no fluorescence signal was detected in the non-inoculated groups. Therefore, GFP-labeled B. axarquiensis TUBP1 exhibited colonization in different parts of cotton plants from the rhizosphere soil.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Verticillium wilt is one of the disastrous diseases affecting cotton, which seriously damages the yield and crop quality [1]. In recent years, with the rise in green and organic agriculture, the use of biological agents to control Verticillium wilt in cotton has attracted much attention. Many biocontrol products such as GB03, MBI600, Baikang and Mai Fengning have been developed [2, 3]. These products mainly including Bacillus subtilis, which control Verticillum wilt through inhibit plant pathogens directly or can also confer induced systemic resistance (ISR) in plants indirectly. Besides, biocontrol products inhibition of pathogen growth by nutrient competition. Bacillus axarquiensis TUBP1 is a Gram-positive biocontrol strain, which was isolated and identified from cotton field soil in Xinjiang, Northwestern China. Studies showed that major active compound was peptide T in B. axarquiensis TUBP1 determined by LC–MS/MS, which led to mitochondria-mediated apoptotic cell death in V. dahliae [4].The biocontrol effects of B. axarquiensis TUBP1 on V. dahliae were 43% in the cotton field [5].
Whether biocontrol bacteria colonization in plant or not is one of the main reasons for the biocontrol effects. There are many factors affecting the colonizing ability of biocontrol bacteria in plant roots, including plant type (different root excretions), soil texture, temperature, and soil moisture [6]. Studying the colonization pattern can not only reveal its microecological characteristics, but also help in evaluating the adaptability and stability of bioprotective activity in plant rhizosphere, thereby providing insights into the mechanism of biological control. As we known that Bacillus has great advantages, such as forming endospores to resist desiccation and UV irradiation, surviving in adverse conditions and producing antimicrobial metabolites to control plant pathogens, potential uses in agriculture. Plant colonization studies revealed that B. pumilus SE34, B. subtilis MB73/2 [7], B. subtilis CB-R05 [8], B. amyloliquefaciens FZB42 have different colonization patterns on various plants [9]. Conventional techniques, such as antibiotic labeling and radioisotope labeling, are normally used to investigate the colonization and biocontrol mechanisms [10]. Moreover, green fluorescent protein (GFP) has also been used to study the colonization of Gram-negative and Gram-positive bacteria including plant pathogens, diazotrophs, and different biocontrol strains [8, 11]. In order to uncover the colonization of B. axarquiensis TUBP1 in cotton plant and rhizosphere soil against V. dahliae, the gfp gene was introduced into wild B. axarquiensis TUBP1 strain by electrotransformation, and the colonization dynamics of B. axarquiensis tagged with gfp was investigated in both gnotobiotic and field systems using confocal laser scanning microscopy (CLSM), scanning electron microscope (SEM), fluorescence spectrophotometer (FL), and flow cytometry (FACS).
Materials and Methods
Strains and Growth Conditions
Bacillus axarquiensis TUBP1 wild strain was originally isolated from the soil of 12 consecutive cropping cotton fields in Alar, Xinjiang province. The strain was cultured in Luria–Bertani (LB) medium at 37 °C and 180 rpm min−1 for 48 h [12].
Evaluation of Antibiotic Resistance of B. axarquiensis TUBP1
To determine the tolerance of B. axarquiensis TUBP1 to a variety of antibiotics, the strain was cultured overnight in LB liquid medium (without any antibiotics). Then, 100 μl of B. axarquiensis TUBP1 was inoculated on a solid plate containing different concentrations of antibiotics (ampicillin, chloramphenicol, streptomycin, kanamycin, chlortetracycline, erythromycin, and apramycin). The inoculated strains were cultured overnight at 37 °C in inverted positions to observe their growth. The concentration of antibiotics used was similar to the optimum concentration gradient of the antibiotic working solution recorded in Takara Catalog (Bowie Biotech Beijing Co., Ltd.).
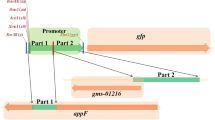
Obtaining pHT-315gfp Plasmid
Plasmid DNA extraction and verification of pHT-315gfp: DH5α carrying pHT-315gfp plasmid was grown at Zhongguo Agricultural University. DH5α competent cells were purchased at Beijing Trans Gen Biotechnology Co., Ltd. The strain was grown in LB medium containing 100 μg ml−1 of ampicillin.
The pHT-315gfp plasmid was extracted according to the following procedure. Single colony of DH5α carrying pHT-315gfp plasmid was inoculated in LB medium. Overnight culture was carried out at 37°C and 180 rpm min−1. The cultured DH5α cells were centrifuged for 10 min at 12,000×g. The precipitate obtained after centrifugation was re-suspended in 200 μl of solution I and mixed. This was followed by the addition of 400 μl of solution II, along with mild shaking. Then, 300 μl of solution III was added, mixed well by shaking, and centrifuged for 10 min at 12,000×g after milky flocculent appearance. The supernatant was absorbed in a fresh 1.5 ml EP tube, to which 5 μl of RNase and 100 μl of chloroform were sequentially added. The supernatant was mixed by oscillation and further centrifuged at 12,000×g for 10 min. This was then absorbed into another fresh 1.5 ml EP tube, and an equal volume of isopropanol was added. The supernatant was kept at – 20 °C for 30 min and centrifuged at 12,000×g for 10 min. The resulting supernatant was discarded and the pellet was washed twice with 70% ethanol. The pellet was dried and dissolved in 30 ml of sterile water. The size of pHT-315 DNA was estimated by gel electrophoresis, and the presence of the gfp gene in the plasmid was verified by double enzyme digestion. The enzyme digestion system had a total volume of 20 μl (including plasmid DNA 10 μl, Hind III 1 μl, EcoR I 1 μl, K buffer 2 μl, ddH2O 6 μl).
Bacillus axarquiensis TUBP1 Electrotransformation
The wild B. axarquiensis TUBP1 competent cells were prepared as follows. Single colony of B. axarquiensis TUBP1 was cultured in 2 ml LB medium and grown overnight at 37 °C with shaking at 180 rpm min−1. 500 μl of the overnight B. axarquiensis TUBP1 was taken into a 250 ml triangular flask containing 50 ml LB medium. This was cultured at 37 °C, with shaking at 200 g for 2–3 h. Subsequently, the bacterial cells were centrifuged at 4 °C and 5000×g for 15 min, followed by addition of a small amount of ultra-pure water (sterilized and pre-cooled). The cells were re-suspended and centrifuged. Then, 10% glycerol solution was added into each centrifuge tube and the total volume was made up to 1 ml. After suspension of the precipitate, 40 μl aliquots of the bacterial cell suspension were placed in each 1.5-ml centrifuge tube. The B. axarquiensis TUBP1 competent cells were stored at − 80 °C for future use. For electrotransformation, 2 μl of the purified pHT-315gfp plasmid was added to the competent cells in a 1-mm electrode cup. These were subjected to an electric shock, with a shock parameter of 1.8 kV for 5–6 ms. 1000 μl SOC liquid medium (room temperature) was added to the electrode cup immediately after the shock, and the cells were resuscitated for 1 h at 37 °C and shaking at 150 g. The 200 μl of transformed product was coated with 300 μg ml−1 erythromycin and cultured overnight in a 37 °C incubator after complete absorption of the bacterial solution.
Screening and Validation of Positive Clones
Single colony was selected and inoculated in LB medium containing 300 μg ml−1 of erythromycin, followed by incubation with shaking at 180 rpm min−1 for 12 h at 37 °C. Cells were collected through centrifugation (12,000×g, 30 s, room temperature) and plasmid DNA was extracted. Transformants were screened and verified with CLSM, PCR (Taq enzyme 0.5 μl, Taq enzyme buffer 2 μl, primer PF+R 2 μl, dNTPS 2 μl, DMSO 22 μl, and ddH2O 11 μl) and sequenced by Sangon Biotech (Shanghai).
Comparative Analysis of the Properties of Wild B. axarquiensis TUBP1 and B. axarquiensis TUBP1-315gfp
The transformed TUBP1-315gfp single colony was inoculated in 10 ml LB medium (containing 300 μg ml−1 erythromycin) and cultured at 37 °C, 180 rpm min−1 for 48 h. 2 ml of the bacterial suspension was aliquoted into two EP tubes. In one tube, the supernatant obtained by centrifugation was mixed in 1 ml sterile water. Then, the emission of green fluorescence from the tube on excitation at 488 nm wavelength was checked with a confocal microscope. The supernatant obtained by centrifuging the second tube of bacterial suspension was used in a dark box ultraviolet analyzer to observe whether the bacteria could emit green fluorescence.
Genetic stability of plasmids was checked by the following protocol: 10 μl TUBP1-315gfp was transferred to 10 ml LB medium (containing 300 μgml−1 erythromycin) and cultured at 37 °C and 180 rpm min−1 for 48 h. 2 ml of this bacterial solution was taken in an EP tube, centrifuged, and the supernatant was discarded. Then, 1 ml sterile water was added, mixed well, and stored at – 20 °C for reserve. The inoculation with 10 μl of TUBP1-315gfp was repeated and transferred to 10 ml LB (containing 300 μg ml−1 erythromycin) for more than 30 generations. The prepared samples were tested for the relationship between fluorescence intensity and passage times with an ultraviolet FL spectrophotometer. The inhibitory activity of B. axarquiensis TUBP1 on Verticillium-induced wilt of cotton before and after transformation was determined by the agar diffusion method.
Colonization of Cotton Rhizosphere
The tested cotton variety was upland cotton Xinluzhong 70. The cotton seeds were disinfected with 75% ethanol surface and the disinfected cotton seeds were soaked for 24 h in TUBP1-315gfp bacterial solution (108 CFU ml−1) cultured for 48 h. V. dahliae was cultured in Potato Dextrose Broth (PDB) liquid medium for 10 days, and the spores were collected and diluted with sterile water to make a pathogenic spore suspension (108 CFU ml−1). The potted soil was divided into three parts after passing through a 40-mesh sieve. The first part was added with tap water as a control group, and the second part was added with an equal volume of TUBP1 medium (108 CFU ml−1) to verify the colonization ability of wild strains on cotton plants in a natural soil environment. In the third, an equal volume of TUBP1-315gfp medium (108 CFU ml−1) was added to verify the colonization ability of the labeled strain on cotton plants in a natural soil environment. The same weight potting soil was passed through a 40-mesh sieve, and the pathogen spore suspension (108 CFU ml−1) was added. After that, the same treatment was performed to verify the colonization ability of wild strains and labeled strains on cotton plants in the environment of pathogenic soil. A total of six treatments were carried out, each treatment was repeated three times. The treated seeds were sown into soils with different treatments with 3 capsules per pot. Seeds were routinely managed after emergence. The cotton plants were regularly irrigated with TUBP1 fermentation broth, TUBP1-315gfp supernatant, and tap water, respectively. A 5–10 cm layer of soil around cotton rhizosphere was collected every 7 days, and sampled a total of 6 times. A total of 108 soil samples were obtained. The experiment was maintained in a greenhouse with a 14-h photoperiod and 22/28 °C day/night cycle. Diluted with sterile water, and 100 μl of soil suspension was absorbed to coat the LB erythromycin-resistant plate. The plate was incubated at 37 °C for 24–48 h, depending on the resistant plate. The colony morphology and fluorescence signal observed in the CLSM were used to identify and count the number of TUBP1-315gfp. Leaves, roots, and stems of cotton plants were collected and mixed with double volume of sterile water. 100 μl of this suspension was used to coat the LB erythromycin-resistant plate, and the plates were cultured at 37 °C for 48 h. The morphology of colonies grown on the resistant plates and the fluorescence signal observed in the fluorescence microscope were used to identify and count the number of TUBP1-315gfp.
Cotton Field Experiment
The tested cotton variety was upland cotton Xinluzhong 70. After the cotton emerged, 720 cotton plants were randomly selected and divided into 4 treatments. Each treatment consisted of 60 cotton plants. Each treatment was repeated 3 times. Different 106, 108, 1010 CFU/ml of B. axarquiensis TUBP1-1662gfp medium and LB medium broth (recording as T1, T2, T3, and CK) were applied to the cotton roots at the cotton seedling stage, budding stage, flowering stage, and bolling stage. The incidence of cotton Verticillium wilt was counted during the bolling stage of cotton and the biocontrol effect of TUBP1-315gfp was calculated.
Statistical Analysis
All data were analyzed in at least three independent assays, and the results were reported as means ± standard deviations. Significant differences between mean values were determined using Tukey test (P < 0.05) following a one-way ANOVA. The statistical analysis was performed using the GraphPad Prism 5 software.
Results
Antibiotic Tolerance in Wild Type B. axarquiensis TUBP1
The results of wild B. axarquiensis TUBP1 sensitivity to antibiotics are shown in Table 1, B. axarquiensis TUBP1 was found to be extremely insensitive to ampicillin and streptomycin and was sensitive to chlortetracycline. With the increasing concentration of chloramphenicol, kanamycin, erythromycin, and apramycin in the medium, B. axarquiensis TUBP1 gradually showed a sensitive state. Erythromycin was selected as the resistance gene carrying the gfp plasmid.
Validation of pHT-315gfp Gene
The full-length pHT-315gfp plasmid was 7705 bp. As shown in Fig. 1a, the size of plasmid strip conformed to the sub-length (taking into account the three forms of the plasmid) and theoretical size of the plasmid obtained by double digestion. The value of 6479 bp + 1226 bp was consistent, and confirmed that the pHT-315gfp plasmid carried gfp gene.
pHT-315gfp plasmid double enzyme digestion verification results and the wild B. axarquiensis TUBP1 was successful tagged with gfp gene by electrotransformation. Lane 1 is the result of double digestion of pHT-315gfp plasmid, lane 2 is pHT-315gfp plasmid. The enzyme used is ECOR I+ Hind III, and the expected band size in lane 8 is 6479 bp + 1226 bp. Lane 3 is the wild strain contrast, lanes 4 and 5 are the B. axarquiensis TUBP1-315gfp plasmid extraction results, lane 6 is the pHT-315gfp plasmid positive contrast, lanes 7 and 8 are the B. axarquiensis TUBP1-315gfp bacterial solution, and lane 9 is pHT-315gfp Plasmid PCR positive contrast. Lane M is Trans 2K Plus II
gfp Tagging of Wild B. axarquiensis TUBP1
Bacillus axarquiensis TUBP1 transformants expressing the pHT-315gfp plasmid were checked for suitability by four methods. First, results of extraction of the TUBP1-315gfp plasmid and colony PCR are shown in Fig. 1b. All the bands corresponding to the theoretical band size were obtained. And, compared to the control group, the pHT-315 plasmid including gfp was predominantly expressed in wild B. axarquiensis TUBP1 transformants. The transformed cells could be easily monitored by confocal microscopy due to the GFP tag (Fig. 2a–f). TUBP1-315gfp was observed to emit bright green fluorescence upon excitation at a wavelength of 488 nm, indicating that the pHT-315gfp gene was successfully transformed and expressed in wild B. axarquiensis TUBP1. The gfp expression level was estimated to be 88.57% in B. axarquiensis TUBP1 based on FACS analysis (Fig. 2f). Finally, to ensure accuracy, the sequence analysis of PCR product of B. axarquiensis TUBP1-315gfp was performed. These results were shown in Fig. 2g, the base sequence of gfp were same in B. axarquiensis TUBP1-315gfp and pHT-315gfp.
The results of monitoring the expression of pHT-315gfp in TUBP1, a, b confocal microscopy and UV irradiation of wild strains, confocal microscopy, and UV irradiation of (d) and e labeled strains. c FACS analysis results of wild strains, f FACS analysis results of labeled strains. g B. axarquiensis TUBP1-315gfp bacterial solution PCR product sequencing results
Stability of B. axarquiensis TUBP1-315gfp
The antifungal activity against V. dahliae and growth kinetics of wild B. axarquiensis TUBP1 and TUBP1-315gfp were measured by the agar diffusion method and ultraviolet (UV) spectrophotometry, respectively. Figure 3a shows that the growth rate of wild ones was higher, compared to the transformed strains. However, there was no significant difference between the two strains in the logarithmic growth phase. Figure 3a shows that the inhibition zone against V. dahliae was approximately 0.14 and 0.17 cm for B. axarquiensis TUBP1 and gfp-tagged TUBP1, respectively. The growth rate and anti-V. dahliae activity of gfp-tagged TUBP1 cells and wild-type strain were similar, indicating that the presence of pHT-315gfp did not interfere with the normal metabolism of B. axarquiensis TUBP1. The stability of plasmid pHT-315gfp in B. axarquiensis TUBP1 was analyzed by plating aliquots of the bacterial culture in LB medium in the presence or absence of antibiotics. The fluorescence intensity or gfp-phenotype in B. axarquiensis TUBP1-tagged cells decreased gradually with the increasing of incubation time. The degree of loss of pHT-315 gfp plasmid following serial subculture in the absence of antibiotics was evaluated for 64 generations in LB, where 40% of the strains were still found to express the pHT-315 gfp plasmid.
Activity comparison and colonization dynamics of B. axarquiensis TUBP1-315gfp and B. axarquiensis TUBP1. a Growth curve of wild strain and labeled strain. Determination of V. dahliae activity by B. axarquiensis TUBP1. Determination of V. dahliae activity by B. axarquiensis TUBP1-315gfp. b Determination of colony number in cotton rhizosphere soil by plate dilution method and indicating the colonization of B. axarquiensis TUBP1 and B. axarquiensis TUBP1-315gfp in cotton rhizosphere soil. c Determination of the genetic stability of pHT-315gfp in B. axarquiensis TUBP1 by the green fluorescence intensity emitted by B. axarquiensis TUBP1-315gfp. d Colonization of B. axarquiensis TUBP1-315gfp in various tissues of cotton plants, The number of colonies isolated from the tissues of cotton by dilution plate method was used to describe the colonization of B. axarquiensis TUBP1-315gfp on cotton plants
Population Dynamics of B. axarquiensis TUBP1-315 gfp in the Rhizosphere Soil of Cotton Plants
The number of B. axarquiensis TUBP1 and B. axarquiensis TUBP1-315gfp in the rhizosphere soil of cotton plants in the presence (inoculated with 6 × 108 CFU ml−1) or absence of V. dahliae was measured by the dilution plate method and fluorescence spectrophotometry, respectively. Approximately 6 × 108 CFU g−1 of B. axarquiensis TUBP1-315gfp cells were detected immediately after inoculation in rhizosphere soil with and without V. dahliae (Fig. 3b). On day 7, the abundance decreased to 106 CFU g−1 rhizosphere soils of cotton plants. At the end of 2 weeks, the cell counts in the rhizosphere peaked at about 105 CFU g−1 soil. After 1 month, the B. axarquiensis TUBP1-315gfp cells counts were about 105 CFU g−1 in the rhizosphere soil. Although the number of bacteria decreased with the increasing of the days of inoculation, the bacterial counts and fluorescence intensity of B. axarquiensis TUBP1-315gfp could be detected till 45 days after inoculating (Fig. 3c). In summary, it was also apparent that the wild-type B. axarquiensis TUBP1 counts were higher than those of B. axarquiensis TUBP1-315gfp in the rhizosphere soil of cotton plants with or without V. dahliae. B. axarquiensis TUBP1-315gfp counts were lower in the rhizosphere soil of cotton plants inoculated with V. dahliae, compared to those without V. dahliae.
Colonization Patterns of B. axarquiensis TUBP1-315gfp Strains in Cotton Plants
As for the colonization model of gfp-tagged B. axarquiensis TUBP1, cotton plant root, stem, and leaf samples were quantitatively analyzed every 7 th day after inoculation by the dilution plate method. At the 7th and 28th day after inoculation, cotton plant root, stem, and leaf samples were observed by CLSM and SEM. Figure 3d shows that the number of gfp-tagged cells colonized at root, stem, and leaf of cotton plants was about 6.1 × 107 CFU g−1 fresh weight after inoculation for 7 days. Compared with the control group, no fluorescent cells were detected in non-inoculated ones. Although fluorescent cells were detected even 45 days after planting, the concentration decreased with the increasing time from 15, 30, to 45 days. CLSM results at the 7th day after inoculation revealed that colonization occurred in the root tip and in defined regions of the elongation and differentiation zones of plant roots.
gfp-expressing B. axarquiensis TUBP1 cells were visible in the form of biofilm or at high densities in roots and root hairs of cotton, in contrast to the control group (Fig. 4a). Stem cross-sections of cotton showing colonization by B. axarquiensis TUBP1-315gfp cells were also detected after 7 days of inoculation (Fig. 4b). Few B. axarquiensis TUBP1-315gfp were observed in leaf tissues after 7 days of inoculation. Although colonization increased 48 h after inoculation with several cells being detected in the leaves, it decreased drastically 72 h after inoculation (Fig. 4c).
CLSM and SEM of B. axarquiensis TUBP1-315gfp colonization on cotton tissue surface. a–c The results of temporal changes in the colonization of B. axarquiensis TUBP1-315gfp on the surface of cotton tissues under CLSM observation. d–f The results of time-varying changes in the colonization of B. axarquiensis TUBP1-315gfp on the surface of cotton tissues under SEM observation. a, d The colonization results of cotton root tissue. b, e, the colonization results of cotton neck tissue. c, f The colonization results of cotton leaf tissue. The left half of a–f is the control group, and the right half is the treatment group
SEM results demonstrated that plants were supervised every 7 days after inoculation and B. axarquiensis TUBP1-315gfp cells were easily monitored and could be distinguished from the background of the root, stem, and leaves. One week after inoculation, B. axarquiensis TUBP1-315gfp cells were mainly found to colonize at the surface and groove of cotton root tip in the form of biofilm or microcolonies, in contrast with the control group (Fig. 4d). One month after inoculation, B. axarquiensis TUBP1-315gfp cells were still observed in the form of biofilm at the root base or in the lateral root junctions at high concentration (Fig. 4d). In the stem section, no B. axarquiensis TUBP1-315gfp cells could be detected in the control group, either 7 or 28 days after inoculation (Fig. 4e). In the treatment group, the number of TUBP1-315gfp cells in the xylem of the longitudinal section of cotton stem was higher at the 7th day after inoculation than that at 28th day after inoculation (Fig. 4e). At the leaf surface, compared to the control group, the number of bacteria around stomata of cotton leaves on the 28th day after inoculation was significantly less than that on the 7th day after inoculation (Fig. 4f).
Biocontrol Effect of B. axarquiensis TUBP1-315gfp in Cotton Field
Figure 5 shows that compared with the control group, the incidence of cotton Verticillium wilt significantly decreased and the biocontrol effect increased significantly with the increase of TUBP1-315gfp concentration applied, and the incidence of T3 treatment group was reduced by 57.63% compared with the control.
Biocontrol effect of B. axarquiensis TUBP1-315gfp on V. dahliae. a Changes in the incidence of cotton after applying B. axarquiensis TUBP1-315gfp, b Biocontrol effects of B. axarquiensis TUBP1-315gfp on V. dahliae in cotton fields. The concentration of B. axarquiensis TUBP1-315gfp administered by T1, T2, and T3 is into 106 CFU ml−1, 108 CFU ml−1, and 1010 CFU ml−1, respectively.
Discussion
The colonization played an important role in the biocontrol efficiency of various antagonistic bacteria against vascular diseases caused by soil-borne pathogens [13]. It was reported that the colonization of B. subtilis SQR9 led to control in Fusarium wilt in cucumber roots [14]. A lipopeptide-producing bacterium B. cereus F-6 colonized at rhizosphere soil could control stem and root rot disease in vanilla plants [15]. B. subtilis B96-II-GFP were detected in high concentration at root and stem and at lower concentration in the leaves of asparagus plants [16]. In our previous study, B. axarquiensis TUBP1 was shown to be a potential candidate agent against V. dahliae via peptide T-inducing mitochondrial damage and mitochondria-mediated apoptotic cell death [4]. In the cotton field, B. axarquiensis TUBP1 showed 43% biocontrol effects on the Verticillium wilt disease [5] while it remained unknown whether the strain could infect and colonize in cotton plant or not. In the present work, the gfp-tagged B. axarquiensis TUBP1 was obtained by electrotransformation of the pHT-315 plasmid carrying the gfp gene, which had a higher copy number in Bacillus, ensuring a higher luminescence intensity of the transformed strain [17]. The plasmid pHT-315 carrying gfp gene could be stably sustained in the wild B. axarquiensis TUBP1 strain for at least 64 generations of TUBP1 strain incubated in LB liquid medium without antibiotics. The pHT-315gfp plasmid was a valuable tool to label Bacillus besides many other plasmids including plasmids pHAPII-gfp in B. subtilis HJ5, pHY300-F1gfp in B. brevis DX01, and pGFP4412-gfp in B. megaterium C4 [18].
The colonization and over 45-day survival of the gfp-tagged TUBP1 strain in rhizosphere soil and different tissues of plants were found to overlap with the entire period of occurrence of Verticillium wilt in cotton. It was showed by CLSM and SEM that B. axarquiensis TUBP1-315gfp colonized the surfaces of the roots of plants in the form of biofilms and microcolonies in the differentiation zones of plant primary roots, cotton root hair zone, and cotton lateral root junctions, which was similar to other results reporting that the root section and root base of plants were easily accessible locations where colonizing bacteria tended to accumulate [16, 19]. Consistent with these studies, the highest concentration of B. axarquiensis TUBP1-315gfp cells was found at the root base of cotton plants during the whole growth phase, which reached up to 6.1 × 107 CFU g−1 fresh weight after inoculation for 7 days. The gfp-tagged B. axarquiensis TUBP1 cells could also be observed in the epidermis, the cortical parenchyma, intercellular spaces, xylem vessels, and pith cells, which maybe pass through cracks formed at the lateral root junctions, followed by a slow migration from roots to stems and leaves [15, 18]. The number of B. axarquiensis TUBP1-315gfp cells gradually decreased from root to stem and leaves along with an increase in the inoculation time length, indicating that B. axarquiensis TUBP1-315gfp strain migrated into the root system from the rhizosphere soil, and then transferred from root to stem and leaves via an unknown pathway [20]. The labeled strain did not lose its activity against Verticillium dahliae. The marked strain showed 55.21% biocontrol effect after being applied to cotton fields, which significantly reduced the incidence of cotton Verticillium wilt. In the soil–plant–microbe complicated environment, different biocontrol strains displayed different colonization patterns depending on their diverse response to components of root secretion [13, 21, 22]. The antagonistic strain of B. axarquiensis TUBP1-315gfp could form biofilms on the surface of cotton root and survive under the suppression of V. dahliae. Future research should be focused on the mechanism of B. axarquiensis TUBP1-315gfp colonization at cotton plant roots and their interaction with V. dahliae in order to uncover the detailed mechanism of bioprotection.
References
Zhang LS, Zhang N, Luo Z, Biao S (2013) Antagonistic Bacillus subtilis HJ5 controls verticillium wilt of cotton by root colonization and biofilm formation. Biol Fertil Soils 49:295–303
Zhen Y (2003) Genetically marking of natural biocontrol bacterium Bacillus subtilis strains with green fluorescent protein gene. Chin J Biotechnol 19:551
Shi YW, Li C, Yang HM, Zhang T (2017) Colonization study of gfp-tagged Achromobacter marplatensis strain in sugar beet. J Microbiol 55:267–272
Zeng H, Li T, Tian J, Zhang LL (2018) TUBP1 protein lead to mitochondria-mediated apoptotic cell death in Verticillium dahliae. Int J Biochem Cell Biol 103:35–44
Zeng H, Chen R, Luo XX, Tian J (2016) Isolation and anti-Verticillium dahliae activity from Bacillus axarquiensis TUBP1 protein. Process Biochem 51:1691–1698
Han QQ, Lü XP, Bai JP, Qiao Y, Paré PW, Wang SM, Zhang JL, Wu YN, Pang XP, Xu WB (2014) Beneficial soil bacterium Bacillus subtilis (GB03) augments salt tolerance of white clover. Front Plant Sci 5:525
Krzyzanowska D, Obuchowski M, Bikowski M, Rychlowski M, Jafra S (2012) Colonization of potato rhizosphere by GFP-tagged Bacillus subtilis MB73/2, Pseudomonas sp. P482 and Ochrobactrum sp. A44 shown on large sections of roots using enrichment sample preparation and confocal laser scanning microscopy. Sensors 12:17608–17619
Ji SH, Gururani MA, Chun SC (2014) Expression analysis of rice pathogenesis-related proteins involved in stress response and endophytic colonization properties of gfp-tagged Bacillus subtilis CB-R05. Appl Biochem Biotechnol 174:231–241
Fan B, Borriss R, Bleiss W, Wu X (2012) Gram-positive rhizobacterium Bacillus amyloliquefaciens FZB42 colonizes three types of plants in different patterns. J Microbiol 50:38–44
Kumar KVK, Yellareddygari SK, Reddy MS, Kloepper JW, Lawrence KS, Zhou XG, Sudini H, Groth DE, Raju SK, Miller ME (2012) Efficacy of Bacillus subtilis MBI 600 against sheath blight caused by Rhizoctonia solani and on growth and yield of rice. Rice Sci 19:55–63
Huang X, Zhang N, Yong X, Yang X, Shen Q (2012) Biocontrol of Rhizoctonia solani damping-off disease in cucumber with Bacillus pumilus SQR-N43. Microbiol Res (Pavia) 167:135–143
Zeng H, Yang SQ (2014) Isolation and identification of antagonistic bacteria TUBP1 against cotton Verticillium dahliae and its disease prevention effect. Cotton J 26(5):445–451 (in Chinese)
Barret M, Morrissey JP, O’Gara F (2011) Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol Fertil Soils 47:729–743
Cao Y, Zhang Z, Ling N, Yuan Y, Zheng X, Shen B, Shen Q (2011) Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol Fertil Soils 47:495–506
Zhao Q, Wang H, Zhu Z, Yu H (2015) Effect of Bacillus cereus f-6 on promoting vanilla (vanilla planifolia andrews.) plant growth and controlling stem and root rot disease. JER 6:1068–1078
Hao BQ, Ma LP, Qiao XW (2015) Quantitative analysis of the migration and accumulation of Bacillus subtilis in Asparagus officinalis. Curr Microbiol 71:357–362
Arantes O, Lereclus D (1992) Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119
Liu X, Zhao H, Chen S (2006) Colonization of maize and rice plants by strain Bacillus megaterium C4. Curr Microbiol 52:186–190
Nybroe O (1999) Green fluorescent protein-marked Pseudomonas fluorescens: localization, viability, and activity in the natural barley rhizosphere. Appl Environ Microbiol 65:4646–4651
Rengpipat S, Wongtangprasert N, Palaga T (2010) The use of green fluorescent protein as a marker for monitoring a probiotic Bacillus S11 in the black tiger shrimp Penaeus monodon. Aquac Nutr 15:297–305
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678
Dennis PG, Miller AJ, Hirsch PR (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72:313–327
Acknowledgements
Our research was supported by the National Science Foundation (31260013), Microbial Resources Utilization Innovation Team in Key Field of XinJiang Production and Construction Corps (2017CB014), and Graduate Innovation Project (TDGRI201810). Hong Zeng and Chuan-xing Wan contributed equally to this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that there have no conflicts of interest.
Research Involving Human and Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, B., Wan, Cx. & Zeng, H. Colonization on Cotton Plants with a GFP Labeled Strain of Bacillus axarquiensis. Curr Microbiol 77, 3085–3094 (2020). https://doi.org/10.1007/s00284-020-02071-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02071-7