Abstract
In the present study five potent rhizobacterial antagonists of Fusarium oxysporum f. sp. ciceris alone and in combination with Mesorhizobium (M) were evaluated for their potential to elicit the defence response reactions to reduce the total loss of plants and enhance the growth of two chickpea cultivars i.e. resistant GPF-2 and susceptible JG-41. Observations revealed that maximum phenolic, peroxidase (PO) and polyphenol oxidase (PPO) activity was induced after 30th day of germination. Maximum phenol concentration of 745.8 and 724.1 μg/gfw root tissues was recorded by Ps45 when co-inoculated with Mesorhizobium in both the varieties i.e. GPF-2 and JG-41 respectively. Isolates Ps45, Ps47 and Ps44 were found most promising to induce PO and PPO activity, in combination with Mesorhizobium and recorded superior over the fungicide with respect to negative control. Similar results were recorded for the phenylalanine ammonia lyase (PAL), maximally induced on 20th day after germination, where dual inoculation of Ps44+M and Ps45+M induced 57.0 and 54.2 nmol of cinnamic acid min−1 gfw−1 in GPF-2. However in case of JG-41, Ps45 and Ba1a exhibited highest PAL activity of 54.2 and 41.4 nmol of cinnamic acid min−1 gfw−1. Malonic aldehyde concentration in stem tissues at 30th day revealed that lipid peroxidation was effectively reduced in rhizobacterial treated plants compared to fungicide and negative control, signifying the role of antagonistic plant growth promoting rhizobacteria in reducing the stress and enhancing the plant’s defence response to reduce the disease incidence and thus improving the plant growth and yield. Moreover the dual inoculations were observed superior over the fungicide treatment as well as single inoculations in terms of growth (root/shoot length and weight), signifying the synergistic effect of screened antagonists and native Mesorhizobium in suppressing the pathogen and thereby enhancing the plant growth.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chickpea is one of the most important grain legume crops in the world, and contributes about 48% of the total pulse production in India [4]. Due to its high nutritive value (25–29% protein, 4–10% fat, 52–71% carbohydrate and 10–23% fibres, minerals and vitamins) chickpea occupy an important position in the largely vegetarian population of the country [3, 21].
Chickpea is usually attacked by Fusarium oxysporum f. sp. ciceris causing wilt, worldwide and is one of the consistent threats to this crop [33]. Fusarium wilt is prevalent in almost all chickpea-growing areas of the world, and resulted loss varies from 14 to 32% in the different states of India [15]. Even this plant disease causes yield losses up to 100% under favourable conditions in chickpea [29, 35].
The most efficient and reliable method of disease control and maximizing crop productivity worldwide to date has been the use of fungicides or resistant cultivars as part of an integrated management approach. However, the high pathogenic variability and development of resistance in different populations of F. oxysporum presents problems for sustainability of resistant cultivars, a major constraint in developing resistant cultivars [7]. The superiority of chemicals over biocontrol agents in terms of effective and quick disease control is well known however, the ill effects of chemicals on human health and environment are major limitations to application of chemical pesticides in the long run [43]. Moreover, use of fungicides is expensive and results in accumulation of toxic compounds which adversely affects the soil biota [20]. Thus, rising public concern about harmful environmental effects of agrochemicals constituted the need for greater sustainability in agriculture with alternate disease control strategies.
Plant disease suppression by soil microorganisms is a possibly effective alternative means of reducing the chemical input in agriculture [12]. Biocontrol of plant pathogenic microorganisms relies on different antagonistic traits including competition for colonization site or nutrients, production of volatile/diffusible antibiotics, enzymes and other biocidal mechanisms [19, 26, 37]. In addition to these among the widely recognized mechanisms of biocontrol mediated by PGPR, induction of systemic resistance (ISR) in host plants to a broad spectrum of pathogens has become a thrust area of research [51]. Some PGPR possess the ability to elicit induced systemic (ISR) resistance against a wide range of pathogens, nematodes and insects [39, 48]. It has been reported that ISR to a particular disease elicited PGPR partly overlaps with that of pathogen-induced systemic acquired resistance (SAR) conferring overall enhanced host resistance to that pathogen [8]. By this overlapping mechanism, PGPR induce or stimulate the production as well as activity of pathogenesis related (PR) proteins in the host plant such as peroxidase (PO), polyphenol oxidase (PPO), phenylalanine ammonia lyase (PAL), chitinases, lipoxygenases and glucanases to enhance the host plant’s self defence line and to suppress the pathogenic effect [52]. Among these, induction of PO along with PPO is regarded as one of the most important mechanism as they not only help in synthesizing lignin for the reinforcement of plant cell walls but are also involved in oxidation of certain metabolites to convert them into toxic compounds for hindering the growth of pathogens, infecting the plants. Moreover the higher phenolic content induced in plants play a key role in the defence mechanism during the exposure to various phytopathogens and even insect pests by generating free radicals and other oxidative species in plants [23]. Prior application of PGPR generally enhances the level of such pathogenesis related proteins/metabolites in the plant tissues to boost the defensive capability of the plants against the respective pathogen. So, the strategy for control of such soil borne pathogens of plants by the use of potential antagonistic microorganisms with the ability to enhance the resistance power has been the focus of intense research throughout the world. This approach is popularly known as biological control of phytopathogens and has been demonstrated to be successful in a number of host pathogen systems.
In context to this, the present investigation was carried out as an alternative strategy to chemical control, with the objective of screening potential antagonists that not only inhibit the test pathogen i.e. Fusarium oxysporum f. sp. ciceris via the production of antimetabolites but can elicit the defence response reactions in the treated plants, so that plants can escape or reduce the pathogenic effects and reduce the yield losses. For this, the most promising rhizobacterial antagonists of Foc, previously isolated and screened by in vitro trials [27] were evaluated for their potential to induce the accumulation or activity of various pathogenesis related compounds and to reduce the plant loss under Foc infected glass house conditions.
Materials and Methods
Bacterial Isolates
Five rhizobacterial isolates from chickpea rhizosphere, previously screened on the basis of antagonistic traits [28] were evaluated for their efficacy to induce various pathogenesis related compounds in plant tissues to reduce the total loss in plants treated with respective cultures, alone and alongside native Mesorhizobium, Fungicide Captan (2 g/kg seeds) and negative control as separate treatments. Mesorhizobium ciceris specific to chickpea was procured from the Department of Microbiology, Punjab Agricultural University, Ludhiana, Punjab, India.
Evaluation of Plant Growth Promoting Potential by Potent Antagonists
The selected antagonists were further evaluated for their potential to enhance the growth of the plants via production of phytohormones and iron chelating agents in vitro conditions. Indole acetic acid production in luria broth by the antagonistic isolates was performed with Van Urk Salkowski reagent using the Salkowski’s method [17]. The method of Gibberellic acid production was determined by Borrow et al. [9]. Siderophore production was detected on Chrome azurol sulphonate agar plate test [41]. Selected bacterial strains were tested by an agar assay using National Botanical Research Institute’s phosphate (NBRIP) medium for phosphate solubilization [16]. The isolates were inoculated into minimal agar medium containing 0.1% insoluble zinc oxide. Twenty four hours fresh grown bacterial isolates were spotted on the zinc containing medium and incubated at 30 °C for 48 h for the clearing zones around the colonies.
Molecular Characterization of Bacterial Antagonists
The selected bacterial antagonists were characterized by PCR amplification method using Bacillus and Pseudomonas specific primers. DNA was extracted as per the procedure followed by Kumari et al. [26].
PCR Amplification
Polymerase chain reaction (PCR) amplification of genomic 16S rRNA was carried using Bacillus and Pseudomonas genus-specific primers.
PCR reaction mixture comprised 2 units Taq DNA polymerase; 1.5 mM Mg2+; 10 × buffer; 0.2 mM each of the four dNTPs and 30 ng of bacterial extracted template DNA and 25 pmol each of the forward and reverse primers. PCR reaction consisted of 35 cycles of amplification with the conditions of denaturation at 94 °C for 1 min, 52 °C for 1 min for primer annealing, 72 °C for 1 min for elongation and final extension for 10 min.
Agarose Gel Electrophoresis
Following PCR amplification, 5 μl of the PCR product was run on 1.2% (w/v) agarose gel, using tris borate ethylene diamine tetra-acetic acid buffer (pH 8.3) at 40 V for 1.5 h. Then the gel was stained with ethidium bromide, and photographed on a UV transilluminator.
Evaluation of Antiphytopathogenic Potential of Antagonistic Rhizobacteria Under Glass House Conditions
Glass House Experiment
A poly-bag (pot) culture experiment was conducted for 50 days to study the influence of the five selected antagonists on the wilt incidence and induction of systemic resistance as enhancement in the production of pathogenesis related proteins viz. Peroxydase, Polyphenol oxidase and phenolic compounds in root tissues of treated chickpea plants (variety GPF-2 and JG-41).
Soil Preparation
Characteristic medium black clayey soil collected from different local chickpea fields of pulses research farm, Department of Plant Breeding and Genetics, Punjab Agricultural University, Ludhiana, was mixed thoroughly and autoclaved thrice at 121 °C and 15 psi pressure for one hour at 24 h intervals.
Pathogen Culture Multiplication and Soil Inoculation
Fusarium oxysporum f. sp. ciceris procured from the Department of Plant Breeding and Genetics was mass multiplied in Potato dextrose broth. Mycelial mat was used to inoculate pathogen in soil i.e. 10 g/kg of the soil. Soil was mixed thoroughly to disperse fungal hyphae and spores properly in the soil.
Bacterial Cultures and Seed Bacterization
Selected rhizobacterial cultures were inoculated at the rate 1% (with fresh 24 h grown cultures) in 100 ml of sterile nutrient broth and were incubated at for 24 h with bacterial count of 107–108 cfu/ml of the broth. The seeds of GPF-2 and JG-41 chickpea varieties were washed with 0.1% mercuric chloride followed by 70% ethanol and then repeatedly with sterile distilled water for surface sterilization. After that, seeds were soaked in selected five bacterial broth cultures (107 ml−1 broth) individually and in combination with native Mesohizobium ciceri (1:1) for 20–30 min before sowing the seeds.
Chickpea Genotypes
Seeds of two chickpea genotypes “GPF-2 (resistant) and JG-41 (susceptible)” were selected and procured from Punjab Agricultural University.
Preparation for Pot Experiment
Selected antagonists and their co-inoculation with Mesorhizobium were examined for their potential to reduce wilt incidence under the glass house conditions, using sterile soil inoculated with pathogen. The experiment was designed with 13 treatments, with 5 selected culture treatments alone and in combination with Mesorhizobium (1:1). The absolute control with pathogen free soil and untreated seeds, negative control with sick soil and untreated seeds and Fungicide treatment with sick soil and Captan treated seeds (2 g/kg seeds) were also maintained as separate treatments. Polyethylene bags (15 × 10 cm) were filled with 250 g sterilized soil inoculated with pathogen mycelial mat. Ten seeds per pot were sown and for each treatment ten replications pots were maintained. Pots were maintained by regular watering up to maturity i.e. 50 days and were examined for seedling emergence during initial 5–10 days. Wilt incidence was recorded up to maturity (till 50th day after sowing) of crop plants and reduction in disease severity was recorded as: % wilt in particular treatment − % wilt in negative control/% wilt in negative control × 100. Plant growth parameters in the terms of root/shoot length and weight were also recorded in all the treatments.
Experimental data were statistically tested by analysis of variance (ANOVA) using CPCS1 software developed by Department of Mathematics, Statistics and Physics, PAU. Differences were considered significant at the p ≤ 0.05 level. Each treatment was analysed with three replicates and standard error (SE) was calculated and expressed in mean ± SE of three replicates.
Analysis of Pathogenesis Related Proteins in Chickpea Under Glass House Conditions
At regular intervals of 10 days, three fresh and healthy plants were uprooted from each treatment. Plant samples were washed thoroughly and then enzymes and phenolics were extracted from the root tissues and were estimated spectrophotometrically.
Estimation of Phenolics
Root samples (1 g) were homogenized in 10 ml of 80% methanol for the extraction of phenolics and then the extract was agitated for 15 min at 70 °C. To 1 ml of the methanol extract, 5 ml distilled water and 250 μl of Folin–Ciocalteau reagent (1 N) were added. Then the reaction mixture was incubated at 25˚C for the development of blue colouration. Total phenolic compounds were estimated spectrophotometerically at absorbance maxima of 725 nm. The amount of phenolic was expressed as μg catechol or phenol mg−1 of fresh root tissue by preparing a standard curve of catechol. Three replications were maintained for each treatment.
Estimation of Peroxidase (PO) Activity
One gram fresh root samples (three replicates for each treatment) were homogenized in 2 ml of 0.1 M phosphate buffer (pH 7.0), and was centrifuged at 16,000 rpm at 4 °C for 15 min and the supernatant was used for PO estimation. For the reaction, to 1.5 ml of 0.05 m pyrogallol, 0.5 ml of enzyme extract and 0.5 ml of 1% H2O2 were added. The absorbance at 420 nm was recorded at 30 s intervals for 3 min [38].
Estimation of Polyphenol Peroxidase (PPO) Activity
The enzyme was extracted in 2 ml of 0.1 M sodium phosphate buffer (pH 6.5) from of the root samples (1 g) and centrifuged at 16,000 rpm for 15 min at 4 °C. For the assay mixture, to 200 µl of the enzyme extract, 1.5 ml of 0.1 M sodium phosphate buffer (pH 6.5) was added and at last 200 µl of 0.01 M catechol was added to start the reaction and the activity was expressed as changes in absorbance at 495 nm.
PO and PPO activities were expressed in mM and μM of purpurogallin produced min–1 mg–1 of fresh weight by using the molar extinction coefficient of 2.47 mM–1 cm–1 for both the enzymes [11].
Estimation of Phenylalanine Ammonia-Lyases (PAL) Activity
The enzyme was extracted in 5 ml of 10 mM sodium phosphate buffer (pH 6.0) from the fresh root tissues (1 g) and the supernatant was recovered by centrifugation at 16,000×g for 15 min at 4 °C. The reaction mixture was prepared by adding 100 μl of enzyme extract to 500 μl of 50 mM Tris HCl and 600 μl of 1 mM phenylalanine and was incubated for 60 min at room temperature. After incubation, 1.5 ml of toluene was added to the reaction mixture and vortexed for 30 s and centrifuged (1000 rpm, 5 min). Then the toluene fraction was separated and was estimated for the presence of transcinnamic acid at wavelength 290 nm against the toluene as blank and the activity of PAL was expressed as n moles of cinnamic acid min−1 mg−1 of fresh root weight [50].
Estimation of Malonic Aldehyde (MDA)
Fresh stem samples (0.2 g) were homogenized in 2 ml 0.1% (w/v) of a trichloroacetic acid (TCA) solution and were centrifuged at 10,000×g for 15 min. For the assay mixture, 250 μL of the supernatant was incubated with 750 μL TBA (0.5% in 20% TCA) for 20 min in a boiling water bath. After incubation, the reaction was stopped by immersing the reaction mixture in an ice bath and then the mixture was centrifuged at 13,000×g for 4 min, and the MDA spectrophotometrically estimated at 532 nm, was calculated as nmol of MDA g−1 of fresh weight by using the extinction coefficient of 155 mM cm−1 [32].
Results and Discussion
The selected antagonists of Fusarium oxysporum f. sp. ciceris i.e. Ba1a and Ba19, presumed to be Bacillus on the basis of blue coloured colonies on Bacillus agar and Ps45, Ps45, Ps47 as Pseudomonas with green coloured colonies on Pseudomonas agar, were further confirmed by PCR amplification reactions using specific primers for both genera. The antagonists were also tested for various mechanism implicated in direct plant growth promotion under in vitro conditions.
Molecular Identification of the Selected Antagonists
Molecular characterization using 16S-rDNA targeted genus-specific PCR primers for Bacillus genus showed amplification only in 2 of the isolates with a product yield of 618 bp (Plate 1a) [49], however positive PCR amplification with Pseudomonas specific primers for the fragments of 16S rDNA of other three isolates (initially morphologically or phenotypically identified on Pseudomonas agar medium), obtained from the pigment producing isolates on pseudomonads specific Kings B medium [25] yielded a product of about 1.5 Kb [11] (Plate 1b) confirming their identity as isolates belonging to Pseudomonas sp. Several reports for rhizobacterial isolates belonging to Bacillus sp. as promising agents for disease management due to their abundance and persistence in soil are well documented [24, 27, 30]. Bacilli are reported to produce a number of volatile, non-volatile and thermostable metabolites which are part of their inhibitory mechanism against the pathogens [44, 47]. However, among PGPR, fluorescent pseudomonads have been well recommended as biofertilizer and biocontrol agent due to their plant growth promoting and antagonistic activity [2].
Plant Growth Promotion Activity of Rhizobacterial Isolates
All the isolates were found positive for the production of plant growth hormones such as indole acetic acid (IAA), gibberellic acid and iron chelating agent, siderophores as their excreations. Studies revealed that plant growth hormones like gibberellins, IAA and cytokinin play important role in bacterial plant interactions [14]. Further they were also found efficient in zinc (Plate 2b) and phosphate solublization (Plate 2a), indicating the production and release of various organic acids responsible for the nutrient solublization, one of the mechanisms by which plant growth promoting rhizobacteria deprive the pathogen from these essential nutrients and enhance the nutrient availability to the plants [10].
Compatibility Test
Rhizobacterial antagonists were evaluated for their compatibility with Mesorhizobium ciceris (recommended culture of Department of Microbiology), specific for chickpea. The overlapping growth to each other on Yeast Mannitol agar plates was determined as compatible interaction between the paired microorganisms. All the antagonists showed positive interaction with Mesorhizobium indicating, their synergistic influence on plant growth promoting performace (Plate 3).
Elucidation of Antiphytopathogenic Potential to Reduce the Disease Severity in Chickpea
Wilt symptoms started after 30 days of sowing, with drooping, decoloured leaves and plants became almost dry and dead in negative control after 50 days. Same as the seed growth, wilt incidence was noticeably reduced by rhizobacterial isolates co-inoculated with Mesorhizobium (M). In this study total loss of plants due to loss in germination and wilt incidence, was recorded and according to observations Ps45+M treatment was most effective as inhibited minimum loss i.ie 23.3% followed by Ps47+M (26.7%) and Ba1a+M (30%) against negative control (80%) in GPF-2 (Plate 4a). However total loss in variety JG-41 90% of loss was recorded in negative control and minimal loss was recorded with Ps47+M (36.6%) (Table 1, Plate 4b). Similar to the present study, bacterial biocontrol agents have also been well explored for their potential to inhibit various other soil borne fungal pathogens. For example, seed treatment with B. thuringiensis B2 (KU158884), B. subtilis B10 (KT921327), B. amyloliquefaciens B13 (KT951658), B. amyloliquefaciens B15 (KT923051) and E. cloacae B16 (KT921429) led to total suppression of cottony rot disease in tomato plants to 20% compared to 100% disease incidence in control, where the soil was infested with S. sclerotium without any seed treatment [1]. This study emphasize on the effective role of plant growth promoting rhizobacteria to control wide range of pathogens, by various antagonistic mechanisms, so as to reduce the disease severity and enhance the germination, growth and thus yield of economically important crops.
Enhancement in Plant Growth by Rhizobacteria
Selected antagonistic rhizobacterial treatments along with Mesorhizobium were also effective in enhancing the plants growth in addition to enhancement in seedling emergence and concision of wilt incidence. Plants were observed for growth in the terms of root/shoot length and at 30th, 40th and 50th day and fresh weight at 20th (10 days before the disease emerged) and 40th day (10 days after the disease symptoms started). Treatment of Ps47+M exhibited highest shoot length in GPF-2 (28.5 ± 1.53 cm) and JG-41 (31.9 ± 1.44 cm) (Table 2, Plate 5a, b). Isolates Ps45, Ba1a and Ba19 alongside Mesorhizobium were also found effective in enhancing the shoot length as well as weight in GPF-2, compared to negative control (Table 2). In a similar report, shoot length and dry weight of chickpea after rhizobacterial inoculation increased up to 92% and 43% respectively in comparison to control Moreover seed inoculation with all isolated bacteria (Pseudomonas fluorescens and Bukholderia spp.) resulted in increased plant height and leaf numbers in chickpea [13]. Supporting reports also revealed the enhancement in plant height in different crops such as sorghum pearl millet, potato and radish plants inoculated with Azospirillum, Pseudomonas and Azotobacter strains [34, 42, 45]. Same as the shoot, root length was also maximally enhanced by Ps47 co-inoculation with Mesorhizobium in GPF-2 (13.8 ± 0.52 cm), however in JG-41 Ba1a + Mesorhizobium inhibited maximum root growth (15.0 ± 0.88 cm) (Table 3). However negative effect on root length was also recorded in some of the treatments due to severity of wilt disease. Observations related to root weight was variably different as Ps45 and Ps47+M inoculation was recorded with maximum root weight i.e. 0.91 g/plant in GPF-2, whereas in Jg-41 Ba19+M inhibited maximum root weight i.e. 0.81 g/plant, followed by Ba1a+M (0.74 g/plant) compared to negative control (0.49 g/plant) (Table 4). Landa et al. [29] report indicated that combined seed and soil treatment with P. macerans RGAF 101 at 20 °C, P. fluorescens RG 26 at 25 or 30 °C or P. fluorescens RGAF 19 at 30 °C significantly (P < 0·05) promoted chickpea growth (as determined by stem length and shoot and root weights) compared with non treated control. Enhancement in root and shoot parameters can be as a result of various plant growth hormones and solubilisation and mobilization of various essential macro and micro nutrients in the soil by these PGPR’s [10, 22].
Evaluation of Bioantagonists to Induce Phenolic Compounds Under Glass House Conditions
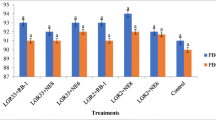
Prior application of PGPR generally enhances the level of phenolics in the plant tissues to boost the defensive capability of the plants against the respective pathogen [23]. Considering their (PGPR’s) role to reduce the pathogenic effect of Foc on the chickpea plants, the selected PGPR were further evaluated for their potential to induce phenolic compounds in chickpea root tissues. Total phenolics were estimated in relation to time i.e. at 10 days interval for 50 days after the germination (i.e. 10 days after sowing). Observations revealed highest accumulation of phenolics at 30th day after germination in both the varieties. Likewise in the seed emergence and disease control, Mesorhizobium used in combination with antagonists enhanced the stimulation effect of the phenolics in chickpea root tissues. Maximum accumulation of phenol was observed in Ps45 (745.84 μg/gfw) followed by Ba1a (673.9 μg/gfw) and Ps47 (621.4 μg/gfw) in combination with Mesorhizobium, compared to captan (507.8 μg/gfw) that was significantly higher than the negative control (484.0 μg/gfw) in chickpea variety GPF-2 (Fig. 1). Likewise in JG-41, Ps44 and Ps45 alongside Mesorhizobium, recorded with highest amount of phenolics i.e. 724.1 and 722.0 μg/gfw in the treated root tissues of chickpea plants, comparatively higher than captan treatment (472.8 μg/gfw) treated plants (Fig. 1). Seed bacterization of Ps45 without Mesorhizobium was also found efficient to induce the production and accumulation of phenolics with the value of 671.4 μg/gfw in JG-41, where seed treatment with other antagonists alone were also found at par the fungicide treatments in both the cultivars. In our previous similar study, out of the five potent antagonists of Fusarium oxysporum f. sp. ciceris, isolate 28P exhibited maximum phenolic value of 408.5 μg/mg, followed by 34P (401.9 μg/mg) and 38P (348.8 μg/mg) in comparison to negative control (189.5 μg/mg) in terms of μg of catechol mg−1 of fresh weight [26]. Phenolic accumulation was also recorded in the collar region of chickpea in the range of 886.70 to 1099.9 μg/gfw and the combined inoculation of Trichoderma and Pseudomonas along with Mesorhizobium was found most effective under Sclerotium rolfsii infected conditions [46]. As the pathogen itself induces the defence response in the plants on exposure, results presented by Patel et al.[36] revealed the highest phenol content in the roots of susceptible genotype (T15-15) of pigeon pea at preinfection stages of Fusarium udum Butler than the other resistant genotypes in contracts to the present study where the susceptible variety JG-41 was recorded with comparatively low phenolics i.e. 385.70 μg/gfw, compared to the resistant GPF-2 genotype (484.27 μg/gfw).
Influence of Bioantagonists on Peroxidase (PO) and Polyphenol Oxidase (PPO) Activity
Similar to phenolics, highest PO and PPO activity was recorded on 30th day after the germination. The PO activity was recorded significantly higher in all the treatments compared to absolute and negative control, indicating the effective role of inducing agents for active defence mechanism. In case of chickpea GPF-2 variety, highest PO activity was recorded in Ps45 (581.9 μmol min−1 mgfw−1) and Ba1a and Ps47 (467.7 μmol min−1 mgfw−1) when used in combination with native Mesorhizobium, compared to fungicide (387.8 μmol min−1 mgfw−1), however Ps45 alone was also recorded with 378.6 μmol min−1 mgfw−1 of PO activity that was found at par the fungicide treatment. On the other hand Ps47+M was maximally recorded with PO activity with 426.1 μmol min−1 mgfw−1, followed by Ps45+M (417.5 μmol min−1 mgfw−1), compared to fungicide (354.4 μmol min−1 mgfw−1) and negative control (297.57 μmol min−1 mgfw−1) (Fig. 2).
Seed bacterization with Ps47+M induced maximum PPO activity i.e. 342.5 μmol min−1 mgfw−1 followed by Ps 45+M (313.1 μmol min−1 mgfw−1) and Ps44+M (279.4 μmol min−1 mgfw−1), significantly higher than the fungicide induced PPO activity of 186.6 μmol min−1 mgfw−1 in root tissues challenged with the pathogen (Fig. 3). Somewhat similar results were found in JG-41, where Ps45, Ps44 and Ba19 alongside Mesorhizobium treated plants were recorded with highest PPO activity of 303.6, 299.5 and 232.3 μmol min−1 mgfw−1, compared to fungicide (194.0 μmol min−1 mgfw−1) and negative control (137.1 μmol min−1 mgfw−1). Likewise PO, comparatively higher PPO activity was recorded in resistant GPF-2 variety, compared to the JG-41 in corroboration to one of the the observations by Sarwar et al. [40], where the inoculation of Ascochyta rabiei although enhanced the level of PPO in all the varieties of chickpea, but comparatively higher activity was recorded in resistant varieties (CM-72 and CM-88), than the susceptible ones (6153 and PB-1). In corroboration to the present findings, Ashraf et al. [6], also reported high PPO levels in resistant chickpea cultivars than the susceptible ones, revealing the role of resistant cultivars to exhibit better interaction efficacy with surrounding microbial flora.
Induction of Phenylalanine Ammonia Lyase (PAL) Activity by Rhizobacterial Antagonists
Activity of PAL enzyme was also significantly affected by the treatments containing the rhizobaterial isolates and the effect of evaluation was recorded highly significant after 20th day of germination. PAL activity was expressed in the terms of nmol of cinnamic acid min−1 mgfw−1 as it catabolizes phenylalanine to cinnamic acid as a product, being involved in cell lignifications process, providing first line of defence for the prevention and establishment of phytopathogenic microflora [18]. Seed priming with Ps44+M, Ps45+M and Ba19 along with Mesorhizobium were evaluated with highest PAL activity with the 57.0, 54.2 and 48.3 nmol of cinnamic acid released min−1 mgfw−1 compared to fungicide (39.1 nmol cinnamic acid min−1 mgfw−1 and negative control (15.3 nmol cinnamic acid min−1 mgfw−1). However in JG-41, Ps45, Ba1a and Ba19 co-inoculated with Mesorhizobium were found to be efficient PAL inducers with the liberation of 54.2, 51.4 and 47.5 nmol of cinnamic acid min−1 mgfw−1 (Fig. 4). All these observations on PAL revealed that its production and activity needs stimulation by the prior application of pathogen or chemical fungicide or inducing PGPR that were absent in absolute control, one of the reason for the low PAL activity in that treatment of both the varieties of chickpea and mungbean. In concurrent with the present finding, Kavino et al. [24], reported a consortium of 2 pseudomonads (Pf and Py15) and a Bacillus culture (Bs16) to significantly enhance the PAL activity along with phenolics, PO, PPO and β-1,3-glucanase in treated tomato plants, compared to the negative control. Likewise phenolics and PO, seed bacterization with Methylobacterium also recorded to enhance the cinnamic acid levels in groundnut leaves during initial hours when challenged with A. niger and S. rolfsii [31].
Malonic Aldehyde (MDA) Concentration
Malon-dialdehyde or Malonic aldehyde (MDA) being a product of lipid peroxydation determines the same in the plant tissues under severe stress conditions due to the generation of reactive peroxides, toxic to the plant metabolic reactions. MDA concentration was observed on 30th day after germination when the plants were in their initial stage of development and the disease severity and the responsive induced resistance was also active in the treated chickpea plants challenged with Fusarium oxysporum f. sp. ciceris. Observation revealed that, MDA concentration was effectively reduced in rhizobacterial treated plants compared to the chemical fungicide and negative control. Moreover co-inoculation with respective Mesorhizobium also significantly reduced the MDA concentrations in the stem tissues of treated chickpea plants of both the varieties. In chickpea variety GPF-2, due to the highly active defence response Ps45 + Mesorhizobium was recorded with the lowest MDA concentration i.e. 39.77 mM gfw−1 (Fig. 5), however in JG-41, Ba19 co-inoculation with Mesorhizobium ciceris, recorded lowest lipid peroxidation with 41.4 mM gfw−1 of MDA in treated chickpea stem tissues, compared to captan treatment with 84.3 and 81.2 mM gfw−1 of MDA concentration in GPF-2 and JG-41 respectively. The plants challenged with Fusarium oxysporum f. sp. ciceris without rhizobacterial treatments recorded comparatively high levels of MDA in GPF-2 (122.0 mM gfw−1) and JG-41 (136.7 mM gfw−1) varieties (Fig. 5), probably due to higher disease severity or biotic stress in these plants, resulting in elevation of reactive peroxides to liberate MDA as a product of lipid peroxidation. The plants in absolute control were observed to exhibit relatively low MDA concentrations than the other treatments due to absence of pathogen infection, indicating the role of pathogens to induce the lipid peroxidation activity due to their pathogenic effects on the respective crops. Further the variability in MDA concentrations had also given an idea about the severity of the disease as well as the resistance efficacy of both the chickpea varieties and their positive interaction with microbial agents to induce or stimulated the defence mechanism and to reduce the respective pathogenic effects. In support, Ferraz et al. [18], reported lower MDA concentarion in Fol infected tomato plants, when sprayed with UFV 618, UFV 252 and UFV 592 antagonists. In a study Mhadhbi et al. [32], reported induction and accumulation of MDA as a result of lipid peroxidation in root nodules of test plant Medicago truncatula, when exposed to osmotic stress, where Sinorhizobium meliloti was observed to reduce the same when implied as seed inoculum. Observations revealed that lipid peroxidation in the cell wall membranes of plant tissues, include one of the mechanisms by which plants respond to various abiotic and biotic stress conditions and can be significantly reduced to low levels by the application of potential biocontrol agents.
Conclusion
In the present study, screened antagonistic isolates alone and in combination with native Mesorhizobium were evaluated for their potential to induce the plants own defence reactions. Results revealed that though antagonists alone were also effective but dual inoculation of antagonists and respective rhizobia induced high levels of pathogenesis related proteins as well as phenolics with much lower Malonic aldehyde concentrations and total loss in plants, inferring their cumulative role in enhancing the defence response in tested plants compared to the fungicide and negative control in both the chickpea varieties under glass house studies. Moreover the effect of co-inoculation on the plant growth parameters i.e. root/shoot length and weight revealed the enhanced defensive capacity of bacterized plants against the Fusarium oxysporum f. sp. ciceris to grow. On the basis of these observations, Ps 45 and Ps47 and Ba1a along with native Mesorhizobium can be used as biofungicides on the condition of their similar effectiveness under field conditions. Further investigations are focussed to even enhance the self defence mechanism in other crops by different combinations of these antagonists against different soil borne pathogens to evaluate their synergistic potential to control a broad range of pathogens to formulate various combinations of these so as to have better results against such soil borne phytopathogens.
References
Abdeljalil NOB, Vallance J, Gerbore J, Rey P, Remadi MD (2016) Bio-suppression of Sclerotinia Stem Rot of Tomato and Biostimulation of Plant Growth Using Tomato associated Rhizobacteria. J Plant Pathol Microbiol. 7(2):1–11
Ahmed M, Khanm MS (2009) Effect of insecticide-tolerant and plant growth-promoting Mesorhizobium on the performance of chickpea grown in insecticide stressed alluvial soils. J Crop Sci Biotechnol 2(4):217–226
Ali M, Kumar S (2005) Chickpea (Cicer arietinum L.) research in India, accomplishment and future strategies. Indian J Agric Sci 75(3):125–133
Anonymous (2015) Commodity profile of pulses—March 2015. Department of Agriculture and Co-operation, Ministry of Agriculture, Government of India
Ardura A, Ana LR, Vazquez EG (2013) Genetic detection of Pseudomonas spp. in commercial amazonian fish. Int J Environ Res Public Health 10:3954–3966
Ashraf SM, Khan TA, Hasan S (2005) Reaction of chickpea varieties to Macrophomina phaseolina and their effect on peroxidase activity. Pak J Bot 37(3):761–767
Bayraktar H, Dolar FS (2012) Pathogenic variability of Fusarium oxysporum f. sp. ciceris isolates from chickpea in Turkey. Pak J Bot 44(2):821–823
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406
Borrow A, Brian PW, Chester VE, Curtis PJ, Hemming HG, Henehan C, Jeffreys EG, Lloyd PB, Nixon IS, Norris GL, Radley M (1955) Gibberellic acids a metabolic product of the fungus Gibberella fujikuroi some observations on its production and isolation. J Sci Food Agric 6:340–348
Castagno LN, Estrella MJ, Sannazzaro AI, Grassano AE, Ruiz OA (2011) Phosphate solubilization mechanism and in vitro plant growth promotion activity mediated by Pantoea eucalypti isolated from Lotus tenuis rhizosphere in the Salado River Basin (Argentina). J Appl Microbiol 110:1151–1165
Chance B, Maheli AC (1995) Assay of catalases and peroxidases. Methods Enzymol 2:764–775
Compant S, Duffy B, Nowak J, Clement C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71:4951–4959
Dasgupta D, Ghati A, Sarkar A, Sengupta C, Paul G (2015) Application of plant growth promoting rhizobacteria (PGPR) isolated from the rhizosphere of Sesbania bispinosa on the growth of chickpea (Cicer arietinum L). Int J Curr Microbiol Appl Sci 4(5):1033–1042
Dobbelaere SJ, Vanderleyden Okon Y (2003) Plant growthpromoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci 22:107–149
Dubey SC, Singh SR, Singh B (2010) Morphological and pathogenic variability of Indian isolates of Fusarium oxysporum f. sp. ciceri causing chickpea wilt. Arch Phytopathol 43:174–190
Edi Premono M, Moawad AM, Vlek PLG (1996) Effect of phosphate-solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indones J Crop Sci 11:13–23
Ehmann A (1977) The Van Urk-Salkowski reagent-a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J Chromatogr 132:267–276
Ferraz HGM, Resende RS, Silveira PR, Andrade CCL, Milagres EA, Oliveira JR, Rodrigues FDÁ (2014) Rhizobacteria induces resistance against Fusarium wilt of tomato by increasing the activity of defense enzymes. Bragantia 73(3):274–283
Jha G, Anjaiah V (2006) Metabolites of rhizobacteria antagonistic towards fungal plant pathogens. Ann Microbiol 1:127–130
Jiménez-Gasco MM, Navas-Cortés JA, Jiménez-Díaz RM (2004) The Fusarium oxysporum f. sp. ciceris/Cicer arietinum pathosystem: a case study of the evolution of plant-pathogenic fungi into races and pathotypes. Int Microbiol 7(2):95–104
Jukanti AK, Gaur PM, Gowda CLL, Chibbar RN (2012) Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br J Nutr 108:11–26
Karnwal A, Kumar V (2012) Influence of plant growth promoting rhizobacteria (pgpr) on the growth of chickpea (Cicer arietinum L.). Ann Food Sci Technol 13(2):1–6
Karthikeyan M, Radhika K, Mathiyajhagan S, Bhaskaran R, Smiyappan R, Vellazhahan R (2006) Induction of phenolics and defence related enzymes in coconut (Cococs nucifera) roots treated with biocongtrol agents. Braz J Plant Physiol 18(3):367–377
Kavino M, Harish S, Kumar N, Saravanakumar D, Domodaran T, Soorianasundaram K, Samiyappan R (2007) Rhizosphere and endophytic bacteria for induction of systemic resistance of banana plantlets against bunchy top virus. Soil Biol Biochem 39:1087–1098
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorecein. J Lab Clin Med 44:301–307
Kumari P, Khanna V, Sharma P, Kumari S (2016) Allelopathic effects of native Bacillus sp. against Fusarium oxysporum causing chickpea wilt. Allelopathy J 38(1):77–90
Kumari S, Khanna V (2014) Effect of antagonistic rhizobacteria coinoculated with Mesorhizobium ciceris on control of fusarium wilt in chickpea (Cicer arietinum L.). Afric J Micro Res 8(12):1255–1265
Kumari S, Khanna V (2018) Biological management of vascular wilt of chickpea (Cicer arietinum L.) incited by Fusarium oxysporum f. sp. ciceris by antagonistic rhizobacteria co-inoculated with native Mesorhizobium. Int J Curr Microbiol Appl Sci 7(1):920–941
Landa BB, Cortés JAN, Díaz RMJ (2004) Influence of temperature on plant–rhizobacteria interactions related to biocontrol potential for suppression of fusarium wilt of chickpea. Plant Pathol 53:341–352
Leon M, Yaryura PM, Montecchia MS, Hernandez AI, Correa OS, Pucheu NL, Kerber NL, Garcia AF (2009) Antifungal activity of selected indigenous Pseudomonas and Bacillus from the soybean rhizosphere. Int J Microbiol 5:1–9
Madhaiyan M, Suresh Reddy BV, Anandham R, Senthilkumar M, Poonguzhali S, Sundaram SP, Sa T (2006) Plant growth-promoting methylobacterium induces defense responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr Microbiol 53:270–276
Mhadhbi H, Fotopoulos V, Djebali N, Polidoros AN, Aouani ME (2009) Behaviours of Medicago truncatula–Sinorhizobium meliloti symbioses under osmotic stress in relation with the symbiotic partner input: effects on nodule functioning and protection. J Agron Crop Sci 195(3):225–231
Moradi H, Bahramnejad B, Amini Siosemarde A, Allahverdipoor KH (2012) Suppression of chickpea (Cicer arietinum L.) Fusarium wilt by Bacillus subtillis and Trichoderma harzianum. Plant Omics J 5(2):68–74
Niranjan SR, Shetty NP, Shetty HS (2004) Seed bio-priming with Pseudomonas fluorescens isolates enhances growth of pearl millet plants and induces resistance against downy mildew. Int J Pest Manage 50(1):41–48
Pande S, Desai S, Sharma M (2010) Impacts of climate change on rainfed crop diseases: current status and future research needs. Natl Symp Clim Change Rainfed Agric Hyderabad 18(20):55–59
Patel NJ, Kandoliya UK, Talati JG (2015) Induction of phenol and defence-related enzymes during wilt (Fusarium udum Butler) infestation in pigeon pea. Int J Curr Microbiol Appl Sci 4(2):291–299
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moenne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361
Ramanathan A, Sammiyapan R, Vidhyasekaran P (2000) Induction of defense mechanisms in greengram leaves and suspension-cultured cells by Macrophomina phaseolina and is elicitors. J Plant Dis Protect 107:245–257
Ryu CM, Murphy JF, Reddy MS, Kloepper JW (2007) A two strain mixture of rhizobia elicits induction of systemic resistance against Pseudomonas syringae and Cucumber Mosaic virus coupled to promotion of plant growth on Arabidopsis thaliana. J Microbiol Biotechnol 17:280–286
Sarwar N, Sarwar Mand Jamil FF (2003) Role of Polyphenol oxidase and catalase in Ascochyta blight resistance in chickpea. Pak J Bot 35(1):111–115
Schwyn B, Neilands JB (1987) Universal chemical assay for detection and determination of siderophore. Anal Biochem 160:47
Shahid M, Singh A, Srivastava M, Sachan CP, Biswas SK (2011) Effect of Seed treatment on germination and vigour in chickpea. Trends Biosci 4:205–207
Sharma P (2011) Evaluation of disease control and plant growth promotion potential of biocontrol agents on Pisum sativum and comparison of their activity with popular chemical control agent-carbendazim. J Toxicol Environ Health Sci 3(5):127–138
Siddiqui ZA, Shakeel U (2009) Biocontrol of wilt disease complex of pigeon pea (Cajanus cajan (L.) Mill sp.) by isolates of Pseudomonas spp. Afr J Plant Sci 3:1–12
Sindhu SS, Suneja S, Goel AK, Parmar N, Dadarwal KR (2002) Plant growth promoting effects of Pseudomonas sp. on coinoculation with Mesorhizobium sp. Cicer strain under sterile and wilt sick soil conditions. Appl Soil Ecol 19:57–64
Singh A, Jain A, Sarma BK, Upadhyay RS, Singh HB (2014) Rhizosphere competent microbial consortium mediates rapid changesin phenolic profiles in chickpea during Sclerotium rolfsii infection. Microbiol Res 169:353–360
Singh D (2013) Exopolysaccharide producing rhizobacteria and their role in moisture stress alleviation. MSc. Thesis. Indian Agricultural Research Institute New Delhi – 110012, India p. 54
Singh UP, Sarma BK, Singh DP (2003) Effect of plant growth-promoting rhizobacteria and culture filtrate of Sclerotium rolfsii on phenolic and salicylic acid contents in chickpea (Cicer arietinum). Curr Microbiol 46:131–140
Spilker T, Coenye T, Vandamme P, LiPuma JJ (2004) PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol 42:2074–2079
Sujatha N, Ammani K (2011) The activities of the pathogenesis related proteins in Vigna mungo. Drug Invention Today 3:86–90
Walters RR, Ratsep J, Havis ND (2013) Controlling crop diseases using induced resistance: challenges for the future. J Exp Bot 5:1–19
Wang N, Liu M, Guo L, Yang X, Qiu D (2016) A Novel Protein Elicitor (PeBA1) from Bacillus amylo liquefaciens NC6 Induces Systemic Resistance in Tobacco. Int J Biol Sci 12(6):757–767
Acknowledgements
The present investigation was conducted in the Pulses section, Department of Plant Breeding and Genetics, Punjab Agricultural University. Further assistance was provided by Department of Microbiology, Punjab Agricultural University, Ludhiana, Punjab, India. First author is also thankful to Department of Science and Technology (DST), New Delhi for providing financial assistance as Doctoral (INSPIRE) fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumari, S., Khanna, V. Induction of Systemic Resistance in Chickpea (Cicer arietinum L.) Against Fusarium oxysporum f. sp. ciceris by Antagonistic Rhizobacteria in Assistance with Native Mesorhizobium. Curr Microbiol 77, 85–98 (2020). https://doi.org/10.1007/s00284-019-01805-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01805-6