Abstract
Stenotrophomonas MB339, a bacterium, which could potentially utilize aromatic compounds and tolerate different heavy metals was isolated from industrial wastewater. Subsequent experiments revealed strains ability to resist antibiotics ofloxacin, streptomycin, rifampicillin, erythromycin, ampicillin, clindamycin, and toxicants including As2+, Hg2+, Cu2+, Ni2+, Pb2+. The shotgun sequencing strategy, genome assembly and annotation uncovered specific features, which make this strain MB339 effectively promising to cope with highly contaminated conditions. This report presents isolate’s assembled genome and its functional annotation identifying a set of protein coding genes (4711), tRNA (69 genes), and rRNA (9 genes). More than 2900 genes were assigned to various Clusters of Orthologous Groups (COGs) and 1114 genes attributed to 37 different Koyoto Encyclopedia of Genes and Genomes (KEGGs) pathways. Among these annotated genes, eighteen were for key enzymes taking part in xenobiotic degradation. Furthermore, 149 genes have been assigned to virulence, disease, and defense mechanisms responsible for multidrug and metal resistance including mercury, copper, and arsenic operons. These determinants comprised genes for membrane proteins, efflux pumps, and metal reductases, suggesting its potential applications in bioremediation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heavy metals such as copper, mercury, arsenic, chromium, when present even in low concentrations, are toxic to microbial cells and other life forms [37]. However, micro-organisms of contaminated environment were found capable to resist relatively high concentration of metals by certain adapted mechanisms. These processes either involve precipitation of heavy metals into complexes, reduction by metal reductases to less toxic metal states or transported out through efflux system [32]. These efflux pumps can effectively extrude many different compounds like organic solvents, dyes, and other mono- and poly-aromatic hydrocarbons [20, 35]. Similarly resistance to antibiotics is generally due to acquired genetic elements carrying several genes, each one encoding specific proteins. These resistance determinants either work for decrease in affinity of microbial target or production of hydrolytic enzymes to inactivate antibiotics [2]. Such microbes are of great interest because of their biotechnological application in bio-mining of precious metals and for removal of heavy metals from contaminated wastewater [13].
The efficient bacterium isolated from contaminated water can minimize the content of metals. Like many others, Stenotrophomonas is ubiquitous in nature, found in extreme environments of both aquatic and terrestrial ecosystems [7, 42]. In terrestrial environments, mostly isolations have been reported from soils (industrial/agricultural), rhizosphere, and endophytic tissues [15, 43]. Stenotrophomonas species are emerging gram-negative multidrug-resistant organisms associated with bone, joint, respiratory, and urinary tract infections in humans [1]. These species possess intrinsic capabilities to resist against antibiotics and toxic metals due to the presence of enzymes and a broad spectrum of efflux pump [38, 49]. Besides being opportunistic pathogen, these strains can act as degrader of xenobiotics, as plant growth promoters and biological control agents [8, 31].
Next generation sequencing is a great step towards contemporary advancement in DNA sequencing [10]. These techniques have enabled us to refine microbial characterization and provide deep understanding of genetic modification in organisms in response to environment which they inhabit [9]. Genetic sequences particularly those of β-lactamase, chloramphenicol acetyltransferase, tetracycline efflux pump, and sulfonamide-resistant dihydropteroate synthase genes have largely been reported in multidrug-resistant Staphylococcus aureus, Pseudomonas aeruginosa, and Acinetobacter baumannii [47]. Similarly in-depth description of many interesting features related to heavy metal resistance, immobilization, and mobilization mechanisms has been made possible through whole genome sequencing (WGS).
Here, we describe the genome of Stenotrophomonas MB339 (Accession No. MSLW00000000), the sequence data of which is accessible in GenBank database. The bacterium was characterized for its abilities to tolerate multiple metals and antibiotics. Moreover, efficient utilization of different mono- and poly-aromatic hydrocarbons as sole source of carbon and energy was also shown by this strain MB339. In this study, we highlighted several genomic features that describe the adaptive response of Stenotrophomonas MB339 to heavy metals and harmful aromatic compounds.
Materials and Methods
Bacterial isolate MB339 was obtained from effluent samples collected from Hattar Industries, Pakistan. Initial isolation was carried out on mixed metal (Cr, Ni, Cd) supplemented nutrient agar plates using spread plate method. Aliquot (50 µl) of effluent samples was plated and plates were incubated at 37 °C for 24 h. Selection of the isolate was done on the basis of its maximum tolerance concentration for some selected metals. Purified each strain was grown in M9 medium supplemented with varying concentrations of metal salts CuSO4·5H2O, K2CrO4, NiCl2, CoCl2, CdCl2, HgCl2, PbCl2, ZnSO4, AgCl2, and As2O3. The maximum tolerance concentrations of each metal were recorded. Furthermore, antibiotic susceptibility test was performed using disk diffusion method [16]. The antibiotic disks used were of different concentrations i.e., Kanamycin (K 30), Ampicillin (AMP 10), Nalidixin acid (NA 30), Ofloxacin (OFX 5), Bacitracin (B 10), Chloramphenicol (C 30), Clindamycin (DA 10), Amoxillin/clavulanic (AMC 30), Erythromycin (E 15), Imepenem (IMP 10), Streptomycin (S 10), Rifampicin (RD 5), Tetracycline (TE 30), Sulfamethoxazole (SXT 25), and Tobramycin (TOB 10).
Minimal agar medium (M9) was prepared by adding Na2HPO4 (6 g), KH2PO4 (3 g), NaCl (0.5 g), NH4Cl (1 g) per liter, and pH was set to 7.04 at room temperature. After autoclaving, caseine hydrolysate (5 g), CaCl2 (0.1 M) and MgSO4 (1 M) were added. Different metal concentrations from stock solutions were supplemented in the medium. Fresh culture was streaked and growth pattern was observed after 24-h incubation at 37 °C. Experiments were carried out until strain’s maximum tolerance concentration against each metal was achieved. Similar experiments were done to assess bacterium utilization potential for aniline, phenanthrene, para nitrophenol (PNP), and pentachlorophenol (PCP).
DNA Extraction and Identification
Bacterium MB339 was cultured in LB broth at 30 °C in rotary shaker at 150 rpm. Genomic DNA was extracted and purified using standard method. Purity of genomic DNA samples (UVA260/A280) was estimated by NanoDrop 2000 UV–Vis spectrophotometer (Thermo Scientific, MA, USA). Identification of strain was achieved by 16S rRNA gene amplification using colony PCR. PCR reaction (25 µl) mixture containing template DNA (1 µl), 12.5 µl of commercially available mastermix (Go Green Matermix, Promega), 0.5 µl of each primer forward (27F 5′-AGAGTTTGATCCTGGCTCAG-3′), reverse (1492R 5′-TACGGCTACCTTGTTACGACTT-3′) and nuclease free water. The reaction was carried out in thermocycler with following conditions: initial denaturation at (i) 95 °C for 2 min, (ii) 35 cycles at 95 °C for 45 s, 56 °C for 45 s, 72 °C for 45 s, and (iii) 1 min at 72 °C. The amplified product was analyzed on 0.7% agarose gel in TAE buffer.
In order to identify the strain, PCR product was purified and reproduced for DNA sequencing. The sequence reads obtained were aligned using BLAST and phylogenetic tree was constructed using maximum likelihood algorithm by MEGA 6.0 software. The sequence was analyzed at NCBI server using BLASTn tool and aligned with sequences of related bacteria from GenBank database using ClustalW. Phylogenetic tree was constructed using Neighbor-Joining method by MEGA 6.0 software with bootstrap analysis on 1000 replicates [44].
Whole Genome Sequence, Assembly, and Annotation
Next generation whole genome shotgun sequencing of strain MB339 was performed using Illumina MiSeq sequencing platform, and the paired end reads were assembled through SPAdes 3.10 [6]. The quality of sequence assembly was verified with QUAST software [18]. Gene prediction and annotation were performed by different pipelines i.e., PGAP Prokaryotic Genome Annotation Pipeline on NCBI (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/), IMG ER Integrated Microbial Genomes Expert Review [29], RAST Rapid Annotation Subsystem Technology [5] and PATRIC [48]. The coding sequences (CDSs) were predicted for various functions using Prodigal [22]. However, rRNA and tRNA genes were located with the help of RNAmmer and tRNA-scan S.E [27, 28]. Enzymes and membrane proteins involved in metal detoxification and aromatic compounds degradation were identified using pfam [34], TIGRfam [39], and InterPro [23] protein databases.
Genome Similarity Assessment
Genome relatedness between Stenotrophomonas sp. MB339 and S. maltphilia WJ66, S. maltophilia K279a, S. maltophilia Ab55555 was assessed using whole genome sequence data of these strains. Average nucleotide identity was calculated by ANIb and ANIm softwares based on BLASTN and MUMmer algorithm using tools JspecieWS v1.2.1 [36] and digital DNA–DNA hybridization dDDH values or Genome-to-Genome distance calculator with web tool GGDC 2.0 server [4].
Furthermore, the general genome features of Stenotrophomonas sp. MB339 were compared with these three closely related genomes available in PATRIC annotation database. This comparison of orthologous coding DNA sequences CDSs shared between our strain (MB339) and. Stenotrophomonas maltophilia strain K279a, WJ66 & Ab55555 were presented as Classic Venn diagram using VennPainter v1.2.0 (https://github.com/linguoliang/VennPainter/).
Results and Discussion
Various industrial processes require certain harmful compounds as, solvents coloring and catalytic agents. Unfortunately, their higher concentrations are continuously being released into the environment. These compounds include various metal salts, phenolic, and chlorinated aromatic compounds. Such materials when released pose serious threats to human population and agriculture [19]. To overcome such conditions, many remediation activities are being practiced in different parts of the world. Among them, bioremediation is the most effective technique based on microbial systems that are applied to transform or destroy toxic materials [17].
In the present study, our main focus was to isolate bacterial strains from polluted sites that can naturally detoxify heavy metals and other xenobiotics. Among many resistant isolates, the gram-negative, aerobic bacterial strain MB339 was able to tolerate higher concentrations of toxicants including inorganic metal salts and organic compounds. Experimental results revealed varying maximum tolerance concentration of metal salts such as 1500 µg/ml Pb2+ (lead), 1000 µg/ml each of CrO42− (chromate), Ni2+ (nickel), As2+ (arsenic), upto 600 µg/ml of Cu2+ (copper), 150 to 200 µg/ml of Hg2+ (mercury), and Ag1+ (silver) in solid M9 minimal medium without glucose. Similar type of studies regarding isolation of metal (Cr, Cd, and Zn) remediating bacteria i.e., Micrococcus sp., Gemella sp., and Hafnia sp. from tannery effluents were carried out in Bangladesh [30]. There is always a strong relationship present between heavy metal and antibiotic resistances. Therefore in the present study, antibiotic susceptibility tests were also performed. Out of fifteen, strain MB339 showed resistance against six antibiotics ofloxacin, streptomycin, rifampicillin, erythromycin, ampicillin, and clindamycin. Many soil bacteria isolated from mine tailing sites were also found resistant to a few heavy metal (Hg, Zn, Ni) and antibiotics mainly beta-lactamase, streptomycin, and amoxicillin [14, 41].
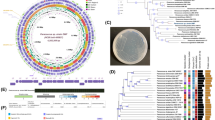
Strain MB339 was able to degrade significant amounts of aniline (1 mM), phenanthrene (6000 mg/l), PNP and PCP (100 mg/l each), on minimal medium. The growth on such compounds depicts a high potential for nitro- and chloro-substituted aromatics. Earlier researchers found bacterial species of different genera capable of consuming aliphatic and aromatic hydrocarbons isolated from polluted soil or water [40]. Isolate MB339 was identified on the basis of its 16S rRNA gene sequence. For analysis, the relevant sequences were downloaded from NCBI database. Phylogenetic tree based on these 16S rDNA partial sequences was constructed using neighbor-joining method in MEGA6 software (Fig. 1). BLASTN analysis revealed the clustering of isolate MB339 with different strains of Stenotrophomonas. The bacterium Stenotrophomonas sp. MB339 showed 99% sequence similarity with Stenotrophomonas sp. CG1. Position of bacterium MB339 in phylogenetic tree indicated close relationship with Stenotrophomonas maltophilia strains TCSS3, FQR3. Thus, MB339 was considered as a member of Stenotrophomonas genus and sequence was deposited in GenBank with accession no. KP723528.1.
Whole Genome Sequence and Phylogenetic Inference
Whole genome sequence of the strain highlighted several distinctive features related to hydrocarbon utilization. To investigate genetic makeup of this potent bacterium, its genomic DNA was sequenced using Illumina MiSeq technique. The sequence reads obtained after refining were assembled into 259 contigs by SPAdes v3.10 [5]. Final draft was of 4898925 bps with 66.2% G + C content and predicted to carry 4711 protein coding sequences (CDSs), 9 rRNA and 69 tRNA with software Prodigal v3.0 [22], RNAmmer [26] and tRNA Scan S.E [28]. The whole genome sequence was annotated by different pipelines i.e., Integrated Microbial Genome Expert Review (IMG ER), PATRIC/RAST and NCBI prokaryotic genome annotation to attain deep understanding of bacterium. The protein coding genes have been assigned to 21 different COG categories.
Average nucleotide identity (ANI) alongwith Genome–Genome Distance Calculator or digital DNA–DNA hybridization (dDDH) is an emerging genome-based criteria for establishing species identity. All the three strains S. maltophilia K279a, S. maltophilia Ab55555, S. maltophilia WJ66 showed Average Nucleotide Identity + blast (ANIb) of 97% and Average Nucleotide identity + Mummer (ANIm) of > 98% (that is above the cut-off of 95% for other strains). Similarly, dDDH value was 75% with strain MB339, which is above the cut-off value 70% for others. However, it is commonly known that an ANI threshold of 95–96% is equivalent to DDH value of 70% for species identification. Therefore, we conducted ANI and dDDH hybridization to confirm the phylogenetic identity of strain MB339 with the representative strains of Stenotrophomonas maltophilia. Hence, both ANI and dDDH results suggested that Stenotrophomonas MB339 belong to Stenotrophomonas maltophilia (Table 1).
Comparative Genome Analysis
Next generation sequencing and bioinformatics analysis have considerable influence on our understanding of microbial communities and phylogenetic relationships among different species. We compared the general features of Stenotrophomonas MB339 genome with other representative genomes S. maltophilia strains K279a, Ab55555 and WJ66 found close to this strain (Table 2).
The genome Stenotrophomonas sp. MB339 (4.8 Mbps) was larger than that of Stenotrophomonas maltophilia strains WJ66 whereas smaller than S. maltophilia Ab55555 of genome 4.9 Mbps. All these four strains have almost the same G + C content i.e., 66%. Similarly the coding sequences (CDSs) of these strains are smaller than our bacterium MB339 with variations in number of tRNA and rRNA genes. This analysis suggested that there is no drastic difference in genome size and other attributes from other strains of Stenotrophomonas maltophilia.
Venn diagram was used to compare the genome pool of these strains. A large number of conserved genes were found in all strains along with some unique feature. A considerable number of coding sequences (13726) are shared that account for almost (30–45%) of all these genomes. According to this analysis, the strain MB339 appeared to be more close to S. maltophilia strain WJ66 (2822similar genes) than to S. maltophilia Ab55555 (199 common genes). When comparison was extended to S. maltophilia K279a strain the number of shared orthologues’ dropped (Fig. 2). This analysis revealed diversity in their genetic makeup. Only small number of coding sequences were shared between our strain Stenotrophomonas sp. MB339 and representatives Stenotrophomonas maltophilia strains WJ66, K279a, Ab55555 these genes 636, 332, 72 (Fig. 2).
Characteristic Features of Stenotrophomonas sp. MB339
The annotation analysis of this genome revealed the involvement of many important genes in various metabolic pathways, possibly essential for survival in harsh environmental condition. As this bacterium was isolated from industrially polluted environment, therefore our main focus was to identify genes that provide resistance against metals and xenobiotics. Among these, 221 genes were assigned to cell wall, membrane, and envelope biogenesis, while 211 genes found associated with signal transduction. We were able to identify 18 genes playing role in degradation pathway of aromatic compounds i.e., benzoate, 1,4- and 2,4-dichlorobenzoate, toluene, xylene, styrene, naphthalene, trinitrotoluene. The genome carried comparatively more protein coding sequences involved in pathway for benzoate degradation by hydroxylation, 1, 4 & 2, 4 dichlorobenzene, toluene, xylene, tetrachloroethylene, biphenol, geraniol and DDT 1, 1-trichloro-2, 2-bis (4-chlorophenyl) ethane degradation. This increase in number of genes are indicative of Stenotrophomonas sp. MB339 adaptability of contaminated environment rather than three closely related genomes Stenotrophomonas maltophilia strains K279a, WJ66 and Ab55555 (Fig. 3).
Genetic Determinants of Specialty Genes
Specialty genes are classes of genes including antibiotic resistance, virulence factor, drug target and human homologs, important for infectious disease. Annotation using PATRIC identified many such genes in Stenotrophomonas sp. MB339 genome. Various antibiotic resistance determinants related to RND efflux pumps and membrane fusion/transporter protein were mapped in accordance with Comprehensive Antibiotic resistance database (CARD) and National Database of Antibiotic resistance (NDARO). Therefore, this MDR-resistant genome carries genes providing resistance to beta lactam compounds i.e., pencillins and carbapenems, aminoglycosides, quinolones, and isonicotinic acids. Less number of efflux pumps are normally found in environmental strains as compared to the clinical strains like the three representative S. maltophilia Ab55555, K279a, and WJ66 used in the present study (Fig. 4).
Detailed investigation of open reading frames highlighted some important genetic determinants for tolerance against heavy metal and metalloids. Such predicted regions included Hg, As, Cu, and Co/Zn/Cd gene operons and some other genes for membrane transport, efflux pumps of metal. Previously significant number of metal (Cu, Cr, As) resistance-associated genes were also reported in Klebsiella quasipneumoniae subsp. similipneumoniae MB373 isolated from this area [3]. According to research reports, these metal resistance determinants are present on operons like ars, mer, and cad representing cluster genes for membrane transporters, metallochaperons, sensors, and enzymes responsible for detoxification of arsenic, mercury, and cadmium, respectively [21, 50]. Transport system analysis was performed by comparing each predicted protein against transporter classification database (http://www.tcdb.org/). Comparably more transporter genes have been found in Stenotrophomonas sp. MB339 than S. maltophilia strain K279a and Ab55555 as illustrated in Fig. 4. Transport proteins generally belong to five different families probably associated with drug and metal resistance like ATP binding cassette (ABC), major facilitator superfamily (MSF), membrane fusion protein (MFP), and resistance nodulation cell division (RND). These systems are important for persistence in contaminated environment and help in utilization of xenobiotics. They facilitate the cells in transportation of amino acids, lipids, carbohydrates, aromatic compounds and nucleotides. The genome Stenotrophomonas sp. MB339 carry more genes for transport proteins involved in metal/drug resistance than S. maltophilia AB55555 and K279a that showed its potential to survive in environments with comparatively high metal concentration.
Genome of Stenotrophomonas sp. MB339 also carried genes related to drug resistance e.g., multidrug and toxin intrusion family efflux pump, RND, AcrB, MexC, MexD, OprJ efflux system (Supplementary File 1). Among 115 genes for cell motility were designated for chemotaxis proteins MotA, MotB, purine binding chemotaxis protein CheW and methyl accepting chemotaxis protein. For motility, pillus assembly (PilO, PilP, PilM, PilN etc) proteins, flagellar biosynthetic proteins FlhG, FlhF, FlhA were also found (Supplementary file 2).
In silico studies regarding Hg removal efficiency in bacteria revealed that merA proteins in the form of MerA-NADPH-FAD complex, not only transforms Hg2+ to Hg0 but also volatilize it [12]. Fortunately Stenotrophomonas MB339 genome possessed gene encoding MerA along with others forming a complete operon of mercury resistance. These genes were located on forward strand as merT gene for mercuric transport protein with subsequent genes merTP (mercuric ion permease family), merP (mercuric ion binding protein), merA (mercuric reductase), merD (mercuric resistance operon regulatory protein), merE (mercuric ion permease and mercuric ion transporter proteins) (Fig. 5a).
Furthermore, two consecutive gene operons supporting Co/Zn/Cd efflux system with the help of membrane fusion proteins were also a part of this genome (Fig. 5b). Cation (Co2+, Cd2+, Zn2+, and Ni2+) extrusion in microbes is driven by transmembrane efflux pumps such as P-type ATPase-mediated CzcCBA system [24]. The presence of Ni and Co resistance genes e.g., CzcD coding for transport system, has been identified in genomes of many methanogenic species that are effectively utilized in wastewater treatment systems [33].
The genes conferring copper resistance were arranged in operon comprising of multi-copper oxidase copA, one of the main component of copper resistance mechanism of gram-negative bacteria (Fig. 5c). Besides, Stenotrophomonas MB339 genome harbor sequences of copB, copC, copD, copM etc for Cu-transporting ATPases and other proteins involved in copper homeostasis. Annotation results depicted many genes coding for efflux pumps for Manganese (mnTP), Arsenic (ArsB), transporter proteins (ACR3), and arsenic reductases (arsC, arsC2) etc. While three to four genes were assigned to membrane proteins TerC for tellurium resistance. Such gene clusters (for copper, arsenate resistance proteins, multi-copper oxidases and metal reductases) were also found in genome of Pseudomonas mendocina S5.2 and Pseudomonas putida strain S13.1.2, a well known copper and heavy metal accumulator [11].
Resistance against metal and antibiotics mechanisms in micro-organisms are generally due to mutation or acquisition of mobile genetic elements [45]. For instance, there are 33 genes for mobile genetic elements with nine transposases and many other for phage components that are generally associated with horizontal gene transfer supporting multidrug/metal resistance, degradation of aromatic compounds, and stress tolerance. The potential role of such genes has been described in many prokaryotes inhabiting harsh environments [25, 46].
This study revealed that both environmental and clinical strains possess similar type of resistance determinants. As antibiotic resistance genes referring multidrug resistance transporter Bcr/CflA family, glyoxalase/bleomycin resistance protein/dioxygenase, class A, B3 beta-lactamase, polymyxin, multidrug (EmrD), fosmidomycin, polymyxin (ArnT), and portable multidrug (norM) resistance protein were also identified in genome of Stenotrophomonas MB339. However, bacterial strains of environmental origin carry more genes for efflux pumps than clinical ones. The distribution of annotated genes for antibiotic and metal resistances in the genome of Stenotrophomonas sp. MB339 is represented in Fig. 6. Briefly, these data suggested the presence of certain complex molecular mechanisms controlling gene expression that help bacteria to survive in extreme environments.
Concluding Remarks
This study concluded that genomic information obtained from annotation strongly supported the experimental data regarding antibiotic and heavy metal resistance of the bacterium. More detailed studies of extra chromosomal elements will provide insight into horizontal gene transfer of these traits. The genes for xenobiotic utilization identified in Stenotrophomonas sp. MB339 will be useful for functional genomic analysis that might result in understanding of different bioconversions. Current findings revealed the role of putative genes in developing comprehensive strategies for remediation of harmful chemicals.
References
Altimira F, Yanez C, Bravo G, Gonzalez M, Rojas L, Seeger M (2012) Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol 12:193
Alvarez-Ortega C, Olivares J, Martinez JL (2013) RND multidrug efflux pumps: what are they good for? Front Microbiol 4:1–11
Aslam F, Yasmin A, Thomas T (2016) Genome Sequence of Klebsiella quasipneumoniae subsp. similipneumoniae MB373, an effective bioremediator. Genome Announcements 4(5):e01068–e01016
Auch AF, Jan M, Klenk HP et al (2010) Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2(1):117
Aziz RK, Bartels D, Best AA et al (2008) The RAST server: rapid Annotations using Subsystems Technology. BMC Genom 9:75
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly alogorithm and its applications to single cell sequencing. J Comput Biol 19:455–477
Berg G, Roskot N, Smalla K (1999) Genotypic and phenotypic relationship between clinical and environmental isolates of Stenotrophomonas maltophilia. J Clin Microbiol 37(11):3594–3600
Caliz J, Montserrat G, Marti E et al (2012) The exposition of calcareous Mediterranean soil to toxic concentrations of Cr, Cd and Pb produces changes in microbiota mainly related to toxic differential metal bioavailability. Chemosphere 89(5):494–504
Cao Y, Fanning S, Proos S, Jordan K, Srikumar S (2017) A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front Microbiol 8:1–16
Chan KG (2016) Whole-genome sequencing in the prediction of antimicrobial resistance. Exp Rev Anti-infect Ther 14(7):617–619
Chong TM, Yin WF, Chen JW et al (2016) Comprehensive genomic and phenotypic metal resistance profile of Pseudomonas putida strain S13. 1.2 isolated from a vineyard soil. ASM Express 6(1):95–101
Dash HR, Sahu M, Mallick B et al (2017) Functional efficiency of MerA protein among diverse mercury resitance bacteria for efficient use in bioremediation of inorganic mercury. Biochimie 142:207–215
Deng H, Li XF, Cheng WD, Zhu YG (2009) Resistance and resilience of Cu-polluted soil after perturbation, tested by a wide range of soil microbial parameters. FEMS Microbiol Ecol 70:137–148
Fashola MO, Ngole-Jeme VM, Babalola OO (2016) Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. Int J Environ Res Public Health 13(11):1047
Franco AR, Calheiros CS, Pacheco CC, De Marco P, Manaia CM, Castro PM (2005) Isolation and characterization of polymeric galloyl-esterdegrading bacteria from a tannery discharge place. Microb Ecol 50:550–556
Gaudreau C, Gilbert H (1997) Comparison of disc diffusion and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni subsp. jejuni and Campylobacter coli. J Antimicrob Chemother 39(6):707–712
Ghoreishi G, Alemzadeh A, Mojarrad M, Djavaheri M (2017) Bioremediation capability and characterization of bacteria isolated from petroleum contaminated soils in Iran. Sustain Environ Res 27(4):195–202
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075
Hasnat A, Rahman I, Pasha M (2013) Assessment of environmental impact for tannery industries in Bangladesh. Int J Environ Sci Develop 4(2):217–220
Hrynkiewicz K, Baum C (2014) Application of microorganisms in bioremediation of environment from heavy metals. In: Environmental deterioration and human health. Springer, Dordrecht, pp. 215–227
Huo YY, Li ZY, Cheng H, Wang CS, Xu XW (2014) High quality draft genome sequence of the heavy metal resistant bacterium Halomonas zincidurans type strain B6T. Stand Genomic Sci 9(1):30–39
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11(1):119
Jones P, Binns D, Chang HY et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240
Karelova E, Harichova J, Stojnev T, pangallo D, Ferianc P (2011) The isolation of heavy-metal resitant culturable bacteria and resiatance determinants from a heavy-metal-contaminated site. Biologia 66(1):18–26
Koonin EV (2016) Horizontal gene transfer: essentiality and evolvability in prokaryotes and roles in evolutionary transitions. F1000Research 5:1805
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
Li XZ, Plesiat P, Nikaido H (2015) The challenge of efflux mediated antibiotics resistance in gram negative bacteria. Clin Microbiol Rev 28(2):337–418
Lowe TM, Eddy SR (1997) tRNA scan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Pillay M, Anderson I (2013) IMG 4 version of integrated microbial genomes comparative analysis system. Nucleic Acids Res 42(1):560–567
Marzan LW, Hossain M, Mina SA, Akhter Y, Chowdhury AMA (2017) Isolation and biochemical characterization of heavy-metal resistant bacteria from tannery effluents in Chittagong city, Bangladesh: bioremediation viewpoint. Egypt J Aquat Res 43(1):65–74
Meyer DD, Santestevan NA, Bucker F et al (2012) Capability of selected bacterial consortium for degrading diesel/biodiesel blends (B20): enzyme and biosurfactant production. J Environ Sci Health Part A 47(12):1776–1784
Moreno R, Rojo F (2013) The contribution of proteomics to the unveiling of survival strategies used by Pseudomonas putida in changing and hostile environments. Proteomics 13:2822–2830
Paulo LM, Ramiro-Garcia J, von Mourik S, Stams AJ, Sousa DZ (2017) Effect of nickel and cobalt on methanogenic enrichment cultures and role of biogenic sulfide in metal toxicity attenuation. Front Microbiol 8:1–12
Punta M, Coggill PC, Eberhardt RY et al (2012) The Pfam families database. Nucleic Acids Res 40:290–301
Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rajas A, Teran W, Segura A (2002) Mechanisms of solvent tolerance in Gram-negative bacteria. Annu Rev Microbiol 56:743–768
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for prokaryotic species definition. Proc Natl Acad Sci USA 106(45):19126–19131
Rodriguez-Rojas F, Tapia P, Castro-Nallar E et al (2016) Draft genome sequence of multi-metal resistant bacterium Pseudomonas putida ATH-43 isolated from Greenwich Islands, Antarctica. Front Microbiol 7:1777
Sanchez MB, Hernandez A, Martinez JL (2009) Stenotrophomonas maltophilia drug resistance. Future Microbiol 4(6):655–660
Selengut JD, Haft DH, Davidsen T et al (2007) TIGRFAMs and genome properties: tools to the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res 35:260–264
Shekhar SK, Godheja J, Modi DR (2015) Hydrocarbon bioremediation efficiency by five indigenous bacterial strains isolated from contaminated soils. Int J Curr Microbiol App Sci 4(3):892–905
Sinegani ASS, Younessi N (2017) Antibiotic resistance of bacteria isolated from heavy metal polluted soils with different land uses. J Glob Antimicrob Resist 10:247–255
Sturz AV, Matheson BG, Arsenault W, Kimpinski J, Christie BR (2001) Weeds as a source of plant growth promoting rhizobacteria in agricultural soils. Can J Microbiol 47(11):1013–1024
Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vangronsveld J, Van der Lelie D (2009) Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl Environ Microbiol 75(3):748–757
Tamura K, Skecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetic analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Tran TT, Munita JM, Arias CA (2015) Mechanisms of antibiotic resistance. Daptomycin resistance. Ann N Y Acad Sci 1354(1):32–53
Vos M, Hesselman MC, Beek TA, van Passel MW, Eyre-Walker A (2015) Rates of lateral gene transfer in prokaryotes: high but why? Trends Microbiol 23(10):598–605
Wang H, Wang J, Yu P, Ge P, Jiang Y, Xu R, Chen R, Liu X (2017) Identification of antibiotic resistance genes in multidrug-resistance Acinetobacter baumannii strain, MDR-SHH02, using whole-genome sequencing. Int J Mol Med 39(2):364–372
Wattam AR, Gabbard JL, Shukla M, Sobral BW (2014) Comparative genomic analysis at the PATRIC, a bioinformatic resource center. Methods and Protocols, Host-Bacteria Interactions. Humana Press, New York, pp 287–308
Youenou B, Favre-Bonte S, Bodilis J, Brothier E, Dubost A, Muller D, Nazaret S (2015) Comparative genomics of environmental and clinical Stenotrophomonas maltophilia strain with different antibiotic resistance profiles. Genome Biol Evol 7:2484–2505
Zhou P, Huo YY, Xu L, Wu YH, Meng FX, Wang CS, Xu XW (2015) Investigation of mercury tolerance in Chromohalobacter isrelensis DSM 6768 T and Halomonas zincidurans B6T by comparative genomics with Halomonas xinjiangensis TRM0175 T. Mar Genomics 19:15–16
Acknowledgements
This work was financially supported by Higher Education Commission of Pakistan under international research support initiative program (IRSIP). The whole genome sequence was done at Romaciotti Centre for Genomics, University of New South Wales, Sydney, Australia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nucleotide sequence accession number: This Whole genome project has been deposited into GenBank under the accession no. MSLW00000000. The version described in this paper is version MSLW01000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aslam, F., Yasmin, A. & Thomas, T. Essential Gene Clusters Identified in Stenotrophomonas MB339 for Multiple Metal/Antibiotic Resistance and Xenobiotic Degradation. Curr Microbiol 75, 1484–1492 (2018). https://doi.org/10.1007/s00284-018-1549-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1549-2