Abstract
A novel bacterium SX-49T with nitrogen-fixing capability was isolated from the rhizosphere soil of maize. Phylogenetic analysis of nifH gene fragment and 16S rRNA gene sequence revealed that the strain SX-49T is a member of the genus Paenibacillus. Values of 16S rRNA gene sequence similarity were highest between SX-49T and P. jamilae DSM 13815T (97.0%), P. brasiliensis DSM 14914T (97.8%), P. polymyxa DSM 36T (97.5%), and P. terrae DSM 15891T (98.8%). The similarity between SX-49T and other Paenibacillus species was < 97.0%. DNA–DNA hybridization values between strain SX-49T and the four type strains were P. jamilae DSM 13815T: 40.6%, P. brasiliensis DSM 14914T: 27.9%, P. polymyxa DSM 36T: 29.2%, and P. terrae DSM 15891T: 66.4%. The DNA G+C content of SX-49T was 46.4 mol%. The predominant fatty acids were anteiso-C15:0, C16:0 and iso-C16:0. The predominant isoprenoid quinone was MK-7. The genome contains 5628 putative protein-coding sequences (CDS), 6 rRNAs and 56 tRNAs. The phenotypic and genotypic characteristics, DNA–DNA relatedness, and genome features suggest that SX-49T represents a novel species of the genus Paenibacillus, and the name Paenibacillus maysiensis sp. nov. is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Paenibacillus was established by Ash et al. [1] based on the analysis of the 16S rRNA gene sequences of group 3 bacilli. Some species of the genus Paenibacillus were transferred from the genus Bacillus, and further descriptions of novel members increased the number of species of the genus Paenibacillus considerably. So far, there are > 110 now (http://www.bacterio.cict.fr/p/paenibacillus.html).
Paenibacillus species have diverse physiological characteristics. They have shown great advantage in agriculture, due to the various enzymes and antimicrobial substances they produced, such as polymyxins and bacitracins [18]. Some strains of the Paenibacillus have the ability of nitrogen fixation [3, 10, 11, 19, 20]. So far, > 20 species of Paenibacillus have been found to be nitrogen-fixers [33]. A nitrogen-fixing strain SX-49T is described in this study, representing a novel species of the genus Paenibacillus.
Materials and Methods
Isolation
A sample of maize rhizosphere soil was collected in Shaanxi province of China (34°48′N, 109°53′E). 1-g sample was suspended in 9-ml sterile water, stirred for 30 min and heated at 80 °C for 15 min. After that, 100-µl suspension was spread on nitrogen-free medium agar plates in triplicate. The nitrogen-free medium consisted 20 g sucrose, 0.1 g K2HPO4, 0.4 g KH2PO4, 0.2 g MgSO4·7H2O, 0.1 g NaCl, 0.01 g FeCl3, and 0.002 g Na2MoO4 per liter water. After incubation at 30 °C for 3 days, single colonies were isolated by streaking plating. Strains were routinely cultured in LD medium (per liter contains 2.5 g NaCl, 5 g yeast, and 10 g tryptone) at 30 °C for further identification and study.
Nitrogen-Fixing Capability
Conserved amino acid sequences within the nifH gene have been exploited to design PCR primers to detect the genetic potential for nitrogen fixation in any environment [2, 21, 26]. A 325 bp fragment of the nifH gene was amplified using two degenerate primers for the nitrogenase Fe protein gene, [forward 5′-GGCTGCGATCC(CGA)AAGGCCGA(CT)TC(CGA)ACCCG-3′, reverse 5′-CTG(GCA)GCCTTGTT(CT)TCGCGGAT(CG)GGCATGGC-3′] and sequenced as described by Ding et al. [10].
Acetylene reduction assays were carried out to test nitrogenase activity of strain SX-49T and reference Paenibacillus strains, including P. jamilae DSM 13815T, P. brasiliensis DSM 14914T, P. polymyxa DSM 36T, P. peoriae DSM 8320T, P. terrae DSM 15891T, and P. durus ATCC 35681T. Strains were cultured in nitrogen-deficient medium (per liter contains: 10.4 g Na2HPO4, 3.4 g KH2PO4, 26 mg CaCl2·2H2O, 30 mg MgSO4, 0.3 mg MnSO4, 36 mg ferric citrate, 7.6 mg Na2MoO4·2H2O, 10 µg p-aminobenzoic acid, 5 µg biotin, 4 g glucose, and 2 mM glutamate). After 24 h at 30 °C, strains were incubated under acetylene for 3 days before acetylene reduction assays were performed on GC-2010 Plus gas chromatograph (SHIMADZU, Japan) as described previously [3, 32] to measure nitrogenase activity. All treatments were in three replicates and experiments were repeated more than three times.
Genome Sequencing, Assembly, and Annotation
The draft genome sequence was produced by using Illumina paired-end sequencing technology at the BGI-Shenzhen. Assembly was conducted using SOAP de-novo v. 1.04 assembler [16]. Gene prediction was made using Glimmer v. 3.0 [9]. Annotation of protein-coding sequence was performed by using the Basic Local Alignment Search Tool (BLAST) against the COG, Kyoto Encyclopedia of Genes and Genomes (KEGG) databases and NCBI nr protein database.
Molecular Characterization
The 16S rRNA gene sequence (1518 bp) of SX-49T was acquired from a PCR product using highly specific forward primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and universal reverse primer 1492R (5′-GGTTACCTTGTTACGACTT-3′). A preliminary phylogenetic analysis was accomplished using EzTaxon database [4]. The phylogenetic tree calculating evolutionary distance matrices was constructed by the neighbor-joining method [27] using MEGA (version 7.0) [15]. Bootstrap analysis was conducted on 1000 replications [12].
Genomic DNA was extracted and purified as described by Yoon et al. [36]. Estimation of the G+C content was accomplished with Escherichia coli K12 as standard using the thermal melting protocol [8]. DNA–DNA relatedness was determined by Ziemke’s method [37].
Phenotypic Characterization
Cell morphology was obtained by scanning electrical microscopy (SEM), after incubated on endospore-forming medium agar plate [yeast extract 0.07%, tryptone 0.1%, glucose 0.1%, (NH4)2SO4, 0.02%, MgSO4·7H2O, 0.02%, K2HPO4, 0.1% (w/v), pH 7.2] for 72 h. Physiological and biochemical characteristics were determined in comparison with P. jamilae DSM 13815T, P. brasiliensis DSM 14914T, P. terrae DSM 15891T, and P. polymyxa DSM 36T. A series of experiments were performed according to Gordon et al. [13], Priest et al. [23], and Rhodes-Roberts [24], including Gram staining, nitrate reduction, production of dextrin, optimal temperature and pH for growth, activities of catalase and oxidase, Voges–Proskauer reaction, and growth inhibition by NaCl and lysozyme. Hydrolysis of casein and starch was tested by Cowan and Steel’s method [7]. Acid production was tested using medium containing 1 g (NH)2HPO4, 0.2 g MgSO4·7H2O, 0.2 g KCl, 0.2 g yeast extract, 10 g sugars or alcohols dissolved in 1 l water [31]. To confirm aerotactic ability, bacterial cells were incubated by mixing with semi-solid medium in test tubes at 40–50 °C, followed by incubation at 30 °C for 3 days.
Chemotaxonomy
Strains were incubated in LD medium at 30 °C for 2 days. The compositions of cellular fatty acid were analyzed according to the method described by Komagata and Suzuki [14] using Sherlock Identification System (MIDI) [28].
Cellular menaquinones and respiratory quinones were extracted, purified, and analyzed by HPLC according to the method described by Collins [5]. Polar lipid was extracted by Minnikin et al.’s method [22], and was identified by two-dimensional TLC by Collins et al.’s method [6]. The type of diamino acid of the cell wall peptidoglycan was determined as described by Schleifer and Kandler [29] and Komagata and Suzuki [14].
Results and Discussion
Nitrogen-Fixing Capability
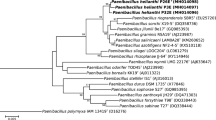
Since bacteria in the soil sample were cultured in nitrogen-free medium on the purpose of isolating nitrogen-fixing strain, strain SX-49T is possible to have nitrogen-fixing capability. To confirm genes encoding nitrogenase are contained in its genome, a 325 bp segment of the nifH gene was amplified and sequenced. Phylogenetic analysis showed that the nifH gene of strain SX-49T was clustered with species of genus Paenibacillus (Fig. 1), revealing that strain SX-49T probably is a nitrogen-fixing Paenibacillus strain. Acetylene reduction assays were performed to verify the nitrogenase activity of SX-49T. As shown in Table 1, strain SX-49T exhibited nitrogenase activity, which was relatively high among nitrogen-fixing Paenibacillus species.
Phylogenetic tree based on partial nifH sequences (292 nt fragment), compared using the neighbor-joining method, showing the position of strain SX-49T. The numbers at branching points stand for bootstrap values from 1000 replicates. Only values > 50% are shown. Bar 0.1 substitutions per nucleotide position
Molecular Characterization
A phylogenetic tree based on 16S rRNA gene sequences was constructed using the neighbor-joining method (Fig. 2). Comparison of 16S rRNA gene sequences showed that SX-49T is clustered with the species of the genus Paenibacillus. While similarities between strain SX-49T and other species of the genus Paenibacillus were below 97.0%, strain SX-49T showed high similarities of 16S rRNA gene sequence with P. jamilae DSM 13815T (97.0%), P. brasiliensis DSM 14914T (97.8%), P. polymyxa DSM 36T (97.5%), and P. terrae DSM 15891T (98.8%).
Neighbor-joining phylogenetic tree based on 16S r RNA gene sequences, positioning strain SX-49T among species of genus Paenibacillus. E. coli KCTC 2441T was used as an out group. Bootstrap analysis was performed with 1000 cycles. Only bootstrap values > 50% are shown at the branch points. Bar 0.02 substitutions per nucleotide positions
Given the high similarity between SX-49T and its closest phylogenetic relatives, DNA–DNA hybridization analysis was conducted. DNA–DNA relatedness between strain SX-49T and P. jamilae DSM 13815T, P. brasiliensis DSM 14914T, P. polymyxa DSM 36T, P. terrae DSM 15891T were 40.6, 27.9, 29.2, and 66.4%, respectively, values below 70%, indicating that strain SX-49T is a novel species [34].
The average nucleotide identity (ANI) between two genomes is the most promising method for prokaryotic species definition [25]. Since 66.4% DNA–DNA hybridization between strain SX-49T and P. terrae DSM 15891T was close to the boundary at 70%, ANI value was calculated. The ANI value between SX-49T and P. terrae HPL-003 is 85.2%; the value below 95% supporting that SX-49T is a novel species [35].
The DNA G+C content of strain SX-49T was 46.4 mol%, ranging between 39 and 54% of validly named Paenibacillus species [30].
Phenotypic Characteristics
Strain SX-49T is Gram-positive and facultatively anaerobic. Colonies on LD agar medium were round, convex, cream white, glossy, typically 1.0–2.0 mm in diameter, and uneven in margins after 72 h of incubation at 30 °C. Ellipsoidal spores were visible, positioned centrally or paracentrally in swollen sporangia (Fig. 3).
In order to determine physiological and biochemical characteristics of SX-49T in comparison with P. jamilae DSM 13815T, P. brasiliensis DSM 14914T, P. polymyxa DSM 36T, and P. terrae DSM 15891T, a series of tests were carried out following the proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria [17]. The NaCl concentration range for the growth of strain SX-49T was 0.1–4% (w/v), with optimal growth at 0.2–0.3%. The pH range for growth was 5.0–9.0, while pH 7.0 was optimal. The temperature range for growth is 10–45 °C, with optimal growth at 30 °C. Strain SX-49T was determined to be catalase negative and oxidase negative. Strain SX-49T has the ability to reduce nitrate to nitrite, positive for the Voges–Proskauer reaction, and negative for the methyl red reaction, which differentiated SX-49T from the most related P. terrae DSM 15891T .
The physiological and biochemical characteristics of SX-49T are shown in Table 2. Compared with type strains of closely related Paenibacillus species, strain SX-49T exhibited nearly identical phenotypic characteristics. However, characteristics on temperature range, hydrolysis of tween 20, and acid production from mannitol differentiated strain SX-49 from type strains of closely related species.
Chemotaxonomic Characteristics
In order to determine the composition of cellular fatty acid, SX-49T and type strains P. jamilae DSM 13815T, P. brasiliensis DSM 14914T, P. polymyxa DSM 36T, P. terrae DSM 15891T were incubated in LD medium at 30 °C for 2 days. As shown in Table 3, major fatty acid constitution of SX-49T was anteiso-C15:0 (61.3%). C16:0 (6.2%), iso-C15:0 (3.9%), iso-C16:0 (5.3), and anteiso-C17:0 (9.6%) are also important fatty acid constitution in strain SX-49T, in conformity to genus Paenibacillus (Table 3). The major menaquinone of SX-49T is MK-7, in accordance with genus Paenibacillus [30]. The polar lipids detected by two-dimensional TLC are diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG) (Supplementary Fig. 1), in agreement with the profile of genus Paenibacillus. The isomer type of diamino acid of the cell wall peptidoglycan was identified as meso-diaminopimelic acid.
Genomic Features
The draft genome of strain SX-49T has been uploaded to GenBank with accession number NZ_ASRY00000000. The genome of strain SX-49T is composed of a single circular molecule of 5.65 Mb with an average G+C content of 46.4%. Totally 5628 putative protein-coding sequences (CDS), 6 rRNAs, and 56 tRNAs were identified. The genome of strain SX-49T contains a nif gene cluster, enables it to fix nitrogen. The nif gene cluster is composed of nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA, and nifV [35].
On the basis of its molecular, phenotypic, and chemotaxonomic characteristics, strain SX-49T is considered to represent a novel species of the genus Paenibacillus, and the name Paenibacillus maysiensis sp. nov. is proposed. The type strain is SX-49T.
Description of Paenibacillus maysiensis sp. nov.
Paenibacillus maysiensis (may. si. en’ sis. L. gen. n. maysiensis of maize, where the type strain SX-49T was isolated).
Cells are motile, Gram-positive. In slightly swollen sporangia, an ellipsoidal spore is formed and located in central or paracentral position of cells. Colonies on LD medium are circular, convex, cream white, with diameter 1.0–2.0 mm. Nitrogen fixation positive. The growth temperature is 10–45 °C, optimal at 30 °C. The growth pH range is 5.0–9.0, optimal at pH 7.0. NaCl concentration of 0.1–4% (w/v) is tolerable for growth, optimal at 0.2–0.3%. Voges–Proskauer reaction is positive and nitrate can be reduced to nitrite. Methyl red reaction is negative. Substrates utilized for growth and acid production are as follows: sucrose, glucose, raffinose, l-arabinose, sodium citrate, d-trehalose, d-galactose, and maltose. Catalase negative and oxidase negative. The major menaquinone is MK-7. The predominant fatty acid is anteiso-C15:0. The major polar lipids are DPG, PE, and PG. Meso-diaminopimelic is the diagnostic diamino acid in the cell wall. The DNA G+C content of type strain SX-49T is 46.4 mol%. The genome consists of a single circular molecule of 5.7 Mb with totally 5628 putative protein-coding sequences (CDS), 6 rRNAs, and 56 tRNAs identified.
The type strain, SX-49T (=CGMCC 1.15332 =JCM 31028), was isolated from rhizosphere soil sample of maize, acquired in Shaanxi Province, P. R. China (34°48′N, 109°53′E). The GenBank (EMBL) accession number for the 16S rRNA gene sequence is JN873138. The Digital Protologue Taxon Number is TA00403.
References
Ash C, Priest F, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64(3):253–260
Auman AJ, Speake CC, Lidstrom ME (2001) nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl Environ Microbiol 67(9):4009–4016
Berge O, Guinebretiere MH, Achouak W, Normand P, Heulin T (2002) Paenibacillus graminis sp. nov. and Paenibacillus odorifer sp. nov., isolated from plant roots, soil and food. Int J Syst Evol Microbiol 52(2):607–616. https://doi.org/10.1099/00207713-52-2-607
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57(10):2259–2261. https://doi.org/10.1099/ijs.0.64915-0
Collins MD (1985) Analysis of isoprenoid quinones. Methods Microbiol 18(08):329–366
Collins MD, Goodfellow M, Minnikin DE (1980) Fatty acid, isoprenoid quinone and polar lipid composition in the classification of Curtobacterium and related taxa. J Gen Microbiol 118(1):29–37. https://doi.org/10.1099/00221287-118-1-29
Cowan ST, Steel KJ (1966) Manual for the identification of medical bacteria. Proc R Soc Med 59(5):468
De Ley J, Park IW, Tijtgat R, Van Ermengem J (1966) DNA homology and taxonomy of Pseudomonas and Xanthomonas. J Gen Microbiol 42(1):43–56. https://doi.org/10.1099/00221287-42-1-43
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23(6):673–679. https://doi.org/10.1093/bioinformatics/btm009
Ding Y, Wang J, Liu Y, Chen S (2005) Isolation and identification of nitrogen-fixing bacilli from plant rhizospheres in Beijing region. J Appl Microbiol 99(5):1271–1281. https://doi.org/10.1111/j.1365-2672.2005.02738.x
Elo S, Suominen I, Kampfer P, Juhanoja J, Salkinoja-Salonen M, Haahtela K (2001) Paenibacillus borealis sp. nov., a nitrogen-fixing species isolated from spruce forest humus in Finland. Int J Syst Evol Microbiol 51(2):535–545. https://doi.org/10.1099/00207713-51-2-535
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Gordon RE, Haynes WC, Pang HN (1973) The genus Bacillus. Agricultural Research Service, U.S. Dept. of Agriculture: for sale by the Supt. of Docs., U.S. Govt. Print. Off.
Komagata K, Susuki K (1987) Lipid and cell-wall systematics in bacterial systematics. Methods Microbiol 19:161–207
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24(5):713–714. https://doi.org/10.1093/bioinformatics/btn025
Logan NA, Berge O, Bishop AH, Busse HJ, De Vos P, Fritze D, Heyndrickx M, Kampfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59(8):2114–2121. https://doi.org/10.1099/ijs.0.013649-0
Ma M, Wang C, Ding Y, Li L, Shen D, Jiang X, Guan D, Cao F, Chen H, Feng R, Wang X, Ge Y, Yao L, Bing X, Yang X, Li J, Du B (2011) Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. J Bacteriol 193(1):311–312. https://doi.org/10.1128/jb.01234-10
Ma Y, Xia Z, Liu X, Chen S (2007) Paenibacillus sabinae sp. nov., a nitrogen-fixing species isolated from the rhizosphere soils of shrubs. Int J Syst Evol Microbiol 57(Pt 1):6–11. https://doi.org/10.1099/ijs.0.64519-0
Ma Y, Zhang J, Chen S (2007) Paenibacillus zanthoxyli sp. nov., a novel nitrogen-fixing species isolated from the rhizosphere of Zanthoxylum simulans. Int J Syst Evol Microbiol 57(Pt 4):873–877. https://doi.org/10.1099/ijs.0.64652-0
Mehta MP, Butterfield DA, Baross JA (2003) Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl Environ Microbiol 69(2):960–970
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47(1):87–95
Priest FG, Goodfellow M, Todd C (1988) A numerical classification of the genus Bacillus. J Gen Microbiol 134(7):1847–1882. https://doi.org/10.1099/00221287-134-7-1847
Rhodes-Roberts M (1981) The taxonomy of some nitrogen-fixing Bacillus species with special reference to nitrogen fixation. In: Berkeley RCW, Goodfellow M (eds) The aerobic endospore-forming bacteria classification and identification. Academic Press, London, pp 315–335
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106(45):19126–19131. https://doi.org/10.1073/pnas.0906412106
Rosch C, Mergel A, Bothe H (2002) Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl Environ Microbiol 68(8):3818–3829
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sasser M, Kunitsky C, Jackoway G, Ezzell JW, Teska JD, Harper B, Parker S, Barden D, Blair H, Breezee J, Carpenter J, Cheek WV, DeMartino M, Evans B, Ezzell JW, Francesconi S, Franko E, Gardner W, Glazier M, Greth K, Harper B, Hart T, Hodel M, Holmes-Talbot K, Hopkins KL, Iqbal A, Johnson D, Krader P, Madonna A, McDowell M, McKee ML, Park M, Parker S, Pentella M, Radosevic J, Robison RA, Rotzoll B, Scott K, Smith M, Syed N, Tang J, Teska JD, Trinh H, Williams LI, Wolcott M (2005) Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J AOAC Int 88(1):178–181
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36(4):407
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47(2):289–298. https://doi.org/10.1099/00207713-47-2-289
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16(3):313–340
Wang L, Liu LZ, Zhao Z, Liu D, Zhang X, Xie B, Hong J, Li Y, Chen P, Dixon S, Li R J (2013) A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet 9(10):e1003865. https://doi.org/10.1371/journal.pgen.1003865
Wang LY, Li J, Li QX, Chen SF (2013) Paenibacillus beijingensis sp. nov., a nitrogen-fixing species isolated from wheat rhizosphere soil. Antonie Van Leeuwenhoek 104(5):675–683. https://doi.org/10.1007/s10482-013-9974-5
Wayne LG, Brenner DJ, Colwell R, Grimont P, Krichevsky M, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evolut Microbiol 37:463–464. https://doi.org/10.1099/00207713-37-4-463
Xie JB, Du Z, Bai L, Tian C, Zhang Y, Xie JY, Wang T, Liu X, Chen X, Cheng Q, Chen S, Li J (2014) Comparative genomic analysis of N2-fixing and non-N2-fixing Paenibacillus spp.: organization, evolution and expression of the nitrogen fixation genes. PLoS Genet 10(3):e1004231. https://doi.org/10.1371/journal.pgen.1004231
Yoon JH, Kim H, Kim SB, Kim HJ, Kim WY, Lee ST, Goodfellow M, Park YH (1996) Identification of Saccharomonospora strains by the use of genomic DNA fragments and rRNA gene probes. Int J Syst Bacteriol 46(2):502–505
Ziemke F, Höfle MG, Lalucat J, Rossello-Mora R (1998) Reclassification of Shewanella putrefaciens Owen’s genomic group II as Shewanella baltica sp. nov. Int J Syst Bacteriol 48(1):179–186. https://doi.org/10.1099/00207713-48-1-179
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest is declared.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Ts., Xie, Jy., Wang, Ly. et al. Paenibacillus maysiensis sp. nov., a Nitrogen-Fixing Species Isolated from the Rhizosphere Soil of Maize. Curr Microbiol 75, 1267–1273 (2018). https://doi.org/10.1007/s00284-018-1519-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1519-8