Abstract
This study first described the composition and characteristics of culturable endophytic bacteria isolated from wild alpine-subnival plant species growing under extreme environmental conditions (i.e., on the border of a glacier with frequently fluctuating and freezing temperatures, strong wind, and high ultraviolet radiation). Using a cultivation-dependent approach and 16S rRNA gene amplification techniques, 93 bacterial isolates showing different phenotypic properties were obtained from 20 different subnival plant species, of which gram-positive bacteria (61.5%), psychrotolerant bacteria (67.3%), and pigmented isolates (70.9%) accounted for a large proportion. All these characteristics of endophytes were closely related to the survival environment of their host plants and were in good agreement with microbes occurring in other cold environments. Phylogenetic analysis indicated that the endophytic isolates consisted of five phylogenetic groups comprising α-proteobacteria, γ-proteobacteria, the high G+C content gram-positive bacteria, the low G+C content gram-positive bacteria, and Flavobacterium–Bacteroides–Cytophaga. The largest generic diversity was found in the HGC group, while Clavibacter, Agreia, Rhodococcus, Sphingomonas, and Pseudomonas were the most prevalent genera. Of all isolates, 46.4% showed a high sequence similarity (98–100%) to strains discovered from other cold environments such as glaciers, tundra, and polar seas. Furthermore, 36.4% of the isolates produced Indole-3-acetic acid and 76.3% were able to solubilize mineral phosphate, which revealed that endophytic bacteria with multiple physiological functions were abundant and widespread in subnival plants. These results are essential for understanding the ecological roles of endophytes and as a foundation for further studying the interactions with plants and environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacteria that reside within plant tissues without doing any harm to the plant are termed endophytic bacteria, and can be isolated from surface-disinfected tissues or extracted from internal plant tissues [20, 21]. These endophytic microorganisms can reside within cells [17], in the intercellular spaces [37] or in the vascular system [7], where their concentrations can vary between 2.0 and 6.0 log10/g fresh weight (FW). Being in a very close association with cells of the host plant, beneficial endophytic bacteria increasingly get scientific and commercial interest for their potential to improve plant quality, e.g., by growth promotion [6, 18, 31, 33], biological control of plant diseases and pests [2, 9], nitrogen fixation [12], and phytoremediation [5, 14]. In addition, interactions between endophytes and their host plant have also been researched in recent years [30].
However, studies mentioned above mainly focused on agricultural crops, economically important crops, and some trees [18, 24, 27, 35]. Of these, cotton, potato, rice, wheat, and sweet corn were extensively studied, whereas plant species growing in extreme conditions and having important officinal values such as Saussurea involucrata (snow lotus), which is a typical alpine plant and also a rare and precious officinal plant growing on the crags of high-altitude mountains covered with snow [10, 34], have not been investigated with respect to their associated microbial community. Very little is known about the diversity, structure, and functions of endophytes within alpine-subnival plants that grow on the border of glacier and can survive under the conditions of frequently fluctuating and freezing temperatures, strong wind, and intense ultraviolet (UV) radiation [1].
The objective of this study was to: (1) isolate endophytes from alpine-subnival plants growing in the freeze–thaw tundra on glacier margins in the Tianshan Mountains, where the average temperature is below zero during the growth period from June to September, using a conventional culture method, and determine physiological characteristics of the isolates; (2) analyze the abundance, diversity, and community structure of bacterial endophytes in this environment by 16S rRNA gene amplification techniques and make comparisons with microbes from other temperate and cold environments; and (3) evaluate the potential ability of isolates to promote plant growth. To our knowledge, this study is the first to describe indigenous endophytic bacteria within alpine-subnival plants, which is essential not only for understanding their ecological roles and the interaction with plants, but also for revealing some phenomena and relationships among glacier, tundra (or soil), plant, microorganisms, and environment.
Materials and Methods
Growth Conditions and Sample Collection of the Alpine-Subnival Plants
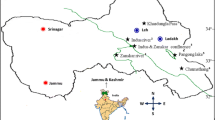
All samples used in this study were collected from an ice-free cirque (43°05′N, 86°49′E; 3800–3900 m) near the No. 1 glacier in the source area of the Urumqi River in the Tianshan Mountains, Xinjiang, China (Fig. 1), where the annual average temperature is less than 5°C during the day and −4°C at night. The temperature fluctuates from almost 4 to −10°C during the favorable growth period, and the plants are able to survive even when covered by snow. In addition, those subnival plants are also subjected to strong wind and high UV radiation [3].
A total of 20 alpine-subnival plant species were sampled in August 2005 (Table 1), of which 4–5 healthy individuals per species were sampled. The plants showed substantial dwarfism, generally being only 3–5 cm in height, and the leaves were usually glomerate; most species had showy or colorful flowers due to the influence of the extremely harsh climate and strong UV radiation.
We collected all individuals’ leaves and stems of each plant species and put them together to survey the presence of endophytic bacteria with consideration of the above-mentioned morphological features of the subnival plants.
Isolation and Physiological Characteristic Determination of Endophytic Bacteria
The isolation and culture conditions of endophytes in subnival plants, including the efficiency of surface-sterilization (which was verified by placing both sterilized tissue pieces and aliquots of water from the final rinse onto plates), growth conditions, incubation temperature, and medium type (e.g., TSA, NA, and R2A), were explored and optimized prior to isolating endophytes from all samples. Only those materials having no contaminants were selected and used for further isolation. The types and population size of isolates were optimal when cultured on TSA medium at 20–25°C for most isolates, so these culture conditions were applied to all isolates. The healthy plant materials (leaves and stem) were surface-sterilized in 70% ethanol solution for 40 s, then rinsed in 3% sodium hypochlorite (NaClO) containing 0.1% Tween20 for 2–3 min, followed by five washes in sterile water. All samples were homogenized with a sterile mortar and pestle, serially diluted with sterile 0.85% NaCl, and plated onto TSA medium. Plates were incubated for 5–7 days at 20–25°C. Colony-forming units (cfu) were counted and the similar bacterial isolates were divided into groups on the basis of morphological characteristics (shape, pigmentation, margin, surface, and texture), the representatives of which were selected for RFLP analysis.
In addition, the Gram reactions were performed and the growth ability at different temperatures including −2, 0, 4, 10, 15, 25, 37, and 45°C were determined.
Screening for IAA-Producing and Mineral Phosphate Solubilizing Endophytic Bacteria
Indole-3-acetic acid (IAA) production was determined using a modification of the qualitative method developed by Bric et al. [8]. Four strains per plate were plated onto TSA medium amended with 5 mM l-tryptophan, overlaid with a nitrocellulose membrane, and incubated at 25°C for 48 h. After bacterial growth the membrane was removed from the plate and treated with Salkowski reagent (2% 0.5 M FeCl3 in 35% perchloric acid) for 15 min at room temperature. Bacteria producing IAA were identified by the presence of a red halo on the membrane corresponding to the position of the IAA-producing colony.
Mineral phosphate solubilization activity was assayed according to references [22, 36]. The isolates were plated onto agar medium containing inorganic phosphate and incubated at 25°C for up to 48 h, solubilization of mineral phosphate being characterized by a clear halo around bacterial colonies with phosphate solubilization capacity.
DNA Extraction and 16S rRNA Gene Amplification
Genomic DNA of bacteria was isolated by using a CTAB method [19]. Amplification of the 16S rRNA gene was performed in a 50 μl final volume containing 1 μl of total genomic DNA, 0.2 μM of 27f primer (5′–3′ AGA GTT TAG TCC TGG CTC AG), 0.2 μM of 1492r primer (5′–3′ TAC GGC TAC CTT GTT ACG ACT T), 200 μM of each dNTP, 5 μl of 10× buffer, and 2 U of Taq DNA polymerase. A negative control (PCR mixture without DNA) was included in all PCR amplifications. The reaction procedure was as follows: one cycle of 5 min at 94°C, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 58°C for 50 s, and primer extension at 72°C for 1 min. Amplification was terminated by a final extension step of 7 min at 72°C and then maintained at 4°C. Amplification products were electrophoresed on a 1.0% (w/v) agarose gel (Promega, USA) in 0.5× TBE buffer. Gels were stained with ethidium bromide and photographed under an UV transilluminator (UVP.com).
RFLP Analysis
For RFLP analysis, 5 μl of PCR products were digested for 12 h at 37°C with AluI, RsaI, and Hin-6I restriction enzymes simultaneously. Restriction fragments were separated in 2.0% agarose gel prepared with 0.5× TBE buffer, stained and photographed as described above, and the band patterns for each sample were compared.
Sequence Analysis and Phylogenetic Tree Construction
The isolates with distinct RFLP profiles were selected for sequencing with the primers used above. The nucleotide sequences obtained in this study and one closest match for each sequence were aligned, and phylogenetic relationships were analyzed with BLAST, ClustalX, and MEGA3 software package using the Kimura two-parameter method for distance matrix calculations and the neighbor-joining method for tree construction [23].
The 16S rRNA gene sequences were deposited in GenBank (accession numbers are listed in Table 2).
Results
Isolation and Characteristic Determination of Culturable Endophytes
Based on the optimized procedures of isolation and cultivation, a total of 93 isolates were obtained after phenotypic analysis, and considerable variation in population size between 103 and 105 cfu/g FW was detected, with the exception that no isolates were obtained from Rhodiola coccinea (Table 1). About 24 genera were identified from 55 endophytic isolates on the basis of RFLP and 16S rRNA gene sequence analysis, of which the highest taxonomic diversity and population density were observed in Polygonum viviparum and Leontopodium leontopodioides, for which 13 and 11 isolates, respectively, were isolated and the population density attained 6.88 × 105 and 9.69 × 105 cfu/g FW, respectively. These findings indicated that endophytic bacteria were ubiquitous and abundant within alpine-subnival plants in an extreme environment, and remarkable differences were observed in the taxonomic diversity and population density of endophytes among different subnival plant species.
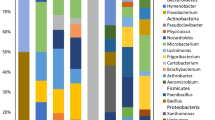
In addition, the Gram reaction, growth temperature range, and some physiological characteristics of isolates were determined (Table 2). These results showed that bacterial endophytes in subnival plants also displayed abundance and diversity in their morphology and properties. Most of the isolates (70.9%) produced pigments of yellow, orange, and pink coloration, and 67.3% of the isolates belonged to psychrotolerant species as they still grew well at −2°C. Moreover, gram-positive bacteria (61.5%) were isolated more frequently than gram-negative bacteria, and most strains (92.9%) were rod shaped.
Taxonomic Identification and Phylogenetic Analysis of Selected Bacterial Endophytes
Based on 16S rRNA gene sequences, a dendrogram representing phylogenetic relationships of 55 endophytic isolates and their closest related matches was constructed. As illustrated in Figs. 2 and 3, the endophytes in subnival plants consisted of members of α-proteobacteria, γ-proteobacteria, Flavobacterium–Bacteroides–Cytophaga (CFB), the high G+C content gram-positive bacteria (HGC), and the low G+C content gram-positive bacteria (LGC). Of the 55 isolates analyzed, 24 bacterial genera were identified, of which the HGC group formed the largest cluster and possessed the highest level of taxonomic diversity comprising 12 genera, and accounting for 55.4% of all isolates. Of these genera, Clavibacter, Rhodococcus, and Agreia were the most prevalent genera. α-Proteobacteria (17.9%) including the genera Methylobacterium, Brevundimonas, Sphingomonas, etc. and γ- proteobacteria (19.6%) containing species of Pseudomonas, Stenotrophomonas, and Xanthomonas were the second largest groups although the genus diversity was relatively low. The members of Sphingomonas (six isolates) in α-proteobacteria and Pseudomonas (eight isolates) in γ-proteobacteria were the most extensive populations. In contrast, only two isolates were obtained in the LGC and CFB groups, which types of isolates and population density were relatively low.
Neighbor-joining tree indicating the phylogenetic relationships of α-proteobacteria and γ-proteobacteria isolated from alpine-subnival plants and their closest related sequences based on 16S rRNA gene sequences. The numbers at the nodes represent bootstrap values >50% based on 100 data replications. Endophytic bacteria and their corresponding accession numbers are shown in bold
Neighbor-joining tree indicating the phylogenetic relationships of the HGC, LGC, and CFB groups isolated from alpine-subnival plants and their closest related sequences based on 16S rRNA gene sequences. The numbers at the nodes represent bootstrap values >50% based on 100 data replications. Endophytic bacteria and their corresponding accession numbers are shown in bold
In the BLAST and phylogenetic analysis, we discovered that some isolates were most closely related to bacteria isolated from plants growing in a mild environment. For example, endophytes of cluster 3 in the γ-proteobacteria grouped with Pseudomonas graminis (GenBank accession no. Y11150) obtained from grass; members of subgroup 2 and subgroup 7 in the HGC group were very similar with strains of Agreia pratensis (GenBank accession no. AJ310412) and Arthrobacter sp. Fa21 (GenBank accession no. AY131225) coming from sward and strawberry, respectively. Most importantly, however, many endophytic bacteria (46.4%) in subnival plants had a high sequence similarity (98–100%) to the strains discovered from other cold environments such as glaciers, tundra, and polar seas. The largest cluster (subgroup 1), including 12 isolates in the HGC group, was closely related to a strain of glacial ice bacterium G50-PD1 (GenBank accession no. AF479348) from the Guliya glacier on the Tibetan Plateau. The next-largest cluster was subgroup 8, which was most closely related to Rhodococcus sp. 5/1 (GenBank accession no. AF181689) obtained from the Antarctic. Furthermore, subgroups 2, 4, and 5 of α-proteobacteria and subgroup 1 of γ-proteobacteria shared 99 to 100% sequence similarity to strains from lakes or seas, Antarctic soil, and deep-sea sediment. It indicated that some microbes, with strong adaptability to low temperature and high UV radiation, surely existed in alpine-subnival plants.
In addition, we also observed that some endophytic bacteria had a wide range of hosts, as they could colonize many different subnival plant species. For example, Clavibacter was distributed in about nine different plants species such as Aster alpinus, Chorispora bungeana, Saussurea involucrata, Thalictrum aquilegifolium, and so on. Species of Pseudomonas (8 host plants) and Sphingomonas (5 host plants) also could reside in a large range of plant species; and some strains isolated from different kinds of subnival plants were highly homologous to each other (99–100% similarity). For instance, the similarity between Enf37 (from Gentiana sp.) and Enf16 (from Saussurea involucrata), Enf55 (from Leontopodium leontopodioides) and Enf8 (from Saussurea superba) attained 100%, whereas certain bacteria colonized only a specific host plant such as Enf10 (from Chorispora bungeana), Enf69 (from Chorispora bungeana), and Enf36 (from Gentiana sp.). Maybe these strains had a strong host specificity or demanded rigorous growth conditions.
Screening for Potential Plant Growth-Promoting Endophytic Bacteria
Predominant culturable endophytic bacteria (which means the concentration of isolate is above 104 cfu/g FW and the frequency of distribution in subnival plants is relatively high) were analyzed for their ability to produce IAA and solubilize phosphate in vitro in order to evaluate the potential of the isolates for plant growth promotion (Table 3, Fig. 4). The results showed that 36.4% of the endophytic bacteria, mainly species of Pseudomonas and Sphingomonas, produced IAA and 76.3% of the bacteria, comprising about ten genera, were able to solubilize mineral phosphate. Of these, the members of Clavibacter and Pseudomonas accounted for a large proportion. Moreover, some strains, such as Enf53 and Enf34, showed the ability to both produce IAA and solubilize mineral phosphate. This indicated that endophytic bacteria possessing the potential for plant growth promotion were common in subnival plants.
Discussion
In recent years, approaches based on amplification of extracted total DNA have been utilized in microbial diversity studies, as they overcome the inability to culture the majority of microorganisms from environmental samples. However, the greatest limitation of this method is that the subsequent work cannot be progressed without isolates or strains, and also the morphological and physiological characteristics and functions of the microbes cannot be adequately known and elucidated. Thus, culturing and characterizing isolates is of particular importance for providing the possibility to explore the diversity with a polyphasic approach and to establish a large collection of organisms. Given our interest in assessing the physiological functions and ecological roles of strains in the future, we therefore used conventional isolation and culture methods in this study to analyze the abundance, diversity, community structure, and the potential ability for plant growth promotion of culturable endophytes within subnival plants growing in an extreme environment. The results demonstrated that a wide variety of bacteria comprising about 24 genera were isolated from within 20 subnival plant species, and the highest population density attained 9.69 × 105 cfu/g FW, which is comparable with other reported endophytes [6]. It also implied that endophytes are distributed ubiquitously in different subnival plant species, and their types and numbers are much abundant and diverse also in the same plant species.
However, differences are apparent in some phenotypic properties and the community structure of indigenous bacteria isolated from subnival plant species growing in an extreme environment and plants or agricultural crops of temperate environments (Table 3). For instance, gram-positive bacteria (61.5%) and cold-tolerant bacteria (67.3%) accounted for a large proportion of all isolates from subnival plants, which agrees well with the results of some studies that mainly focused on microorganisms of glaciers and tundra [4, 26, 38, 39]. In contrast, endophytic bacteria obtained from mesophilic plants, such as corn and cotton, were 75 and 100% gram-negative, respectively [16, 24]. These results imply that microorganisms from similar environments show similar characteristics. This is most likely owing to their regional characteristic and environmental selectivity. Moreover, most isolates (70.9%) produced a variety of pigments in culture and represented the majority of the culturable bacterial community in subnival plants. Some studies [29, 32] have demonstrated that bacteria producing certain pigments show stronger resistance to UV radiation and low temperature than those of achromatous bacteria. Predominantly pigmented organisms have been isolated from other cold environments [11, 26], and Fong et al. [15] found a correlation between the production of carotenoids and the cold adaptation of microorganisms, possibly due to increased rigidity of the membranes. This might permit endophytic bacteria to survive in an extreme environment, and also possibly have a positive effect on the UV radiation and cold tolerance mechanisms of subnival plants. As Di Fiore and Del Gallo [13] described, endophytic bacteria are a natural component of the microecology system of plants, and their existence improves the adaptability of the host plant to adversity and enhances the balance of ecosystems.
Interestingly, the percentage of isolates with the potential for plant growth promotion was higher than those of other plant-associated bacteria in this study. Kuklinsky-Sobral et al. [22] showed that 34 and 49% of endophytic bacteria isolated from soybean possessed the capability to produce IAA and solubilize immobilized mineral phosphates, respectively. This suggested that endophytic bacteria could enhance the host plant’s absorption of phosphate and improve the fitness of the plant–bacterium interaction [36]. In contrast, 36.4 and 76.3% of isolates from subnival plants were able to produce IAA and solubilize mineral phosphates, respectively, which revealed that in subnival plants the endophytic bacteria with multiple physiological functions were more abundant and widespread than in plants from other environments. This would be very propitious for vigorous plant growth in a harsh climate and infertile soil.
Regarding the population structure, endophytes in subnival plants principally belonged to five large groups comprising α-proteobacteria, γ-proteobacteria, CFB, HGC, and LGC. The largest generic diversity and population density was found in the HGC group, consisting of 12 genera and accounting for 55.4% of the isolates analyzed, followed by the α-proteobacteria and γ-proteobacteria groups, which included 17.9 and 19.6% of all isolates, respectively. Of the latter two groups, species of Sphingomonas and Pseudomonas were the most extensive population whereas the types of isolates and population density in the LGC and CFB groups were relatively lower. Many of our isolates were related to genera such as Methylobacterium, Rhodococcus, Sphingomonas, Arthrobacter, and Frigoribacterium, representatives of which have been found in other permanently cold environments [25, 38]. This is consistent with the suggestion that the occurrence of related phenotypes in geographically diverse cold environments is possibly due to similar strategies to survive freezing and remain active at low temperature [28]. Microbes associated with plants that grow in more mild environmental conditions mainly consisted of α-proteobacteria, β-proteobacteria, γ-proteobacteria, HGC, LGC, and CFB groups and were dominated by the genera Pseudomonas, Bacillus, Enterobacter, and Agrobacterium [22, 41]. These obvious differences also indicated that the environmental conditions have a strong selective effect on the population structure of microbes. Furthermore, potentially new bacterial species or new strains with important functions might be discovered from this study, given that the sequence similarity was lower than 96% for Enf15 and Enf54, for example.
The source or origin of endophytic bacteria isolated in this study is uncertain as their host plants grow in a complicated terrain belt with a changeable climate. Several studies reported that the primary source of endophytic bacteria was from soil, and most of these bacteria comprised a variety of edaphons such as Pseudomonas, Bacillus, Enterobacter (which is also a typical component of the human microbiota), and Agrobacterium [24, 41]. However, no species of Enterobacter and Agrobacterium were isolated among our endophytes. Bai et al. [4], who investigated the bacterial diversity and population structure of subnival soil in the Tianshan Mountains, suggested that the variety and population density of bacteria were relatively low in soil. Their findings show remarkable differences to our results, because isolates in subnival soil mostly belonged to the HGC and CFB groups, and members of Arthrobacter accounted for a large proportion. Moreover, sequence alignment showed that many endophytes (46.4%) exhibited high sequence homology to bacteria from other cold environments such as glaciers, Antarctic soil, lakes, and polar seas. Therefore, some relationships or special transportation route might exist in these microbes and environments. Most importantly, however, the plant species included in this study experience frequently fluctuating and freezing temperatures and, therefore, are repeatedly subjected to the freeze–thaw process, together with strong wind and hailstones, which may cause wounding of leaves and stems; at the same time, frequent precipitation during the growth season can create a humid and hydric environment. All of these factors may facilitate the deposition and invasion of bacteria transported by wind and particles in the atmosphere. Thus, based on the above conclusions and some previous findings, we speculate that the endophytic bacteria within subnival plant tissues are of mixed origin and most likely come from the external environment of plant. Of course, additional investigation is needed in order to verify this hypothesis.
In addition, when the microorganisms transported by wind and dust in the atmosphere are deposited on substrates including a glacier (No. 1 glacier), soil, or subnival plant, plant tissues would be a more suitable environment for survival and propagation of the microbes than a glacier and soil, as they provide not only sufficient nutrition but also protection from the impact and damage from unfavorable environmental conditions. Consequently, we consider that microbial information contained by subnival plants should be very rich, and would better reflect and reveal some information in a certain period at this area if combined with a survey of glacial and edaphic microorganisms.
In conclusion, our study proved that alpine-subnival plants host large populations of endophytic bacteria from a diverse range genera and species, which not only enriches the microorganism resource, but also provides a sound foundation for further research on the relationship between plants and microbes. However, many aspects, including the ecological roles of endophytic bacteria, the relationship between endophytes and plant resistance mechanisms to extreme environmental stress, and the relationship of microbes among glaciers, tundra, and subnival plants, need to be further explored and investigated, as this study about endophytes in alpine-subnival plants represents just the beginning.
References
An LZ, Liu YH, Feng GN, Feng HY, Chen GD (2000) Studies on characteristics of communities in the source region of Urumqi River. Acta Bot Boreal Occident Sin 20:80–86
Backman PA, Sikora RA (2008) Endophytes: an emerging tool for biological control. Biol Control 46:1–3
Bai ZY, Ohata T, Higuchi K (1989) Calculation results of radiational climate in glacierized cirque and glacier-free cirque at the headwater of Urumqi River in Tianshan Mountains. J Glaci Geocry 11(4):336–349
Bai Y, Yang DQ, Wang JH, Xu SJ, Wang XX, An LZ (2006) Phylogenetic diversity of culturable bacteria from alpine permafrost in the Tianshan Mountains, northwestern China. Res Microbiol 157(8):741–751
Barac T, Tagahavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV, Vangronsveld J (2004) Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol 22(5):583–588
Barbieri P, Zanelli T, Galli E, Zanetti G (1986) Wheat inoculation with Azospirillum brasilense Sp6 and some mutants altered in nitrogen fixation and indole-3-acetic acid production. FEMS Microbiol Lett 36:87–90
Bell CR, Dickie GA, Harvey WLG, Chan JWYF (1995) Endophytic bacteria in grapevine. Can J Microbiol 41:46–53
Bric JM, Bostock RM, Silverstone S (1991) Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol 57:535–538
Brooks DS, Gonzalez CF, Appel DN, Filer TH (1994) Evaluation of endophytic bacteria as potential biological control agents for oak wilt. Biol Control 4:373–381
Chen YZ, Li FL (2005) Research progress of chemical components and their medicine roles in snowdrop. Chin Wild Plant Resour 24(3):1–4
Christner BC (2002) Recovery of bacterial from glacial and subglacial environments. Thesis, Ohio State University, Columbus
Dalton DA, Kramer S, Azios N, Fusaro S, Cahill E, Kennedy C (2004) Endophytic nitrogen fixation in dune grasses (Ammophila arenaria and Elymus mollis) from Oregon. FEMS Microbiol Ecol 49(3):469–479
Di Fiore S, Del Gallo M (1995) Endophytic bacteria: their possible role in the host plants. In: Fendrik I, Del Gallo M, Vanderleyden J, De Zamaroczy M (eds) Azospirillum VI and related microorganisms. Springer Verlag, Berlin, pp 169–187
Doty SL (2008) Tansley review: enhancing phytoremediation through the use of transgenics and endophytes. New Phytol 179:318–333
Fong NJC, Burgess ML, Barrow KD, Glenn DR (2001) Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl Microbiol Biotechnol 56:750–756
Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW (1997) Bacterial endophytes in agricultural crops. Can J Microbiol 43:895–914
Jacobs MJ, Bugbee WM, Gabrielson DA (1985) Enumeration, location, and characterization of endophytic bacteria within sugar beet roots. Can J Bot 63:1262–1265
Jha P, Kumar A (2009) Characterization of novel plant growth promoting endophytic bacterium Achromobacter xylosoxidans from wheat plant. Microb Ecol 58(1):179–188
Johnson JL (1981) Genetic characterization. In: Gerhardt P, Murray RGE, Costilaw RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, pp 450–472
Kado CI (1992) Plant pathogenic bacteria. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes. Springer-Verlag, New York, pp 659–674
Kobayashi DY, Palumbo JD (2000) Bacterial endophytes and their effects on plants and uses in agriculture. In: Bacon CW, White JF (eds) Microbial endophytes. Marcel Dekker, New York, pp 199–233
Kuklinsky-Sobral J, Araujo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL (2004) Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ Microbiol 6:1244–1251
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
McInroy JA, Kloepper JW (1994) Studies on indigenous endophytic bacteria of sweet corn and cotton. In: O’Gara F, Dowling DN, Boesten B (eds) Molecular ecology of rhizosphere microorganisms: biotechnology and the release of GMOs. VCH, New York, pp 19–28
Miteva VI, Sheridan PP, Brenchley JE (2004) Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl Environ Microbiol 70:202–213
Miteva V, Teacher C, Sowers T, Brenchley J (2009) Comparison of the microbial diversity at different depths of the GISP2 Greenland ice core in relationship to deposition climates. Environ Microbiol 11(3):640–656
Naik BS, Shashikala J, Krishnamurthy YL (2009) Study on the diversity of endophytic communities from rice (Oryza sativa L.) and their antagonistic activities in vitro. Microbiol Res 164(3):290–296
Priscu JC, Christner BC (2003) Earth’s icy biosphere. In: Bull AT (ed) Microbial diversity and bioprospecting. ASM Press, Washington, pp 130–145
Rajogopal L, Sundari CS, Balasubramanian D, Sonti RV (1997) The bacterial pigment xanthomonadin offers protection against photodamage. FEBS Lett 415:125–128
Rosenblueth M, Martinez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 19:827–837
Sturz AV, Christie BR, Nowak J (2000) Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit Rev Plant Sci 19:1–30
Sundin GW, Jacobs JL (1999) Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.). Microb Ecol 38:27–38
Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Van Der Lelie DD (2009) Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar. Appl Environ Microbiol 75:748–757
Tan DY, Zhu JW, Yao F (1998) Studies on reproduction ecology in Saussurea involucrata 1 habitat and the analyses on botanic and phenological characters. J Xinjiang Agric Univ 21(1):1–5
Ulrich K, Ulrich A, Ewald D (2008) Diversity of endophytic bacterial communities in poplar grown under field conditions. FEMS Microbiol Ecol 63:169–180
Verma SC, Ladha JK, Tripathi AK (2001) Evaluation of plant growth promotion and colonization ability of endophytic diazotrophs from deep water rice. J Biotechnol 91:127–141
Verma SC, Singh A, Chowdhury SP, Tripathi AK (2004) Endophytic colonization ability of two deep-water rice endophytes, Pantoea sp. Ochrobactrum sp. using green fluorescent protein reporter. Biotechnol Lett 26:425–429
Xiang SR, Yao TD, An LZ, Xu BL, Wang JX (2005) 16S rRNA sequences and differences in bacteria isolated from the Muztag Ata Glacier at increasing depths. Appl Environ Microbiol 71:4619–4627
Yang DQ (2008) Phylogenetic diversity and distribution of microorganisms in the Tianshan Mountains. Thesis, School of Life Science, Lanzhou University, Lanzhou, China
Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P (2002) Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208
Zou WX, Tan RX (1999) Biological and chemical diversity of endophytes and their potential applications. In: Li CS (ed) Advances in plant sciences. China Higher Education Press, Beijing, pp 183–190
Acknowledgments
This research was supported by the National Natural Science Foundation (30700082); the National Outstanding Youth Foundation of China (30625008); the International Cooperation Project (2009DFA61060), the Agricultural Science and Technology Achievements Transformation Funding Project (0910XCNA066) and the Major Project of Cultivating New Varieties of Transgenic Organisms (2009ZX08009-029B).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sheng, H.M., Gao, H.S., Xue, L.G. et al. Analysis of the Composition and Characteristics of Culturable Endophytic Bacteria Within Subnival Plants of the Tianshan Mountains, Northwestern China. Curr Microbiol 62, 923–932 (2011). https://doi.org/10.1007/s00284-010-9800-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9800-5