Abstract
This study aimed to adopt MPN-PCR (most probable number-polymerase chain reaction) for rapid detection of the quantity of Vibrio parahaemolyticus in seafood. V. parahaemolyticus in seafood could be quantitated by MPN statistics according to PCR products. The sensitivity of MPN-PCR was 100 times higher than that of direct PCR. Of 225 seafood samples from Qingdao, 165 were positive for the presence of V. parahaemolyticus, with an MPN value of >719 per gram, and about 41.5% of samples were positive for tdh gene-possessing cells. Eighty muscle tissues from the 225 seafood samples were investigated by direct PCR and MPN-PCR, but no V. parahaemolyticus was detected. The MPN-PCR test could be completed in less than 16 h from the time of sample preparation. It was rapid, sensitive, and reliable for comprehensive detection and quick quantitative determination of V. parahaemolyticus in seafood and it revealed the potential risk of illness associated with their consumption.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vibrio parahaemolyticus is a causative agent of acute gastroenteritis in humans who consume raw or undercooked fish and shellfish. It is an important food-poisoning pathogen in coastal countries [7]. Rapid detection and identification of pathogens are the key issues in controlling food-borne infections. It has been demonstrated that the polymerase chain reaction (PCR) technique can detect a low number of specific bacteria against a large background of other prokaryotic and eukaryotic cells and of organic material which may be present in the samples [1, 13]. Therefore, the PCR technique is a particularly suitable method for analyzing environmental samples. The most probable number (MPN) concept was developed for estimation of organisms based on probability. Detection methods based on MPN-PCR have been described for enumeration of specific microorganisms in soil samples [11, 14] and of V. parahaemolyticus [4, 9, 10]. This method can be readily applied using any primer system, without extensive developmental work [14].
Specific primers of V. parahaemolyticus reported by Bej et al. [2] were chosen for PCR amplification, which was combined with the MPN method for estimation of abundance, to realize reliable, rapid, and quantitative detection of small amounts of live target microorganisms in seafood samples. The incidence of V. parahaemolyticus in various ready-to-eat seafoods from Qingdao in China could contribute to contamination with or growth of V. parahaemolyticus.
Materials and Methods
Preparation of Samples and Culture Enrichment
Seafood was obtained at the local retail market. Different seafood samples were harvested following aseptic techniques, and samples of 0.01–1 g were processed in different aseptic homogenizers containing 1.8 ml sterile physiological saline. Three serial 10-fold dilutions—500, 50, and 5 μl—following the MPN culture method (three tubes, three dilutions), were carried out to determine the quantity of V. parahaemolyticus in the seafood. The final volume of each of the nine tubes was 5 ml, which was enriched at 25°–30°C for 8–12 h, with shaking, at 160 rpm for DNA extraction.
Conventional MPN Culture Analysis
After culture enrichment, samples (10 μl) were subcultured onto thiosulfate-citrate-bile salt-sucrose (TCBS) agar and incubated at 28°C for 18–24 h. Thereafter, green colonies were screened and examined for V. parahaemolyticus [1]. For identification of V. parahaemolyticus, pure cultures of sucrose nonfermenting (green) colonies were subcultured and tested for cytochrome oxidase and gelatinase. Suspected V. parahaemolyticus isolates were then finally identified using the API 20 E system (bioMérieux, Marcy I’ Etoile, France).
Preparation of DNA Templates, Multiplex PCR Assay, and MPN for Target Cell Quantification
The rest of the culture was collected by centrifugation and the sediment was then resuspended in 5 ml sterile physiological saline and centrifuged. The sediment was resuspended in 1 ml TE buffer and heated to 100°C for 10–15 min to facilitate cell lysis and DNA release for the PCR assay.
Three primer pairs based on tl, tdh and trh genes were used for multiplex PCR amplification [2]. According to the results of agarose gel electrophoresis, an MPN value per gram of seafood sample could be determined.
Sensitivity of MPN-PCR for Detection of Artificially Contaminated V. parahaemolyticus in Seafood
A 1.0-g sample of fish muscle was homogenized for 5 min in 9 ml sterile physiological saline to produce a uniform food homogenate for all experiments. Nine serial 10-fold dilutions of V. parahaemolyticus FYZ8621.4-enriched culture were used as diluents, with final concentrations of 0, 0.12, and 1.2 × 107 CFU/ml. Five-milliliter diluents included 50 μl food homogenate each in these artificially contaminated food homogenate microcosms. One-milliliter diluents of 0, 0.12, and 1.2 × 107 CFU/ml, respectively, were extracted as DNA templates to determine the sensitivity of direct PCR.
Statistical Analysis
Eight groups of shrimp and 10 groups of clams were analyzed, and there were 10 seafood samples in each group. Each seafood sample was used for each MPN examination. The MPNs of 180 shrimp and clam samples in different seasons were calculated for statistical analyses. Statistical analyses were performed using the SigmaPlot 10.0 statistical software. The average MPN of shrimp and clam samples in different seasons was also determined. Differences (p ≤ 0.05) between any two seasons and the amounts of V. parahaemolyticus existing in different seasons were determined by analysis of the MPN.
Results and Discussion
The Sensitivity of MPN-PCR
When the food homogenate was incubated for 8 h in enrichment culture at 28°C, an initial inoculum of 12 CFU of V. parahaemolyticus per milliliter amplified the desired PCR product. However, the sensitivity of direct PCR was about 100 times lower than that of MPN-PCR. The results of MPN-PCR showed that the positive detection rate was high and the detection limit was low (Table 1), while the results of the direct PCR method were usually negative. The results of MPN-PCR showed that some samples, such as crab viscus or gill and fish gill, were positive for tl- and tdh gene-possessing cells, which indicated that these seafoods were a potential reservoir for V. parahaemolyticus in the study area. In our study, pathogens in ≥400 μg seafood tissue could be detected by direct PCR without culture enrichment. However, pathogens in ≤11 μg of the same seafood tissue could be detected using enriched cultures.
Statistical Analysis of Quantitative Results
According to the overall test results, there were statistically significant differences among seasons. Differences between winter and spring (p < 10−6), between winter and summer (p < 10−4), between winter and autumn (p < 10−4), and between spring and autumn (p = 0.029) were statistically significant (p ≤ 0.05). Based on the tlh gene, the amounts of V. parahaemolyticus existing in different seasons is listed in Fig. 1, which shows that V. parahaemolyticus grew better at a higher temperature.
Results of Quantitative Analysis of Seafood Samples
In the samples tested, significant numbers of total, tl gene-possessing, and tdh gene-possessing V. parahaemolyticus were detected by the MPN-PCR technique but not by the direct PCR method or conventional MPN culture procedure. Only one sample from shrimp liver and pancreas was positive for tdh gene-possessing cells by the direct PCR method (Fig. 2). Ten of the suspected V. parahaemolyticus isolates were confirmed using the API 20E kit and PCR amplification, and two tdh gene-positive strains were isolated by MPN culture. However, selective media are known to have a decreased sensitivity of detection in natural populations, for a variety of reasons, including poor or no growth on selective media. Moreover, during subculture of enriched cultures on TCBS agar, target cells may not produce isolated colonies because of the background of other bacteria, including other Vibrio species. Such practical difficulties or limitations may also contribute to the lower recovery of V. parahaemolyticus by the MPN culture method [1]. Compared to routine MPN culture methods, detection of V. parahaemolyticus using MPN-PCR avoids the limitation mentioned above, and it is not restricted to time-intensive testing of individual isolates by phenotypic assays [6].
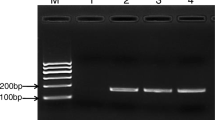
Comparison of PCR products from enriched and nonenriched culture samples. Nonenriched culture: lane 1, clam water pipe and border; lane 3, shrimp liver and pancreas; lane 5, fish gill; lane 7, crab viscus; lane 9, scallop water pipe and border. Enriched culture: lane 2, clam water pipe and border; lane 4, shrimp liver and pancreas; lane 6, fish gill; lane 8, crab viscus; lane 10, scallop water pipe and border. M: DNA marker
In this study, 225 samples of seafood from Qingdao were examined in different seasons and V. parahaemolyticus was recovered from 165 (73.3%) samples (Table 1). By the MPN-PCR technique, about 41.5% of samples were positive for tdh gene-possessing cells. In addition, the occurrence of V. parahaemolyticus in 80 samples of muscle tissue from various seafoods was investigated by direct PCR and MPN-PCR, but no V. parahaemolyticus was detected.
It is important to recognize that amplification products are only signals which show the presence of the appropriate target DNA sequences in the samples. The presence of amplification products does not imply that the target organism is viable. In fact, it has been demonstrated that PCR can amplify DNA from dead cells [1, 5]. In addition, it is difficult to enumerate a bacterial population in a complex microbial community. MPN-PCR overcame this limitation and realized the rapid quantitative detection of live target bacteria in seafood. It was considered more informative to demonstrate the bacterium, rather than testing only for the presence of hemolysin. Testing only for the presence of hemolysin can provide a false sense of security, because even if hemolysin is not produced, there can be significant contamination with V. parahaemolyticus. Therefore, undifferentiated total V. parahaemolyticus, instead of virulent V. parahaemolyticus, has long been used as an indicator for control of food contamination, with the aim of preventing infection.
Monitoring and routine screening for the presence of V. parahaemolyticus are necessary, especially as the levels of this pathogen are increasing. Doing so may make it possible to reduce the incidence of infections with the organism [3]. However, seafood with only a small number of V. parahaemolyticus organisms can reach an infectious dose in only a few hours [1, 3]. MPN-PCR is a useful detection method because of its demonstrated combination of speed and sensitivity [8], both of which are critical to any assay for the detection of bacteria. The test could be completed in less than 16 h from the time of sample preparation. Its speed and facility will make it adaptable for identification of many bacterial pathogens and provide the potential for its adaptation for direct detection in other types of seafood. In addition, development of genotypic tools for detecting and tracking pathogenic factor genes in a variety of environmental components is also essential for understanding the ecology of pathogenic V. parahaemolyticus [1].
References
Alam MJ, Tomochika KI, Miyoshi SI (2002) Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol Lett 208:83–87
Bej AK, Patterson DP, Brasher CW, Vickery MCL, Jones DD, Kaysner CA (1999) Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Meth 36:215–225
Bilung LM, Radu S, Bahaman AR, Rahim RA, Napis S, Ling MWCV (2005) Detection of Vibrio parahaemolyticus in cockle (Anadara granosa) by PCR. FEMS Microbiol Lett 252:85–88
Hara-Kudo Y, Sugiyama K, Nishibuchi M et al (2003) Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in Seafood and the coastal environment in Japan. Appl Environ Microb 69:3883–3891
Josephson KL, Gerba CP, Pepper RL (1993) Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microb 59:3513–3515
Kaysner CA, Abeyta CJ, Scott RF, Krane MH, Wekell MM (1996) Enumeration of Vibrio species, including V. cholerae from samples of an oyster growing area, Grays Harbor, Washington. J Food Protect 53:300–302
Lee KK, Liu PC, Huang CY (2003) Vibrio parahaemolyticus infectious for both humans and edible mollusk abalone. Microbes Infect 5:481–485
Martín B, Jofré A, Garriga M, Hugas M, Aymerich T (2004) Quantification of Listeria monocytogenes in fermented sausages by MPN-PCR method. Lett Appl Microbiol 39:290–295
Miwa N, Nishio T, Arita Y, Kawamori F, Masuda T, Akiyama M (2003) Evaluation of MPN method combined with PCR procedure for detection and enumeration of Vibrio parahaemolyticus in seafood. Medline Abstr 44(6):289–293
Miwa N, Kashiwagi M, Kawamori F, Masuda T, Sano Y, Hiroi M, Kurashige H (2006) Levels of Vibrio parahaemolyticus and thermostable direct hemolysin gene-positive organisms in retail seafood determined by the most probable number-polymerase chain reaction (MPN-PCR) method. Medline Abstr 47(2):41–45
Picard C, Ponsonnet C, Paget E, Nesme X, Simonet P (1992) Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microb 58:2717–2722
Scott E (1996) Foodborne disease and other hygienic issues in the home. J Appl Bacteriol 80:5–9
Tsai YL, Olson BH (1992) Detection of low number of cells in soils and sediments by polymerase chain reaction. Appl Environ Microb 58:754–757
Vesa M, Seppo N, Seppo K, Tuula P, Kristina L (1997) MPN-PCR-quantification method for staphylococcal enterotoxin c1 gene from fresh cheese. Int J Food Microbiol 36:135–143
Acknowledgments
This work was supported by Grant 30371108 from the National Natural Science Foundation of China and National High-Tech R&D Program 2003AA622070.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luan, X., Chen, J., Liu, Y. et al. Rapid Quantitative Detection of Vibrio parahaemolyticus in Seafood by MPN-PCR. Curr Microbiol 57, 218–221 (2008). https://doi.org/10.1007/s00284-008-9177-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-008-9177-x