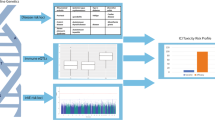
Abstract
Background
Immune checkpoint inhibitors (ICIs) can cause profound immune-related adverse events (irAEs). The host genetic background is likely to play a role in irAE susceptibility because the presentation of toxicity varies among patients and many do not develop irAEs despite continued ICI use. We sought to identify potential genetic markers conferring risk for irAEs.
Methods
We conducted a pilot exploratory study in 89 melanoma patients who received ICIs (44 with irAEs, and 45 without irAEs after at least 1 year from starting treatment). Genotyping was performed using the Infinium Multi-Ethnic Global-8 v1.0 Bead Chip. The genotype data were extracted using PLINK (v1.90b3.34) and processed for quality control. Population structure-based clustering was carried out using IBS matrix, pairwise population concordance test (p < 1 × 10–3), and phenotype distribution for all study participants, resulting in seven population structure-based clusters. In the analytical stage, 599,931 variants in autosomal chromosomes were included for the association study. The association test was performed using an additive genetic model with exact logistic regression, adjusted for age, sex, and population cluster.
Results
A total of 30 variants or single-nucleotide polymorphisms with p < 1 × 10–4 were identified; 12 were associated with an increased risk of irAEs, and the remaining 18 were associated with a decreased risk. Overall, nine of the identified single-nucleotide polymorphisms mapped to eight unique genes that have been associated with autoimmunity or inflammatory diseases.
Conclusion
Several genetic variants associated with irAEs were identified. Additional larger studies are needed to validate these findings and establish their potential functional relevance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of immune checkpoint inhibitors (ICIs) targeting cytotoxic T-cell lymphocyte-associated protein-4 (CTLA-4) and programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) has led to a major breakthrough in the treatment of cancer [1,2,3]. Nevertheless, the therapeutic benefits of ICIs are limited by toxicity arising from off-target inflammatory and/or autoimmune responses that can be life threatening and may require high-dose immunosuppressive therapies and permanent discontinuation of ICIs. The presentation of toxicity varies widely among patients; although many patients develop immune-related adverse events (irAEs) in multiple organs, some develop irAEs limited to one organ and others never develop irAEs despite continued ICI use. Notably, onset of well-defined autoimmune diseases has also been reported in some patients following initiation of ICI therapy; these diseases include rheumatoid arthritis, myasthenia gravis, cryoglobulinemic vasculitis, and Sjogren syndrome, among others [4,5,6,7]. Conceivably, the genetic background of patients receiving ICIs could play a role in susceptibility to irAEs, contributing to the observed variation. Elucidating which individual genetic determinants may predispose patients to the development of irAEs can contribute to understanding of the pathogenesis of irAEs, support clinical decision-making, and improve the management of cancer patients receiving ICIs.
We conducted a pilot genome-wide study of patients with melanoma who received ICIs, comparing the genotypes of those who developed irAEs with the genotypes of those who did not, using a comprehensive, multi-ethnic microarray. Our objective was to agnostically interrogate the entire genome to identify genetic markers conferring risk for irAEs, to inform future, larger studies on the choice of potential candidate genes.
Materials and methods
Study design
We designed a case–control study with a convenience sample of cancer patients who received ICIs at The University of Texas MD Anderson Cancer Center between January 2012 and September 2017 identified from melanoma and specialty clinics for diagnosis and management of irAEs. Patients who developed irAEs after initiation of ICI therapy were considered cases, and those who did not develop irAEs for at least 1 year after initiating ICI therapy were considered controls. Common inclusion criteria for both cases and controls were as follows: (1) age > 18 years; (2) histologic diagnosis of melanoma; and (3) treatment with a US Food and Drug Administration-approved ICI. The study was approved by The Institutional Review Board at The University of Texas MD Anderson Cancer Center.
Specific inclusion criteria for cases included the following: (1) ≥ grade 2 irAEs occurring at any time during ICI therapy, or within 1 month of the last ICI infusion; (2) use of corticosteroids or other immunosuppressive therapy for a minimum of 2 weeks for treatment of an irAE (except for endocrinopathies); (3) no other conditions (e.g., infection) that could potentially cause similar symptoms, as per medical record; and (4) no prior history of inflammatory or autoimmune disease in the same affected organs. Specific organ irAEs were identified based on the Common Terminology Criteria for Adverse Events [8]. Patients who developed irAEs in multiple organs and those who had irAEs limited to one organ were included. Skin irAEs are the most common ICI-related toxicity, typically mild, and do not require systemic immunosuppressive therapy making diagnosis sometimes difficult, so patients with isolated rash were excluded from the study.
Specific inclusion criteria for controls included the following: (1) for ipilimumab-treated patients: at least 12 weeks of treatment without any irAEs during therapy and within the 9 months subsequent to the last treatment dose; and (2) for patients treated with anti-PD-1/PD-L1: at least 1 year of treatment without irAEs. To maximize similarities between cases and controls other than the presence or absence of irAEs, controls were matched to the cases by type of ICI agent. Eligible patients were approached during their follow-up visits in the melanoma clinic and those who agreed to participate in the study signed an informed consent form. Patients were interviewed to evaluate past medical history and their medical records were reviewed for additional pertinent information (patient demographics, prior autoimmune diseases, type of ICI agent, duration of ICI treatment, and occurrence of toxicity). The final inclusion of patients as cases or controls was agreed upon by two investigators (N.A. and A.D.).
Genotyping
Genotyping was performed at the Sequencing and Microarray Facility at MD Anderson using the Infinium Multi-Ethnic Global-8 v1.0 Bead Chip, which covers all five super-continental populations, account for subtle genetic variations that exist even among Caucasians, is based on the most current genomic information (phase 3 of the 1000 Genomes Project), and is augmented with significant clinical database content and functional content. Briefly, genomic DNA was isolated from whole blood samples and was assessed for quality and quantified; 200 ng of dsDNA was denatured, neutralized, and isothermally amplified. The amplified DNA was enzymatically fragmented, precipitated, and re-suspended in hybridization buffer before being loaded to the chip, which was incubated overnight. Following hybridization, the chip was washed to remove unhybridized and non-specifically hybridized DNA and then stained by single-base extension of the primers, which uses the captured DNA as a template and incorporates a fluorescently labeled complementary nucleotide. Chips were scanned on the Illumina iScan, which uses a laser to excite the incorporated fluorophore and records high-resolution images of the light emitted from the fluorophores.
The genotype data were processed using PLINK (v1.90b3.34). The initial set of 1,523,415 raw variants on autosomal chromosomes (1 through to 22) and sex chromosomes (23 and 24) was processed for quality control, including call rate (> 99%), non-missing genotyping rate (> 95%), minor allele frequency (no less than 5%), and Hardy–Weinberg equilibrium (p < 1 × 10–6). Then, the pairwise genetic distance was calculated using identity-by-state (IBS matrix) implemented in the genome option of PLINK, and the population structure-based clustering was carried out using IBS matrix, pairwise population concordance test (p < 1 × 10–3), and phenotype distribution for all patients, resulting in seven structure groups.
Statistical analysis
This is a pilot study and our sample size included 44 cases and 45 controls selected from the populations described above. Based on the proposed sample size, we have more than 80% power to detect odds ratios (OR) of 3.58, 3.38, 3.66 or above as significant, depending on the prevalence of the single-nucleotide polymorphisms (SNPs) in the population (50, 30, and 20%, respectively). Descriptive statistics were used to summarize the data, with median and range for continuous variables and frequencies and percentages for categorical variables. Chi-square and Fisher exact tests were used to compare categorical variables, and Wilcoxon or Kruskal–Wallis tests were used to compare continuous variables between groups.
A total of 599,931 variants in autosomal chromosomes remained after quality control processes were complete, and these chromosomes were included in the analysis. Testing was performed using an additive genetic model with exact logistic regression adjusting for sex, age, and seven population clusters identified from the identity-by-state analyses to account for race/ethnicity. Population cluster analyses identify homogeneous groups to account for underlying subpopulations beyond the self-reported race and ethnicity. Because this was a pilot study and the aims were primarily hypothesis generating, within a small sample of patients, we were interested in identifying only a small subset of SNPs that are suggestive of association. Therefore, we used p ≤ 1 × 10–4 as the threshold for selecting SNPs, and testing for multiple comparisons was not possible.
Literature search
To highlight the most up-to-date evidence in the literature, we conducted a comprehensive Medline search from inception to December 2019 and identified previous studies that evaluated the association between autoimmune SNPs or variants and occurrence of irAEs and/or tumor response to ICI therapy. We then recognized the full list of SNPs within these previously reported genes and evaluated their potential association with irAEs in our sample.
Results
Patient characteristics
A total of 90 melanoma patients were included in our analysis; 45 were cases (i.e., patients who developed irAEs) and 45 were controls (i.e., no irAEs). However, one patient with both melanoma and renal cell carcinoma was excluded from the case group in our final analysis because the patient was receiving ICI therapy for renal cell carcinoma and did not have active melanoma. Patient demographics and baseline characteristics, including the distribution of age, sex, population clusters, and class of ICI therapy, are provided in Table 1. The overall median age of the patients was 64 years (range 23–92 years); 63 (71%) were male and 85 (96%) were white. Eighty-four patients (94%) had unresectable stage III/IV metastatic melanoma and the remaining five were receiving adjuvant ICI after surgical resection. Single-agent anti-PD-1/PD-L1 therapy was the most frequently used ICI therapy, in 49 patients (55%). Colitis (10%) and pneumonitis (9%) were the most frequently reported irAEs in our sample population. Other irAEs included hypophysitis, thyroiditis, arthritis, and multiple irAEs (≥ 2 irAEs). Among the cases, the median duration of follow-up after initiation of ICI therapy was 2.0 years (range 0.31–4.3 years). In the control group, the median duration of follow-up for patients treated with ipilimumab was 2.5 years (range 1–3 years), and for those receiving anti-PD-1/PD-L1 therapy, 1.5 years (range 1–5 years).
Association of SNPs with irAEs
The full list of all SNPs or variants from chromosome 1–22 for our sample is shown in Supplemental Table 1. We identified a total of 30 variants or SNPs that were significantly associated (p ≤ 1 × 10–4) with irAEs development.
Twelve SNPs were associated with significantly increased odds of developing irAEs (Table 2). Four of these were mapped to genes that have been previously associated with inflammatory and autoimmune diseases. Carriers of at least one copy of a minor allele A for rs11743438 were more likely to develop irAEs (OR 4.3; 95% confidence interval [CI] 2.3–8.0; p = 5.56 × 10–6). rs11743438 was mapped to the GABRP (gamma-aminobutyric acid type A receptor subunit pi) gene, which has been associated with autoimmune movement disorders [9]. Carriers of at least one copy of a minor allele A for JHU_20.57183980 were more likely to develop irAEs (OR 6.9; 95% CI 2.7–17.6; p = 8.85 × 10–6). JHU_20.57183980 was mapped to the DSC2 (desmocollin 2) gene, which has been associated with autoimmune bullous disorders of the pemphigus group [10]. Carriers of at least one copy of a minor allele G for rs56328422 were more likely to develop irAEs (OR 4.2; 95% CI 2.1–8.4; p = 4.14 × 10–5). rs56328422 was mapped to BAZ2B (bromodomain adjacent to zinc finger domain 2B) gene, which has been associated with Murray Valley encephalitis. Carriers of at least one copy of a minor allele T for rs3026321 were more likely to develop irAEs (OR 19.8; 95% CI 2.6–152.7; p = 6.31 × 10–5). rs3026321 was mapped to the SEMA5A (semaphorin 5A) gene, which has been associated with systemic lupus erythematosus, rheumatoid arthritis, and idiopathic thrombocytopenic purpura [11,12,13]. The other eight variants or SNPs associated with increased risk of developing irAEs along with their base-pair position, gene location (when available) and associated odds ratios with confidence intervals are listed in Table 2 top panel.
We also identified 18 SNPs that were associated with significantly reduced odds of developing irAEs (Table 2). Five of these SNPs were mapped to four genes that were previously associated with inflammatory and autoimmune diseases. rs2117997 was mapped to the ANKRD42 (ankyrin repeat domain 42) gene, associated with systemic inflammatory response syndrome [14]. rs55733913 was mapped to the PACRG (parkin coregulated) gene, associated with autoimmune thyroid diseases [15]. rs162263 and kgp3960064 were both mapped to the ROBO1 (roundabout guidance receptor 1) gene, associated with autoimmune diabetes [16]. rs10814859 was mapped to the GLIS3 (GLIS family zinc finger 3) gene, which has also been associated with autoimmune diabetes, and possibly with rheumatoid arthritis [17, 18]. These variants or SNPs associated with decreased risk of developing irAEs along with their base-pair position, gene location (when available) and associated odds ratios with confidence intervals are listed in Table 2 bottom panel.
In addition, our literature search identified a list of 36 genes that were previously evaluated in selected studies of irAEs and/or for tumor response with ICI therapy [19,20,21,22] (Supplemental Table 2). The full list of SNPs within these genes (n = 296) and their association parameters with irAEs in our sample is shown in Supplemental Table 3. Six SNPs showed nominal statistical significance (p < 0.05), although above the value we predetermined for cutoff (Table 3). The carriers of at least one copy of a minor allele A for exm1425699 were more likely to develop irAEs (OR 2.643; 95% CI 1.3–5.3; p = 0.0067); this SNP maps to the TYK2 (tyrosine kinase 2) genes, which encode for tyrosine kinases that are instrumental in the coding of various inflammatory cytokines associated with autoimmune diseases. The other five SNPs were mapped to the HLA (human leukocyte antigen), TNFAIP3 (TNF alpha induced protein 3), and TYK2 genes, and all were associated with significantly reduced odds of developing irAEs.
Discussion
The current study, to the best of our knowledge, is the first exploratory genome-wide study to identify genetic variants that may be associated with the risk of developing irAEs in melanoma patients treated with ICIs. Our objective was to agnostically examine a broad array of genetic markers to evaluate their possible association with immune toxicity in patients with irAEs, to provide information on potential candidate genes to be examined in subsequent larger studies. Because the current study was exploratory, we did not aim to ascertain genetic associations with specific organ toxicity and included patients with various irAEs.
We identified 12 SNPs that were associated with an increased risk of developing an irAE, and 18 others appeared to be protective. Overall, nine of the identified SNPs were mapped to eight unique genes that have previously been associated with inflammatory and autoimmune diseases. Our data suggest that patients harboring at least one variant allele A in the GABRP SNP rs11743438, one variant allele A in the DSC2 SNP JHU_20.57183980, one variant allele G in the BAZ2B SNP rs56328422, or one variant allele T in the SEMA5A SNP rs3026321 have an increased risk of developing irAEs. Our findings align with earlier studies that identified the association between these genes and susceptibility to autoimmune diseases, which share some clinical features with ICI-induced irAEs such as rheumatoid arthritis, lupus erythematosus, and blistering skin diseases, among others [9,10,11,12,13]. Although the functional consequences of the identified SNPs need to be elucidated, one could speculate that the use of ICI therapy in patients harboring these SNPs triggered the autoimmune phenotypes to clinically manifest in patients with the predisposing genotype. Several other SNPs were associated with a reduced risk of immune toxicity; five of these SNPs map to genes that are known to be associated with autoimmune diseases, including systemic inflammatory response syndrome, autoimmune thyroid disease, and type I diabetes [14,15,16,17,18]. Our analysis reported a “protective” effect, which may indicate that these SNPs may have a regulatory role in the underlying gene transcription and, thus, help suppress the target immune-adverse event. Whether these variants help gene expression or suppression depends on the biological pathways, which are beyond the scope of this study but provide the suggestive information for scientific community to carry out further investigation to better understand underlying biological etiology. The small number of patients in this pilot study does not allow for definitive inferences, and larger studies are warranted to validate our findings.
The pathogenesis of irAEs in patients receiving ICI therapy is thought to be driven by increased inflammation and/or autoimmunity [23,24,25]. Whether germline genetic variants that are known to contribute to susceptibility to autoimmune diseases [26] could also play a role in susceptibility to irAEs remains an intriguing question. Preclinical studies have shown that mice deficient in CTLA-4 die from massive lymphoproliferative disorders, and those with genetic deletion of PD-1/PD-L1 have an increased risk of autoimmunity [27,28,29,30]. In humans, selected polymorphisms in CTLA-4- and PD-1-related alleles have been linked to a number of autoimmune diseases such as thyroiditis, neurologic disorders, inflammatory bowel disease, rheumatoid arthritis, ankylosing spondylitis, lupus erythematosus, and type I diabetes [28, 31,32,33,34,35,36,37].
Queirolo and colleagues evaluated the association between six SNPs associated with the CTLA-4 gene (− 1661A > G, − 1577G > A, − 658C > T, − 319C > T, + 49A > G, and CT60G > A) and the occurrence of endocrine irAEs in 173 melanoma patients treated with ipilimumab; only the CTLA-4 gene variant − 1661A > G (rs4553808) appeared to predispose patients to endocrinopathies following treatment initiation [38]. Similarly, Bins and colleagues assessed the association between seven SNPs in four genes involved in the PD-1 pathway, including programmed cell death 1 (PDCD1), zeta chain of T cell receptor-associated protein kinase 70 (ZAP70), interferon gamma (IFNG), and PTPN11, and occurrence of irAEs in 161 patients with non-small cell lung cancer treated with nivolumab [19]. In that study, carriers with at least one copy of a minor allele G in the PTPN11 333–223A > G SNP (rs2301756) had increased odds of developing elevated transaminases, and those with a homozygous variant for the IFNG − 1616 T > C SNP had an increased risk for rheumatologic irAE. They also found that patients with the TT genotype in the PDCD1 804C > T SNP (rs2227981) had decreased odds of developing any grade treatment-related adverse events. However, these findings could not be replicated in their validation cohort that included another 161 patients. In our sample of melanoma patients, genetic variants in CTLA-4, PDCD1, or PD-L1 were not associated with irAE risk.
Genetic variations in human leucocyte antigens (HLA) have previously been associated with a number of autoimmune diseases such as rheumatoid arthritis and type 1 diabetes. Recently, Cappelli and colleagues evaluated whether HLA class I and II alleles were more prevalent among patients with ICI-induced arthritis (n = 26) compared with the general population (n = 726) [20]. In that study, HLA DRB1*04:05 was increased in patients with ICI-induced arthritis (OR 8.6; 95% CI 1.7–43.4; p = 0.04). Nonstatistically significant associations with other alleles were also reported including an increase in HLA A*03:01, and HLA C*12:02 and a decrease in DQB1*03:01 (OR 0.4; 95% CI 0.1–1.1; p = 0.06). Patients with ICI-induced arthritis were compared with patients with primary rheumatoid arthritis (n = 220) and no differences were observed in the probability of having at least one HLA shared epitope allele (which confers risk for rheumatoid arthritis), but rheumatoid arthritis patients were more likely to be homozygous. A single-center prospective cohort study of rheumatic syndromes induced by ICI therapy reported two patients with ICI-induced arthritis harboring HLA-DRB1*01:01; both instances of irAEs manifested as arthritis resembling rheumatoid arthritis, and one of the patients had positive antibodies against citric citrullinated peptide [39]. Another small series reported that four of five melanoma patients with type I diabetes diagnosed following initiation of anti-PD-1 therapy were found to have HLA DRB01*03 or 04, which are known to be associated with an increased risk of type I diabetes in the general population [40]. In our sample population, which included patients with different irAE phenotypes, carrying a specific genetic variant within the HLA region—HLA DRB1 (rs3830135), HLA-DQA1 (rs28654242), and HLA-DQA2 (rs17500468)—conferred reduced odds of developing irAEs.
Specific SNPs within the PTPN22 (protein tyrosine phosphatase non-receptor type 22) gene have been associated with several autoimmune diseases [41]; however, no significant association with irAEs was identified in our sample population.
As our sample was small, we did not attempt to evaluate genetic associations with specific organ irAE. Our goal was to identify signals for potential genetic markers associated with developing any irAE, as many patients present with immune toxicity across various organs.
Growing evidence suggests that patients who develop irAEs could have better tumor response to ICI therapy [42,43,44], raising a question about the possibility of shared genetic associations between treatment-related toxicity and efficacy. HLA genotyping was recently performed in 1535 patients with melanoma and non-small cell lung cancer treated with ICIs; extended survival was observed in patients with HLA-B44, whereas those with HLA-B62, including (HLA-B*15:01), had worse disease outcomes [45]. Another study evaluated the association between autoimmune germline genetic variants and tumor response to ICI therapy in 436 patients with melanoma, but that study did not examine the association of the genetic variants with irAEs [22]. In that study, the authors performed a comprehensive search of the published literature to select genetic variants associated with autoimmune diseases in genome-wide association studies. The authors considered 25 variants; each variant is known to be associated with at least three autoimmune traits that could manifest clinically as part of irAEs, and at least one of these associations exceeded the genome-wide association study level of significance (p = 1E − 07). The authors reported that rs17388568 (IL2 and IL21 genes) was associated with an increased response to anti-PD-1 therapy. In the current study, no attempt was made to evaluate tumor response, as our study had a case–control design and controls were chosen on the basis of continuing their ICI therapy, and therefore were likely to be responders, so it was not adequate for such an aim.
Our study was exploratory and preliminary in nature. It was limited by the small sample size; therefore, it was powered only to evaluate large ORs given the set threshold for statistical significance at p ≤ 1 × 10–4, and smaller effect sizes were unlikely to be detected. Because of the exploratory nature of our study, we did not use the genome-wide threshold of 5.0 × 10–8 for significance. Additionally, because of the small sample, we included all types of irAEs as one group which could have lowered the statistical power of detecting an association, and we were not able to evaluate if differing associations exist among specific organ irAEs which are thought to have different underlying mechanisms. Although colitis was the most frequent irAE in our sample population, we did not identify any SNPs that were previously known to be associated with colitis [46, 47]. This could be partially explained by the small sample size (only nine patients had colitis) and the fact that ICI-related colitis represents a distinct form of colitis, which has similarities and differences with ulcerative colitis and Crohn’s disease [48,49,50,51]. Moreover, the small sample size did not allow for testing or adjusting for other factors that could affect the occurrence of irAEs such as the recent use of cancer vaccines. However, our study provides the first exploratory genome-wide evaluation in melanoma patients who have various irAEs compared with other patients who have received ICI therapy without developing irAEs. Larger number of patients received ipilimumab and ICI combination among the case group (12 out of 44 cases = 27%; 95% CI 15–43%); however, the difference among cases and controls (5 out of 45 controls = 11%; 95% CI 2–24%) did not reach statistical significance (overlapping confidence intervals). Future larger studies should include cases and controls matched by therapy received and other relevant factors. Since ipilimumab and anti-PD-1/PD-L1 agents target different immune regulatory mechanisms, conducting a study specifically focused on the anti-PD-1/PD-L1 treated cases will further clarify the etiology of irAEs.
In conclusion, in our exploratory study, we identified 30 genetic variants associated with the risk of irAEs in patients with melanoma receiving ICIs; 12 SNPs were associated with an increased risk, and 18 others seemed to be protective. The genes associated with some of these SNPs were previously known to be associated with inflammatory and autoimmune diseases. Our findings suggest a contributing role of polygenic predisposition in the occurrence of ICI-induced irAEs. Our study was exploratory, and as our next step we plan a prospective validation cohort study of melanoma patients initiating anti-PD1 agents to further evaluate the association between the identified autoimmune SNPs, immune toxicity, and efficacy of ICI therapy. Given the large number of potential alleles that may be associated with immune toxicity, the wide variation in clinical phenotypes, and the increasing number of cancer types for which ICIs are used, it will be necessary to establish large prospective registries to further define risk profiles. Understanding the effect of germline genetic variants on the risk of irAEs may lead to the development of polygenic risk scores that could be implemented in clinical settings to identify patients who are at risk and who require additional surveillance and prompt treatment when receiving ICI therapy, primarily those who receive treatment in the adjuvant setting.
Data availability
Not applicable.
Abbreviations
- CI:
-
Confidence interval
- CTLA-4:
-
Cytotoxic T-cell lymphocyte-associated protein-4
- ICI:
-
Immune checkpoint inhibitor
- irAE:
-
Immune-related adverse event
- OR:
-
Odds ratio
- PD-1/PD-L1:
-
Programmed cell death-1/programmed cell death-ligand 1
- SNP:
-
Single-nucleotide polymorphism
References
KEYTRUDA- Pembrolizumab [package insert]. County Cork IM, 2019. https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/125514s067lbl.pdf. Accessed 1 May 2020
YERVOY-ipilimumab [package insert]. Princeton NBS, 2019. https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/125377s104lbl.pdf. Accessed 1 May 2020
Opdivo-nivolumab [package insert]. Princeton NBMS, 2019. https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/125554s075lbl.pdf. Accessed 1 May 2020
Abdel-Wahab N, Shah M, Suarez-Almazor ME (2016) Adverse events associated with immune checkpoint blockade in patients with cancer: a systematic review of case reports. PLoS ONE 11(7):e0160221
Bertrand A, Kostine M, Barnetche T, Truchetet ME, Schaeverbeke T (2015) Immune related adverse events associated with anti-CTLA-4 antibodies: systematic review and meta-analysis. BMC Med 13:211
Wang Y, Zhou S, Yang F, Qi X, Wang X, Guan X et al (2019) Treatment-related adverse events of PD-1 and PD-L1 inhibitors in clinical trials: a systematic review and meta-analysis. JAMA Oncol 5(7):1008–1019
Le Burel S, Champiat S, Routier E, Aspeslagh S, Albiges L, Szwebel TA et al (2018) Onset of connective tissue disease following anti-PD1/PD-L1 cancer immunotherapy. Ann Rheum Dis 77(3):468–470
Thompson JA, Schneider BJ, Brahmer J, Andrews S, Armand P, Bhatia S et al (2019) Management of immunotherapy-related toxicities, version 1.2019. J Natl Compr Cancer Netw 17(3):255–289
Balint B, Bhatia KP (2016) Stiff person syndrome and other immune-mediated movement disorders—new insights. Curr Opin Neurol 29(4):496–506
Muller R, Heber B, Hashimoto T, Messer G, Mullegger R, Niedermeier A et al (2009) Autoantibodies against desmocollins in European patients with pemphigus. Clin Exp Dermatol 34(8):898–903
Du Y, Wu X, Chen M, Wang W, Xv W, Ye L et al (2017) Elevated semaphorin5A in systemic lupus erythematosus is in association with disease activity and lupus nephritis. Clin Exp Immunol 188(2):234–242
Lyu M, Li Y, Hao Y, Sun T, Liu W, Lyu C et al (2015) Elevated semaphorin 5A correlated with Th1 polarization in patients with chronic immune thrombocytopenia. Thromb Res 136(5):859–864
Gras C, Eiz-Vesper B, Jaimes Y, Immenschuh S, Jacobs R, Witte T et al (2014) Secreted semaphorin 5A activates immune effector cells and is a biomarker for rheumatoid arthritis. Arthritis Rheumatol 66(6):1461–1471
Labiad Y, Venton G, Farnault L, Baier C, Colle J, Mercier C et al (2018) A transcriptomic signature predicting septic outcome in patients undergoing autologous stem cell transplantation. Exp Hematol 65:49–56
Martinez-Hernandez R, Serrano-Somavilla A, Ramos-Levi A, Sampedro-Nunez M, Lens-Pardo A, Munoz De Nova JL et al (2019) Integrated miRNA and mRNA expression profiling identifies novel targets and pathological mechanisms in autoimmune thyroid diseases. EBioMedicine 50:329–342
Glawe JD, Mijalis EM, Davis WC, Barlow SC, Gungor N, McVie R et al (2013) SDF-1-CXCR4 differentially regulates autoimmune diabetogenic T cell adhesion through ROBO1-SLIT2 interactions in mice. Diabetologia 56(10):2222–2230
Aslam MM, John P, Fan KH, Bhatti A, Jahangir S, Feingold E et al (2019) Exploration of shared genetic susceptibility loci between type 1 diabetes and rheumatoid arthritis in the Pakistani population. BMC Res Notes 12(1):544
Inshaw JRJ, Cutler AJ, Crouch DJM, Wicker LS, Todd JA (2020) Genetic variants predisposing most strongly to type 1 diabetes diagnosed under age 7 years lie near candidate genes that function in the immune system and in pancreatic beta-cells. Diabetes Care 43(1):169–177
Bins S, Basak EA, El Bouazzaoui S, Koolen SLW, Oomen-de Hoop E, van der Leest CH et al (2018) Association between single-nucleotide polymorphisms and adverse events in nivolumab-treated non-small cell lung cancer patients. Br J Cancer 118(10):1296–1301
Cappelli LC, Dorak MT, Bettinotti MP, Bingham CO, Shah AA (2019) Association of HLA-DRB1 shared epitope alleles and immune checkpoint inhibitor-induced inflammatory arthritis. Rheumatology 58(3):476–480
Khan Z, Hammer C, Guardino E, Chandler GS, Albert ML (2019) Mechanisms of immune-related adverse events associated with immune checkpoint blockade: using germline genetics to develop a personalized approach. Genome Med 11(1):39
Chat V, Ferguson R, Simpson D, Kazlow E, Lax R, Moran U et al (2019) Autoimmune genetic risk variants as germline biomarkers of response to melanoma immune-checkpoint inhibition. Cancer Immunol Immunother 68(6):897–905
Dulos J, Carven GJ, van Boxtel SJ, Evers S, Driessen-Engels LJ, Hobo W et al (2012) PD-1 blockade augments Th1 and Th17 and suppresses Th2 responses in peripheral blood from patients with prostate and advanced melanoma cancer. J Immunother 35(2):169–178
von Euw E, Chodon T, Attar N, Jalil J, Koya RC, Comin-Anduix B et al (2009) CTLA4 blockade increases Th17 cells in patients with metastatic melanoma. J Transl Med 7:35
Bailey SR, Nelson MH, Himes RA, Li Z, Mehrotra S, Paulos CM (2014) Th17 cells in cancer: the ultimate identity crisis. Front Immunol 5:276
Gutierrez-Arcelus M, Rich SS, Raychaudhuri S (2016) Autoimmune diseases—connecting risk alleles with molecular traits of the immune system. Nat Rev Genet 17(3):160–174
Okazaki T, Honjo T (2006) The PD-1-PD-L pathway in immunological tolerance. Trends Immunol 27(4):195–201
Scalapino KJ, Daikh DI (2008) CTLA-4: a key regulatory point in the control of autoimmune disease. Immunol Rev 223:143–155
Schildberg FA, Klein SR, Freeman GJ, Sharpe AH (2016) Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 44(5):955–972
Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP et al (1995) Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 270(5238):985–988
Barreto M, Santos E, Ferreira R, Fesel C, Fontes MF, Pereira C et al (2004) Evidence for CTLA4 as a susceptibility gene for systemic lupus erythematosus. Eur J Hum Genetics 12(8):620–626
Lee YH, Bae SC, Kim JH, Song GG (2015) Meta-analysis of genetic polymorphisms in programmed cell death 1. Associations with rheumatoid arthritis, ankylosing spondylitis, and type 1 diabetes susceptibility. Z Rheumatol 74(3):230–239
Lee YH, Kim JH, Seo YH, Choi SJ, Ji JD, Song GG (2014) CTLA-4 polymorphisms and susceptibility to inflammatory bowel disease: a meta-analysis. Hum Immunol 75(5):414–421
Lee YH, Woo JH, Choi SJ, Ji JD, Song GG (2009) Association of programmed cell death 1 polymorphisms and systemic lupus erythematosus: a meta-analysis. Lupus 18(1):9–15
Li G, Shi F, Liu J, Li Y (2014) The effect of CTLA-4 A49G polymorphism on rheumatoid arthritis risk: a meta-analysis. Diagn Pathol 9:157
Liu JL, Zhang FY, Liang YH, Xiao FL, Zhang SQ, Cheng YL et al (2009) Association between the PD1.3A/G polymorphism of the PDCD1 gene and systemic lupus erythematosus in European populations: a meta-analysis. J Eur Acad Dermatol Venereol 23(4):425–432
Wu J, Zhang L, Zhou Y (2016) The association between CTLA-4 (+49 A/G) polymorphism and susceptibility to ankylosing spondylitis: a meta-analysis. Int J Rheum Dis 19(12):1237–1243
Queirolo P, Dozin B, Morabito A, Banelli B, Carosio R, Fontana V et al (2018) CTLA-4 gene variant − 1661A>G may predict the onset of endocrine adverse events in metastatic melanoma patients treated with ipilimumab. Eur J Cancer 97:59–61
Kostine M, Rouxel L, Barnetche T, Veillon R, Martin F, Dutriaux C et al (2018) Rheumatic disorders associated with immune checkpoint inhibitors in patients with cancer-clinical aspects and relationship with tumour response: a single-centre prospective cohort study. Ann Rheum Dis 77(3):393–398
Magis Q, Gaudy-Marqueste C, Basire A, Loundou A, Malissen N, Troin L et al (2018) Diabetes and blood glucose disorders under Anti-PD1. J Immunother 41(5):232–240
Brownlie RJ, Zamoyska R, Salmond RJ (2018) Regulation of autoimmune and anti-tumour T-cell responses by PTPN22. Immunology 154(3):377–382
Judd J, Zibelman M, Handorf E, O’Neill J, Ramamurthy C, Bentota S et al (2017) Immune-related adverse events as a biomarker in non-melanoma patients treated with programmed cell death 1 inhibitors. Oncologist 22(10):1232–1237
Downey SG, Klapper JA, Smith FO, Yang JC, Sherry RM, Royal RE et al (2007) Prognostic factors related to clinical response in patients with metastatic melanoma treated by CTL-associated antigen-4 blockade. Clin Cancer Res 13(22 Pt 1):6681–6688
Haratani K, Hayashi H, Chiba Y, Kudo K, Yonesaka K, Kato R et al (2018) Association of immune-related adverse events with Nivolumab efficacy in non-small-cell lung cancer. JAMA Oncol 4(3):374–378
Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V et al (2018) Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359(6375):582–587
Liu QF, Li Y, Zhao QH, Wang ZY, Hu S, Yang CQ et al (2015) Association of STAT4 rs7574865 polymorphism with susceptibility to inflammatory bowel disease: a systematic review and meta-analysis. Clin Res Hepatol Gastroenterol 39(5):627–636
Ng SC, Tsoi KK, Kamm MA, Xia B, Wu J, Chan FK et al (2012) Genetics of inflammatory bowel disease in Asia: systematic review and meta-analysis. Inflamm Bowel Dis 18(6):1164–1176
Siakavellas SI, Bamias G (2018) Checkpoint inhibitor colitis: a new model of inflammatory bowel disease? Curr Opin Gastroenterol 34(6):377–383
Dougan M (2017) Checkpoint blockade toxicity and immune homeostasis in the gastrointestinal tract. Front Immunol 8:1547
Geukes Foppen MH, Rozeman EA, van Wilpe S, Postma C, Snaebjornsson P, van Thienen JV et al (2018) Immune checkpoint inhibition-related colitis: symptoms, endoscopic features, histology and response to management. ESMO Open 3(1):e000278
Wang Y, Abu-Sbeih H, Mao E, Ali N, Qiao W, Trinh VA et al (2018) Endoscopic and histologic features of immune checkpoint inhibitor-related colitis. Inflamm Bowel Dis 24(8):1695–1705
Acknowledgements
We are grateful to Erica Goodoff, from the Department of Scientific Publications, Research Medical Library at The University of Texas MD Anderson Cancer Center for her valuable contribution. We are also grateful to Haidee Chancoco, from the Biospecimen Extraction Resource of MD Anderson Cancer Center for samples processing and DNA extraction.
Funding
This study was funded by a Thrive grant from the Health and Environmental Science Institute to perform this study (Grant Thrive FP00000078). The funding agency had no role in the study’s design; collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit this paper for publication. The University of Texas MD Anderson Cancer Center is supported in part by the National Institutes of Health through Cancer Center Support Grant P30CA016672 and CA016672 (ATGC).
Author information
Authors and Affiliations
Contributions
MES-A and SSS had full access to all of the data in the study and take responsibility for the integrity and the accuracy of the data analysis. Study concept and design: NA, SSS, MES-A. Acquisition of data: NA, AD. Analysis and interpretation of data: NA, RKY. Quality assessment: RKY. Drafting of the manuscript: NA. Critical revision of the manuscript for important intellectual content: AD, RKY, AF, LAC, JHT, RD, VS, SSS, MES-A. Statistical analysis: NA, RKY. Administrative, technical, or material support: SSS, MES-A. Study supervision: SSS, MES-A. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Institutional Review Board at The University of Texas MD Anderson Cancer Center. IRB number: PA16-0928. All patients signed an informed consent prior to study participation.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The is the first pilot genome-wide study to identify genetic variants that may be associated with the risk of developing immune-related adverse events in melanoma patients treated with checkpoint inhibitors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abdel-Wahab, N., Diab, A., Yu, R.K. et al. Genetic determinants of immune-related adverse events in patients with melanoma receiving immune checkpoint inhibitors. Cancer Immunol Immunother 70, 1939–1949 (2021). https://doi.org/10.1007/s00262-020-02797-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-020-02797-0