Abstract
We previously reported that hairy cell leukemia (HCL) patients have high percentages of CD56+/CD57+/CD3+ large granular lymphocytes consistent with cytotoxic T-lymphocytes (CTLs), and other investigators have reported skewing of the T-cell repertoire. In previous studies of up to seven HCL patients, many of the 22 established T-cell receptor (TCR) beta variable region (TRBV) families showed mono- or oligoclonal restriction. To determine whether percentages of CTLs are correlated with TRBV clonal excess, we studied 20 HCL patients with flow cytometry, PCR of TCR gamma and TRBV regions, and fractional gel electrophoresis of PCR-amplified TRBV CDR3 domains (CDR3 spectratyping). Increased percentages of CD3+/CD8+/CD57+ CTLs correlated with more mono/oligoclonal and fewer polyclonal TRBV families (r=0.53; P=0.016). Age correlated with number of mono/oligoclonal TRBV families (r=0.51; P=0.022). Time since last purine analog therapy correlated with number of polyclonal TRBV families (r=0.46; P=0.040), but treatment with the anti-CD22 recombinant immunotoxin BL22 was not related to clonal excess. We conclude that abnormalities in the T-cell repertoire in HCL patients may represent deficient immunity, and may be exacerbated by purine analogs. Increased CD3+/CD57+ T-cells may be a useful marker of abnormal TRBV repertoire in HCL patients, and might prove useful in deciding whether patients should receive biologic antibody-based treatment rather than repeated courses of purine analog for relapsed disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hairy cell leukemia (HCL) is a B-cell lymphoproliferative malignancy representing 2% of all leukemias [1]. Hairy cells are mature though not terminally differentiated [2], and display surface CD19, CD20, CD22, immunoglobulin (Ig), CD79A, CD40, CD103, and CD11c [3, 4]. CD25 is expressed in 80% of patients and the CD25-negative cases usually belong to a poor-prognosis HCL variant (HCLv) group [5–8]. The highly effective purine analogs cladribine (chlorodeoxyadenosine, or CdA) and pentostatin (deoxycoformycin or DCF), are not curative, but induce long-term complete remissions in most patients until repeated courses fail to induce responses [9–16]. Purine analogs cause significant decreases in CD4+ and CD8+ lymphocytes [17, 18]; the median time to recovery of CD4+ T-cells is up to 4 years, longer than the time to recovery of CD8+ T-cells. Immunobiologic targeted treatments include the anti-CD20 monoclonal antibody (MAb) Rituximab [19–22] and the anti-CD22 recombinant immunotoxin BL22 [23, 24], neither of which targets T-cells. Complete and partial remissions have been achieved with both agents in chemoresistant HCL [19–24].
Large granular lymphocytes (LGL) have been described in HCL as either CD3-negative NK cells or CD3+ cytotoxic T-lymphocytes (CTLs) which are clonal but not malignant [25–28]. Kluin-Nelemans et al. reported T-cell restriction in 25 HCL patients by TCR gamma (TCRG) PCR [29], and in seven patients by RT-PCR of each of 22 TCR beta variable (TRBV) families [29, 30]. In one case, expansion of CD8+/CD57+ CTLs was documented [31]. HCL-specific T-cell clones could be isolated from HCL patients [32, 33], but clonally expanded T-cells from two HCL patients were not specific for hairy cells [31]. Thus, it is not clear that T-cell restriction in HCL represents an immune response to the tumor. We have reported that HCL patients after purine analogs frequently have increased percentages of CTLs, defined by CD8+/CD3+, CD56+/CD3+, and CD57+/CD3+ T-cells [34]. In that study, six of six patients analyzed had TCRG restriction by PCR. It has not been possible, due to the limited number of patients previously studied, to determine whether CTL antigens, TCRG PCR, or clinical factors correlate with mono/oligoclonal skewing of the α/β T-cell repertoire.
To explore the relationship between increased percentages of CTL cells and skewing of the T-cell repertoire, we studied 22 purine analog-pretreated HCL patients for TCRG and TRBV restriction, and TRBV expression. We performed flow cytometry to quantify circulating CTLs, NK cells, or CD4+ cells. In 20 of these patients we performed CDR3 spectratyping to determine if each of 22 established TRBV families was expressed in a mono/oligoclonal or polyclonal fashion. To begin to investigate the clinical significance of T-cell restriction in HCL, we studied patients with a wide range of clinical situations with respect to disease burden and prior treatment.
Material and methods
Patients and controls
Blood collected in heparin and EDTA tubes was obtained as part of sample acquisition protocols with informed consent approved by the NCI Investigator’s Review Board. All patients had clinical presentation and morphology consistent with HCL and had a complete peripheral blood mononuclear cell (PBMC) immunophenotypic analysis as part of their standard evaluation. For each patient, dates of blood collection for flow cytometry and PCR results were the same or up to 3 weeks apart. Patients #14 and 18 had hairy cell leukemia variant (HCLv) and the other patients had classic HCL. The two patients with HCLv each had HCL cells negative for CD25 but otherwise consistent with HCL, including positive for CD103 and bright positive for CD22, CD11c and CD20. The diagnosis of HCL by flow cytometry also required the demonstration of monoclonality based on lambda or kappa surface light chains. Peripheral blood specimens from eight normal age-matched donors were also collected in EDTA tubes. The clinical status and response to BL22 of most of the patients was recently reported [23].
Isolation and purification of genomic DNA from whole blood samples
Whole blood was fractionated by centrifugation in CTP Cell Preparation Tubes (BD, Franklin Lakes, NJ). Nucleated cells were removed and washed with phosphate-buffered saline (without Mg2+ or Ca2+). The pelleted, washed cells were lysed with DNAzol (Invitrogen, Carlsbad, CA) and genomic DNA was isolated according to the manufacturer’s methods. The concentration of DNA was determined by optical density at 260 nm. One microgram of purified DNA was placed in 0.5 ml PCR tubes, mixed with 20 μl of Gene Releaser resin (Bioventures Inc., Murfreesboro, TN) and layered with mineral oil. The resin/DNA mixtures were preincubated in a thermocycler (Model 9700, Applied Biosystems, Foster City, CA) according to the Gene Releaser protocol, prior to the addition of the specific polymerase chain reaction (PCR) master mixes.
T-cell receptor gamma chain gene rearrangement studies
To assess clonality of the TCRG gene, consensus primers directed to conserved sequences in the junctional regions (Jγ12, 5′-CAAGTGTTGTTCCACTGCC-3′ and Jp12, 5′-GTTACTATGAGC(T/C)TAGTCC-3′) and framework determinants of the variable regions (Vγ101, 5′-CTCACACTC(C/T)CACTTC-3′ and Vγ11, 5′-TCTGG(G/A)GTCTATTACTGTGC-3′) of the γ chain were used to PCR amplify genomic DNA of each sample as previously described [35]. PCR products were separated and analyzed by 16% polyacrylamide gel electrophoresis and visualized by staining with ethidium bromide.
Purification and separation of peripheral blood CD4+ and CD8+ cells
CD4+ and CD8+ T-cells were positively selected from whole peripheral blood by CD4 and CD8 isolation kits using monoclonal antibodies coupled to magnetic beads (Dynal Biotech, Oslo, Norway), according to the manufacturer’s instructions. Separated CD4+ and CD8+ populations were tested by anti-CD4, anti-CD8, and anti-CD19 monoclonal antibodies (BD Biosciences, San Jose, CA).
Quantitative PCR studies (general)
Reverse transcription was performed followed by quantitative PCR (RT-PCR and RQ-PCR) of total RNA samples from peripheral blood of HCL patients and age-matched control donors. Family-specific forward and reverse primers were used as well as SYBR Green I for reaching high sensitivity. Each family specific reaction was performed in triplicate. A melting curve analysis was done to discriminate between specific and nonspecific PCR products. Intensity values of TCR beta constant region PCR products from tenfold dilutions were used for a reference standard for relative quantification of TRBV family expression.
RNA extraction and cDNA synthesis
Total RNA was extracted from PBMC, CD4+ or CD8+ T-cell populations, using the QIAamp RNA blood Mini kit (QIAGEN, Valencia, CA), according to the manufacturer’s instruction. The 25 μl reaction mixture, containing 1–3 μg total RNA, 2 μl 10 mM dNTP mix (Invitrogen), and 2 μl 0.5 μg/μl Oligo(dT)12–18 primer (Invitrogen) was denatured at 65°C for 5 min and immediately chilled on ice. First strand cDNA synthesis was performed in a 40 μl reaction mixture also containing 8 μl 5 First-Strand Buffer (Invitrogen), 4 μl 0.1 M DTT (Invitrogen), 2 μl of 40 units/μl RnaseOUT (Invitrogen), and 0.5 μl of 200 U/μl SuperScript II RnaseH- Reverse Transcriptase (Invitrogen). The reaction mixture was incubated at 42°C for 50 min, followed by 15 min at 70°C to inactivate Reverse Transcriptase, and then stored at −20°C. The reaction mixture was treated with 1 μl of 2 U/μl Ribonuclease H (Invitrogen) before amplification.
Analysis of TCR beta chain variable gene usage
Quantitative PCR was performed in triplicates in 20 μl final volumes containing 10 μl QuantiTect SYBR Green PCR Kit (QIAGEN GmbH, Hilden, Germany), and 0.5 μl (20 pmole) of each primer in a DNA Engine Opticon 2 Continuous Fluorescence Detector (MJ research, MA). Cycling conditions were 15 min at 95°C, followed by 40 cycles at 10 s at 94°C, 15 s at 58°C, 30 s at 72°C, signal reading at the end of this step, then final elongation for 10 min at 72°C. After the last cycle and final elongation, standard and melting curves were obtained. Amplification efficiencies of different TRBV families were normalized relative to TCR beta chain constant region amplification. For TRBV families [36], specific primers are listed in Table 1 and sequences are presented.
Normalization of TRBV specific cDNA concentration
The TRBV chain-specific cDNA concentration was normalized relative to specific real-time PCR by amplifying part of TCR beta constant genes [37]. Briefly, a 5′ CB-con-F primer (5′-GAG GGT CTC GGC CAC CTT-3′ and 5′-GGC AGA CAG GAC CCC TTG-3′) and a 3′ CB-con-R primer (5′-CCACTGTGCACCTCCTTCC-3′), both specific for TRBC1 and TRBC2 genes, were used in real-time PCR with serial tenfold dilutions of cDNA. The TRBC PCR product intensities were then used by Opticon Monitor 201.10 quantitation software (MJ Research Inc., Waltham, MA) as standards for the relative determination of TRBV chain-specific concentration.
Analysis of CDR3 length spectratyping
CDR3 length polymorphism analysis was performed as described [38]. Briefly, cDNA samples were amplified in 35 cycles of PCR using primers specific for the corresponding TRBV-families as 5′ primers and 3′ CB-con-R3-HEX as the 3′ primer. The products were purified by QIAquik PCR Purification Kit (QIAGEN, Valencia, CA) and checked on 2% agarose gels (Bio-Rad Laboratories, Hercules, CA) stained with ethidium bromide. 10 μl containing TRBV chain-specific PCR product and 15:1 deionized formamide (Applied Biosystem, Foster City, CA) GeneScan-500 ROX Size Standard (Applied Biosystem, Warrington, UK) mixture, was loaded onto 12% denaturing polyacrylamide sequencing gels and the electrophoresis was run in an ABI Prism 3100 sequencing machine. The data were collected by computer and analyzed by the GeneScan 3.1 software. A dominant CDR3 peak (or peaks) was defined as monoclonal (high-intensity signal) when there was a dramatic reduction of other CDR3 signals within that particular TRBV family, when the area under a single peak was >50% of the total area under the curve area [38, 39].
Statistical analysis
Differences in continuously measured parameter values between two groups of patients were evaluated for statistical significance using the Wilcoxon rank sum test. Differences in binomial (dichotomous) parameters were compared using Fisher’s exact test. Correlations were performed using Spearman (nonparametric) rank correlation analysis. Correlations such that |r|>0.70 were interpreted as being strong, those such that 0.50<|r|<0.70 were moderately strong, 0.3<|r|<0.5 were weak to moderately strong, and those with |r|<0.30 were considered weak. The P-value associated with a correlation coefficient is for a test of r=0, and thus is less important in interpretation than the actual magnitude of the coefficient.
All P-values are two-sided and have not been formally adjusted for multiple comparisons. In this exploratory analysis, when many related comparisons were made, in particular when individual evaluations are made for each TRBV family, only P-values such that P<0.005 should be interpreted as being statistically significant, while those such that 0.005<P<0.05 could be considered trends.
Results
To characterize the previous finding that HCL patients frequently have increased percentages of lymphocytes with CTL phenotype and appear oligoclonal with respect to TCRG rearrangements [34], we evaluated HCL patients with and without CTL elevations for oligoclonal restriction in both TCRG and TRBV domains. Blood from patients was used to produce both genomic DNA for TCRG PCR analysis, and cDNA from total RNA to evaluate TRBV usage. In addition to amplification of TRBV chains by PCR for quantitation, fragments were subjected to CDR3 spectratyping to investigate clonality.
Patient characteristics
To examine a wide range of variations in T-cell repertoires, we purposefully used patients with a wide variety of ages, disease burden, type of treatment last used, and time since last treatment. Peripheral blood specimens from 22 patients with a confirmed diagnosis of HCL were examined. As shown in Table 2, the age was 35–83 (median 56), and the male–female ratio was 4.5:1. There were 18–105 (median 51) months since the last purine analog treatment. All patients were treated with BL22 except for patient P00, and two patients were assessed prior to beginning BL22. The remaining 19 patients were tested 10–45 (median 23) months after beginning BL22 and had received 1–23 (median 4) cycles. White blood counts (WBC) were 1.6–61.2 (median 4.4) and circulating hairy cell (HCL) counts were 0–56.9 (median 0.002) cells/l×10−9. Half of the patients had prior splenectomy. Serious infections were uncommon except for Aspergillus pneumonia in P47 just prior to beginning BL22. Patient P14 had Pneumocystis carinii pneumonia and patient 17 had an unspecified pneumonia 33 and 41 months prior to assessment by PCR. Thus, the HCL patients were a diverse group expected to vary widely in T-cell repertoire.
PBMC analysis by flow cytometry
As previously reported [34], patients with HCL had an increase in percent lymphocytes phenotypically consistent with CTLs. As shown in Table 3, which lists % lymphocytes relative to total T-cells, 50, 50, and 41% of patients had high % CD56+/CD3+, CD57+/CD3+, and CD8+/CD3+ cells, respectively. Thus, 15 (68%) of the patients had T-cells with abnormally high percentages of cells phenotypically consistent with CTLs.
TCR gamma rearrangement studies
To detect TCRG restriction, which occurs in the chromosomes of / T-cells, PCR was performed on genomic DNA prepared from PBMCs of HCL patients, and results are shown in Table 3. Examples of restricted and polyclonal gamma chain rearrangement cases are shown in Fig. 1. A restricted (oligoclonal) TCRG pattern was seen in 17 of 22 patients (77%) and the remaining five patients (23%) had a polyclonal pattern. By an exact-Wilcoxon rank sum test, patients with TCRG restriction had higher percentages of CD3+/CD57+ (P=0.005), CD3+/CD56+ (P=0.048), and a trend toward higher % CD3+/CD8+ (P=0.055) cells, but lower percentages of CD3+/CD4+ (P=0.004) cells. TCRG restriction was associated with a lower number of BL22 cycles (P=0.003). Thus, TCRG restriction was not associated with heavier BL22 treatment, and was associated with higher percentages of cells phenotypically consistent with CTLs.
Results of gamma chain T-cell receptor (TCR) rearrangement studies by polymerase chain reaction (PCR). a Example of polyclonal rearrangement, P14. b (P41) and c (phase II patient) represent examples of clonal (restricted) rearrangements. There are two samples at two different time points 9 months apart, demonstrating the stability of the clonal bands. The asterisks represent 118 and 72 bp. The 72 bp marker was used to align the gels
TRBV expression
Since the vast majority of T-cells in our patients, as in most T-cells in the normal population, expressed α/β rather than γ/δ TCRs, we analyzed TRBV gene usage. Relative usage of different TRBV genes in total T-cell populations for 22 HCL patients and eight control donors is shown in Fig. 2a. Compared to normal controls, HCL patients had higher usage of TRBV14, TRBV20-1, and TRBV29-1 and lower usage of TRBV10-3, TRBV19, and TRBV28 (all P<0.01). To determine whether TCRG restriction is associated with over- or underexpression in certain TRBV families, TRBV expression was compared for the 17 patients who had TCRG restriction and the five with TCRG polyclonality (Fig. 2b). Observed trends included higher TRBV18 and TRBV29-1 and lower TRBV11 with restriction (all P=0.030), but these differences were only trends in the context of an analysis of 21 TRBV family subsets.
T-cell receptor beta variable (TRBV) gene family usage. a Comparison between mean values (±SD) of all investigated hairy cell leukemia (HCL) patients and control donors. b Comparison between TRBV usage by HCL patients with restricted (n=17) and polyclonal (n=5) T-cell receptor gamma (TCRG). *, P<0.05, identifying trends or better
Effect of BL22 on T-cells and TRBV gene usage
To determine whether the anti-CD22 recombinant immunotoxin BL22 would decrease circulating T-cells, as was reported for purine analogs [13, 17, 18], the 21 patients treated with BL22 were assessed by flow cytometry before and at 1, 2, 4, 6, 12, 18, and 24 months after beginning BL22. As shown in Fig. 3, median T-cell counts decreased only slightly at 1 month and after 2 months were higher than baseline. A pairwise comparison of individual values for the patients showed no significant difference from baseline for any of the time points (all P>0.05 by Wilcoxon signed rank test). Thus, BL22 showed no evidence of T-cell toxicity which is common with purine analogs, which was expected based on its selective targeting of B-cells.
Changes in T-cells after treatment with BL22. Median T-cell concentrations (cells/mm3) quantified by flow cytometry in the peripheral blood. Mean ± SD (range) values were 862±1,236 (54–5,328) prior to BL22 (n=21), and 287±432 (120–1,897) at 1 month ± 10 days (n=20), 259±306 (119–1,064) at 2 months ± 15 days (n=20), 509±615 (19–2,125) at 4 months ± 20 days (n=16), 371±493 (116–2,062) at 6 months ± 30 days (n=19), 644±1,040 (140–4,847) at 1 year ± 60 days (n=20), 458±714 (265–3,379) at 1.5 years ± 75 days (n=18), and 388±505 (29–1,882) at 2 years ± 90 days (n=15) after beginning BL22. The horizontal dashed line indicates the lower limits of normal (650 cells/mm3)
TRBV CDR3 spectratyping in total T-cell population
To determine whether expression in each TRBV family was polyclonal or mono/oligoclonal, the CDR3 length distribution pattern was determined. The tests were performed on the same T-cell samples as in the TRBV gene usage analyses. As shown in Fig. 4a, the CDR3 distribution patterns, normally Gaussian for families with polyclonal T-cell expression, were compared for HCL patients with or without TCRG restriction, and also with normal age-matched controls. Data were classified for each family of each patient with respect to whether the peaks were monoclonal/oligoclonal, polyclonal, or mixed/indeterminate. The number of families with mono/oligoclonal expression varied from 0 in patients 27 and 29 to 21 in patients 18 and 44 (Table 3). TCRG restriction was associated with evidence of TRBV restriction, including fewer total TRBV peaks/family (P=0.005), higher number of mono/oligoclonal families (P=0.004), and lower number of polyclonal families (0.005).
Relationship of T-cell restriction to clinical factors
Next, the association of TRBV restriction to clinical factors was investigated. By Spearman correlation analysis, it was found that age correlated positively with the number of mono/oligoclonal TRBV families (r=0.51, P=0.022). Time since last purine analog therapy correlated with number of polyclonal families (r=0.46, P=0.040). In contrast, time since last course of BL22 (all |r|<0.15, P>0.65) and number of cycles of BL22 (all |r|<0.25, P>0.35) were not correlated to TRBV PCR results. Thus, purine analog therapy and age but not BL22 therapy were associated with TRBV restriction.
TRBV CDR3 spectratyping in sorted T-cell populations
To further explore whether the mono/oligoclonal CDR3 patterns shown in Fig. 4a were originating from cells consistent with CTLs or from a different type of T-cell, PBMCs were sorted by magnetic beads into CD4+ and CD8+ populations before producing material for PCR. As shown in Fig. 4b, in each of three patients tested, peaks from CD8+ T-cells had much more restriction than did peaks from CD4+ T-cells. Since other markers of CTLs (CD56 and CD57) were present on CD8+ cells but not on CD4+ cells in these patients (data not shown), these data confirm that CTLs were the source for T-cell restriction in these patients.
Relationships between restriction and flow cytometry data
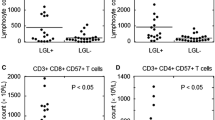
As shown in Table 3 and Fig. 5, there was a moderately strong correlation between percent (%) CD3+/CD57+ and the number of mono/oligoclonal TRBV families per patient (Fig. 5a, r=0.53, P=0.016). There was a moderate inverse correlation between percent (%) CD3+/CD57+ and the number of polyclonal families (Fig. 5b, r=−0.51, P=0.023). The average number of peaks per family, which has been used previously as an index of monoclonality [40], varied from 3.21 to 8.05 (Table 3), correlating inversely with percent (%) CD3+/CD57+ (Fig. 5c, r=−0.42, P=0.067). Overall, the HCL patients evaluated for TRBV CDR3 (n=20) had much lower total expression from polyclonal TRBV families compared to controls (n=8) (Fig. 5d, P<0.0001). No moderately strong correlations were observed between mono/oligoclonality and CD56, CD8, or CD4 (all |r|<0.35, P>0.10).
Correlations and comparisons using percents of T-cells positive for CD57. The number of mono/oligoclonal (filled circle) and polyclonal (open circle) TRBV families/patient are shown in (a) and (b). c Average number of peaks per TRBV family (TRBV complexity score) (filled triangle). Total TRBV expression (open triangle) in polyclonal CDR3 families is shown in (d), contrasting patients (n=20) with normal (controls n=8), P<0.0001 for the difference. Coefficients and P-values in the text are from Spearman correlations, while the lines in the figures show simple linear regression of the actual data
Discussion
Previous TRBV studies in two to seven HCL patients reported TRBV skewing [29–31, 41]. In the present manuscript, we studied enough patients to investigate quantitative relationships between flow cytometry data and restriction in the TRBV repertoire. We found that the percent of T-cells positive for CD57 was moderately strongly correlated with both TCRG and TRBV restriction. Sorting experiments confirmed that the restriction was from the CD8+ CTLs and not from CD4+ T-cells. Our data is able to show for the first time that patients with increased CTLs had loss of polyclonality, worse with more recent purine analog therapy, but not BL22 immunotoxin therapy.
We found that CD3+/CD57+ T-lymphocyte percentage correlated with more restricted and fewer polyclonal TRBV families, and that fewer polyclonal TRBV families correlated with recent purine analog but not with BL22 therapy. Correlations were not as strong for the percent of T-cells positive for CD56, CD8, or CD4 and restriction. Although clonally skewed T-cells in HCL have been observed in some cases to be CD4+ [28, 29], CTLs are generally CD8+/CD4-. The lack of correlation with CD56 is probably because a lower percentage of CTLs are CD56+ than CD57+. Finally, CD8 is a marker expressed by many types of T-cells not classified as CTLs [42]. We used the % of T-cells which were CD3+/CD57+ rather than the % of total lymphocytes so the results would not be distorted by patients with high numbers of circulating malignant lymphocytes.
Monoclonal and oligoclonal expansion have been reported for T-cells in patients with other malignancies [43–45]. In CLL, myeloma, and chronic myelogenous leukemia, oligoclonally expanded CTLs have been reported to be directed against autologous tumor antigens [43, 46, 47]. After allogeneic stem cell transplant, a low number of TRBV CDR3 peaks/family (restriction) correlated with the development of chronic graft vs host disease [40]. We originally speculated that elevations of CTLs in HCL may reflect an immune response to the tumor, since in several cases CTLs increased during follow-up [34]. The more recent study [31] showed that the CTLs in two HCL patients were not autoreactive. Our data show a substantial decrease in polyclonality with TRBV restriction, which suggests such patients may have more limited cell-mediated immunity.
Either cladribine or pentostatin can induce profound T-cell toxicity with reductions in CD4+ lymphocytes until a median of 4 years [13, 17, 18]. Reductions in CD8+ lymphocytes are significant but less prolonged. Median nadirs after cladribine of 139/μl and 92/μl for CD4 and CD8+ lymphocytes have been reported [17]. It is likely that purine analogs, particularly with repeated courses, may eliminate normal T-cell clones and exacerbate restriction in the T-cell repertoire. In the future, it may be advantageous to use biologic therapies like BL22 as front line therapy for HCL due to their lack of T-cell toxicity. However, at the current time, BL22 is being tested only in patients who are resistant to purine analogs. Currently, there are no firm guidelines for when to retreat HCL patients with purine analogs rather than T-cell sparing biologic agents. It is possible that mono/oligoclonal skewing of the T-cell repertoire, as easily measured by CD3+/CD57+ percentage, could be useful to assess immune damage due to prior purine analog therapy. Relationships of these measures to infectious morbidity and mortality will require analysis of several-fold higher numbers of patients. Over the next several years, we anticipate that a high percentage of HCL patients pretreated with purine analogs will receive anti-CD22 immunotoxin and/or ritixumab. To determine when these T-cell sparing agents should be used instead of repeat courses of purine analog for relapsed HCL, it may be useful to follow patients carefully for markers of immune function, and our data suggest that the percent of CD57+/CD3+ T-cells could be a useful marker.
References
Goodman GR, Bethel KJ, Saven A (2003) Hairy cell leukemia: an update. Curr Opin Hematol 10:258–266
Polliack A (2002) Hairy cell leukemia: biology, clinical diagnosis, unusual manifestations and associated disorders discussion (449–350). Rev Clin Exp Hematol 6:366–388
Robbins BA, Ellison DJ, Spinosa JC, Carey CA, Lukes RJ, Poppema S, Saven A, Piro LD (1993) Diagnostic application of two-color flow cytometry in 161 cases of hairy cell leukemia. Blood 82:1277–1287
Matutes E, Morilla R, Owusu-Ankomah K, Houliham A, Meeus P, Catovsky D (1994) The immunophenotype of hairy cell leukemia (HCL). Proposal for a scoring system to distinguish HCL from B-cell disorders with hairy or villous lymphocytes. Leuk Lymphoma 14(Suppl 1):57–61
Juliusson G, Liliemark J (1992) Rapid recovery from cytopenia in hairy cell leukemia after treatment with 2-chloro-2’-deoxyadenosine (CdA): relation to opportunistic infections. Blood 79:888–894
Hoffman MA, Janson D, Rose E, Rai KR (1997) Treatment of hairy-cell leukemia with cladribine: response, toxicity, and long-term follow-up. J Clin Oncol 15:1138–1142
Blasinska-Morawiec M, Robak T, Krykowski E, Hellmann A, Urbanska-Rys H (1997) Hairy cell leukemia-variant treated with 2-chlorodeoxyadenosine–a report of three cases. Leuk Lymphoma 25:381–385
Matutes E, Wotherspoon A, BritoBabapulle V, Catovsky D (2001) The natural history and clinico-pathological features of the variant form of hairy cell leukemia. Leukemia 15:184–186
Flinn IW, Kopecky KJ, Foucar MK, Head D, Bennett JM, Huchison R, Corbett W, Cassileth P, Habermann T, Golomb H, Rai K, Eisenhauer E, Appelbaum F, Cheson B, Grever MR (2000) Long-term follow-up of remission duration, mortality, and second malignancies in hairy cell leukemia patients treated with pentostatin. Blood 96:2981–2986
Maloisel F, Benboubker L, Gardembas M, Coiffier B, Divine M, Sebban C, Blanc M, Abgrall JF, Lederlin P, Harousseau JL, Blaise AM, Grosbois B, Morice P, Ghandour C, Castaigne S (2003) Long-term outcome with pentostatin treatment in hairy cell leukemia patients. A French retrospective study of 238 patients. Leukemia 17:45–51
Goodman GR, Burian C, Koziol JA, Saven A (2003) Extended follow-up of patients with hairy cell leukemia after treatment with cladribine. J Clin Oncol 21:891–896
Tallman MS, Peterson LC, Hakimian D, Gillis S, Polliack A (1999) Treatment of hairy-cell leukemia: current views. Semin Hematol 36:155–163
Saven A, Burian C, Koziol JA, Piro LD (1998) Long-term follow-up of patients with hairy cell leukemia after cladribine treatment. Blood 92:1918–1926
Cheson BD, Sorensen JM, Vena DA, Montello MJ, Barrett JA, Damasio E, Tallman M, Annino L, Connors J, Coiffier B, Lauria F (1998) Treatment of hairy cell leukemia with 2-chlorodeoxyadenosine via the Group C protocol mechanism of the National Cancer Institute: a report of 979 patients. J Clin Oncol 16:3007–3015
Filleul B, Delannoy A, Ferrant A, Zenebergh A, Van Daele S, Bosly A, Doyen C, Mineur P, Glorieux P, Driesschaert P, Sokal C, Martiat P, Michaux JL (1994) A single course of 2-chloro-deoxyadenosine does not eradicate leukemic cells in hairy cell leukemia patients in complete remission. Leukemia 8:1153–1156
Carbone A, Reato G, Di Celle PF, Lauria F, Foa R (1994) Disease eradication in hairy cell leukemia patients treated with 2- chlorodeoxyadenosine [letter]. Leukemia 8:2019–2020
Seymour JF, Kurzrock R, Freireich EJ, Estey EH (1994) 2-chlorodeoxyadenosine induces durable remissions and prolonged suppression of CD4+ lymphocyte counts in patients with hairy cell leukemia. Blood 83:2906–2911
Seymour JF, Talpaz M, Kurzrock R (1997) Response duration and recovery of CD4+ lymphocytes following deoxycoformycin in interferon-alpha-resistant hairy cell leukemia: 7- year follow-up. Leukemia 11:42–47
Lauria F, Lenoci M, Annino L, Raspadori D, Marotta G, Bocchia M, Forconi F, Gentili S, La Manda M, Marconcini S, Tozzi M, Baldini L, Zinzani PL, Foa R (2001) Efficacy of anti-CD20 monoclonal antibodies (Mabthera) in patients with progressed hairy cell leukemia. Haematologica 86:1046–1050
Hagberg H, Lundholm L (2001) Rituximab, a chimaeric anti-CD20 monoclonal antibody, in the treatment of hairy cell leukaemia. Br J Haematol 115:609–611
Nieva J, Bethel K, Saven A (2003) Phase 2 study of rituximab in the treatment of cladribine-failed patients with hairy cell leukemia. Blood 102:810–813
Thomas DA, O’Brien S, Bueso-Ramos C, Faderl S, Keating MJ, Giles FJ, Cortes J, Kantarjian HM (2003) Rituximab in relapsed or refractory hairy cell leukemia. Blood 102:3906–3911
Kreitman RJ, Squires DR, Stetler-Stevenson M, Noel P, Fitzgerald DJ, Wilson WH, Pastan I (2005) Phase I trial of recombinant immunotoxin RFB4(dsFv)-PE38 (BL22) in patients with B-cell malignancies. J Clin Oncol 23:6719–6729
Kreitman RJ, Wilson WH, Bergeron K, Raggio M, Stetler-Stevenson M, FitzGerald DJ, Pastan I (2001) Efficacy of the Anti-CD22 Recombinant Immunotoxin BL22 in Chemotherapy-Resistant Hairy-Cell Leukemia. New Engl J Med 345:241–247
Ruco LP, Procopio A, Maccallini V, Calogero A, Uccini S, Annino L, Mandelli F, Baroni CD (1983) Severe deficiency of natural killer activity in the peripheral blood of patients with hairy cell leukemia. Blood 61:1132–1137
Smith BR, Rosenthal DS, Ault KA (1985) Natural killer lymphocytes in hairy cell leukemia: presence of phenotypically identifiable cells with defective functional activity. Exp Hematol 13:189–193
Bassan R, Rambaldi A, Allavena P, Abbate M, Marini B, Barbui T (1988) Association of large granular lymphocyte/natural killer cell proliferative disease and second hematologic malignancy. Am J Hematol 29:85–93
Airo P, Rossi G, Facchetti F, Marocolo D, Garza L, Lanfranchi A, Prati E, Brugnoni D, Malacarne F, Cattaneo R (1995) Monoclonal expansion of large granular lymphocytes with a CD4+ CD8dim+/− phenotype associated with hairy cell leukemia. Haematologica 80:146–149
Kluin-Nelemans JC, Kester MG, Melenhorst JJ, Landegent JE, van de Corput L, Willemze R, Falkenburg JH (1996) Persistent clonal excess and skewed T-cell repertoire in T cells from patients with hairy cell leukemia. Blood 87:3795–3802
Kluin-Nelemans HC, Kester MG, van deCorput L, Boor PP, Landegent JE, van Dongen JJ, Willemze R, Falkenburg JH (1998) Correction of abnormal T-cell receptor repertoire during interferon-alpha therapy in patients with hairy cell leukemia. Blood 91:4224–4231
Spaenij-Dekking EH, Van der Meijden ED, Falkenburg JH, Kluin-Nelemans JC (2004) Clonally expanded T cells in hairy cell leukemia patients are not leukemia specific. Leukemia 18:176–178
van de Corput L, Kluin-Nelemans HC, Kester MG, Willemze R, Falkenburg JH (1999) Hairy cell leukemia-specific recognition by multiple autologous HLA-DQ or DP-restricted T-cell clones. Blood 93:251–259
Spaenij-Dekking EH, Van Delft J, Van Der Meijden E, Hiemstra HS, Falkenburg JH, Koning F, Drijfhout JW, Kluin-Nelemans JC (2003) Synaptojanin 2 is recognized by HLA class II-restricted hairy cell leukemia-specific T cells. Leukemia 17:2467–2473
Xie XY, Sorbara L, Kreitman RJ, Fukushima PI, Kingma DW, Stetler-Stevenson M (2000) Development of lymphoproliferative disorder of granular lymphocytes in association with hairy cell leukemia. Leukemia & Lymphoma 37:97–104
McCarthy KP, Sloane JP, Kabarowski JH, Matutes E, Wiedemann LM (1992) A simplified method of detection of clonal rearrangements of the T-cell receptor-gamma chain gene. Diagn Mol Pathol 1:173–179
Nishimura MI, Kawakami Y, Charmley P, O’Neil B, Shilyansky J, Yannelli JR, Rosenberg SA, Hood L (1994) T-cell receptor repertoire in tumor-infiltrating lymphocytes. Analysis of melanoma-specific long-term lines. J Immunother Emphasis Tumor Immunol 16:85–94
Choi YW, Kotzin B, Herron L, Callahan J, Marrack P, Kappler J (1989) Interaction of Staphylococcus aureus toxin “superantigens” with human T cells. Proc Natl Acad Sci USA 86:8941–8945
Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P (1993) The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci USA 90:4319–4323
Serrano D, Monteiro J, Allen SL, Kolitz J, Schulman P, Lichtman SM, Buchbinder A, Vinciguerra VP, Chiorazzi N, Gregersen PK (1997) Clonal expansion within the CD4+CD57+ and CD8+CD57+ T cell subsets in chronic lymphocytic leukemia. J Immunol 158:1482–1489
Tsutsumi Y, Tanaka J, Miura Y, Toubai T, Kato N, Fujisaw F, Toyoshima N, Ota S, Mori A, Yonezumi M, Chiba K, Kondo T, Hasino S, Kobayasi R, Masauji N, Kasai M, Asaka M, Imamura M (2004) Molecular analysis of T-cell repertoire in patients with graft-versus-host disease after allogeneic stem cell transplantation. Leuk Lymphoma 45:481–488
Van De Corput L, Falkenburg JH, Kluin-Nelemans JC (1998) T-cell dysfunction in hairy cell leukemia: an updated review. Leuk Lymphoma 30:31–39
Rocha B, Tanchot C (2004) CD8 T cell memory. Semin Immunol 16:305–314
Farace F, Orlanducci F, Dietrich PY, Gaudin C, Angevin E, Courtier MH, Bayle C, Hercend T, Triebel F (1994) T cell repertoire in patients with B chronic lymphocytic leukemia. Evidence for multiple in vivo T cell clonal expansions. J Immunol 153:4281–4290
Rezvany MR, Jeddi-Tehrani M, Osterborg A, Kimby E, Wigzell H, Mellstedt H (1999) Oligoclonal TCRBV gene usage in B-cell chronic lymphocytic leukemia: major perturbations are preferentially seen within the CD4 T-cell subset. Blood 94:1063–1069
Alatrakchi N, Farace F, Frau E, Carde P, Munck JN, Triebel F (1998) T-cell clonal expansion in patients with B-cell lymphoproliferative disorders. J Immunother 21:363–370
Brown RD, Yuen E, Nelson M, Gibson J, Joshua D (1997) The prognostic significance of T cell receptor beta gene rearrangements and idiotype-reactive T cells in multiple myeloma. Leukemia 11:1312–1317
Molldrem JJ, LeePP, Wang C, Felio K, Kantarjian HM, Champlin RE, Davis MM (2000) Evidence that specific T lymphocytes may participate in the elimination of chronic myelogenous leukemia. Nat Med 6:1018–1023
Acknowledgments
We thank technicians, nurses, patient care coordinators, and data managers from the Clinical Immunotherapy Section of the Laboratory of Molecular Biology, including Karen Bergeron, Kelly Cahill, Linda Ellison, Sonya Duke, Rita Mincemoyer, Barbara Debrah, and Inger Margulies. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arons, E., Sorbara, L., Raffeld, M. et al. Characterization of T-cell repertoire in hairy cell leukemia patients before and after recombinant immunotoxin BL22 therapy. Cancer Immunol Immunother 55, 1100–1110 (2006). https://doi.org/10.1007/s00262-005-0099-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-005-0099-1