Abstract
Purpose
We report findings from the first-in-human study of [11C]MDTC, a radiotracer developed to image the cannabinoid receptor type 2 (CB2R) with positron emission tomography (PET).
Methods
Ten healthy adults were imaged according to a 90-min dynamic PET protocol after bolus intravenous injection of [11C]MDTC. Five participants also completed a second [11C]MDTC PET scan to assess test-retest reproducibility of receptor-binding outcomes. The kinetic behavior of [11C]MDTC in human brain was evaluated using tissue compartmental modeling. Four additional healthy adults completed whole-body [11C]MDTC PET/CT to calculate organ doses and the whole-body effective dose.
Results
[11C]MDTC brain PET and [11C]MDTC whole-body PET/CT was well-tolerated. A murine study found evidence of brain-penetrant radiometabolites. The model of choice for fitting the time activity curves (TACs) across brain regions of interest was a three-tissue compartment model that includes a separate input function and compartment for the brain-penetrant metabolites. Regional distribution volume (VT) values were low, indicating low CB2R expression in the brain. Test-retest reliability of VT demonstrated a mean absolute variability of 9.91%. The measured effective dose of [11C]MDTC was 5.29 μSv/MBq.
Conclusion
These data demonstrate the safety and pharmacokinetic behavior of [11C]MDTC with PET in healthy human brain. Future studies identifying radiometabolites of [11C]MDTC are recommended before applying [11C]MDTC PET to assess the high expression of the CB2R by activated microglia in human brain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endocannabinoid signaling in human brain stems from endocannabinoid (endogenous anandamide, 2-arachidonoylglycerol) binding to cannabinoid receptors such as the cannabinoid receptors type 1 (CB1R) and type 2 (CB2R). Unlike the CB1R that is expressed in relatively high levels across the human brain, the CB2R is most highly expressed in peripheral lymphatic tissues and is expressed at relatively low level by glial, neuronal, and endothelial cells in healthy brain [1]. In contrast, high CB2R expression in brain tissue has been reported in cases of neurologic conditions, including postmortem cases of Alzheimer’s disease (AD) and Parkinson’s disease (PD) [2]. While microglia may be the cellular source of high CB2R expression in AD and PD [3, 4], the availability of CB2R on dopaminergic midbrain neurons in PD may diminish over the course of neuronal loss [5]. Beyond research to understand the role of CB2R across various neurodegenerative conditions, early efforts are emerging to target CB2R therapeutically for neuroprotection [6]. Imaging CB2R may yield further insight into its role in shaping neurologic diseases, with potential also for informing and monitoring relevant, therapeutic interventions.
We recently developed [11C]MDTC [7] with high binding affinity for the CB2R (ki = 0.7–1.2 nM) for PET imaging in vivo. In contrast to low [11C]MDTC uptake in healthy mouse brain, lipopolysaccharide (LPS)-treated mice with robust neuroinflammation showed specific uptake of [11C]MDTC across the brain. Furthermore, specific [11C]MDTC uptake was found in areas of amyloid-β plaque burden in a mouse model of AD (APPswe/PS1dE9 mice) [7]. [11C]MDTC is a radioactive analogue of A-936339, a selective CB2R agonist that has been demonstrated to have high affinity for the CB2R [8]. A-936339 was developed by Abbott Laboratories for studying CB2R activation in the central nervous system and for investigating pharmaceuticals targeting the CB2R [8].
Here, we report pharmacokinetic data from first-in-human use of [11C]MDTC PET in healthy brain as well as test-retest reproducibility of regional brain binding. We also report [11C]MDTC whole-body human dosimetry data.
Materials and methods
Murine radiometabolite analysis
Male CD-1 mice (31–35 g) were injected via the lateral tail vein with 7.4 MBq of [11C]MDTC. The mice were sacrificed by cervical dislocation at 10 and 30 min after injection, and a terminal blood sample was obtained. The mouse brains were rapidly removed, homogenized in 2 mL of ACN:H2O 50–50% (vol) with 0.06-M-ammonium formate to extract parent radiotracer and radiometabolites, and centrifuged at 20,000 g at 4 °C. Resulting blood and brain samples were analyzed by reverse phase high-performance liquid chromatography (RP-HPLC).
Human subjects
This study received approval from a Johns Hopkins Institutional Review Board and was conducted under a US investigational new drug application (IND 131367). All participants provided written informed consent to one of the following study portions: (1) brain pharmacokinetics, (2) test-retest reproducibility of brain pharmacokinetics, or (3) whole-body dosimetry.
Each subject completed clinical interview and physical assessment, with screening blood work, electrocardiogram, and urine toxicology. Adult volunteers in stable health were included. Exclusion criteria included any evidence of an unstable medical condition; current or past psychiatric or neurologic illness (including history of head injury with loss of consciousness); history of recreational substance abuse including marijuana (assessed by self-report and urine toxicology); current psychotropic medication use; contraindication to magnetic resonance imaging (MRI) (i.e., implanted metal, claustrophobia); or contraindication to PET imaging with arterial line placement (i.e., clotting factor deficiency, pregnancy/breastfeeding). The brain pharmacokinetics and test-retest reproducibility portions also excluded individuals with any clinically relevant, structural abnormality found on screening structural brain MRI.
Ten individuals [six men and four women, ages 32.7 ± 11.4 years, body mass index (BMI) 25.5 ± 2.8] underwent [11C]MDTC brain PET. Five (three men, two women) of these 10 participants completed a second [11C]MDTC brain PET within 1 week of the baseline scan in order to assess test-retest reproducibility. Four unique, additional participants (two men, two women; ages 27.8 ± 3.0 years, BMI 21.9 ± 2.5) underwent [11C]MDTC PET/CT for whole-body dosimetry.
Brain MRI
MR structural T1-weighted imaging was acquired on a Siemens MAGNETOM Prisma or on a Siemens MAGNETOM Trio 3 Tesla scanner (Malvern, PA, USA) with a Trio head matrix 12-channel head coil or Prisma head/neck 20-channel head coil, to obtain a 3-dimensional (3D) magnetization-prepared rapid gradient-echo (MP-RAGE) sequence. Images were reconstructed to 320 × 320 × 192 matrix, with voxel size 0.8 × 0.8 × 0.8 mm.
Radiosynthesis
[11C]MDTC was prepared as previously described [7], with molar activity of 309.1 ± 96.1 GBq/μmol at the time of injection. The radiochemical purity was 98.1 ± 1.3 % at the end of synthesis.
Dynamic brain PET
[11C]MDTC dynamic PET neuroimaging was performed on a High-Resolution Research Tomograph scanner (HRRT, Siemens Healthcare, Knoxville, TN, USA), which is an LSO-based, dedicated brain PET scanner with 2.5-mm reconstructed image resolution [9]. Each emission scan started at the time of bolus intravenous injection of [11C]MDTC. The average injected mass across all [11C]MDTC brain PET studies was 0.8 ± 0.3 μg, with injected radioactivity of 639.0 ± 65.2 MBq. Ninety minutes of continuous listmode data were acquired from each participant. All participants were fitted with a thermoplastic facemask to minimize head movement.
The PET images were reconstructed into 30 frames (four 15-s, four 30-s, three 1-min, two 2-min, five 4-min, and twelve 5-min frames) using the iterative ordinary-Poisson ordered-subset expectation-maximization algorithm (6 iterations, 16 subsets, post-reconstruction Gaussian filtering with 2-mm full width at half maximum), with correction for radioactive decay, dead time, attenuation, scatter, and randoms [9]. The attenuation maps were generated from a 6-min transmission scan performed with a 137Cs point source prior to each emission scan. The reconstructed image volume spanned 31 cm × 31 cm transaxially and 25 cm axially. The image matrix consisted of 256 × 256 × 207 voxels and a voxel size of 1.22 × 1.22 × 1.22 mm.
Arterial blood measurement
Arterial blood sampling was conducted over the course of each [11C]MDTC brain PET for generation of a metabolite-corrected arterial input function. Blood samples were obtained at the following intervals after bolus radiotracer injection (p.i.): From time of injection to 1.5 min p.i., samples were collected as fast as possible (average, 16 samples); from 1.5 to 3 min p.i, samples were collected every 30 s; from 3 to 5 min p.i, samples were collected every 2 min; from 5 to 30 min p.i., samples were collected every 5 min; and from 30 to 90 min p.i., samples were collected every 15 min. Plasma was immediately isolated from each whole blood sample using centrifugation, and radioactivity was counted in a cross-calibrated well-counter.
The fraction of parent [11C]MDTC and radiometabolites in plasma was determined by RP-HPLC analysis from collection of 3–6 mL blood samples before radiotracer injection (background) and at 5, 10, 20, 30, 45, 60, and 90 min p.i. The RP-HPLC system was standardized using [11C]MDTC, and its nonradioactive analog before blood plasma sample analysis. The RP-HPLC method [10] was used with an Agilent Technologies system containing a 1260 Infinity Quaternary Pump, a 1260 Infinity Column Compartment module, a 1260 Infinity UV Detector, and a Raytest GABI Star radiation detector that was controlled by OpenLab CDS EZChrom (A.01.04) software. The HPLC system was equipped with a 4 × 20 mm capture column packed with Phenomenex Strata™-X 33-μm Polymeric Reverse Phase sorbent and a Waters reverse phase XBridge BEH C18 5 μM 4.6 × 150-mm analytical column. Plasma samples (0.1–1.2 mL) were loaded onto a 2-mL Rheodyne injector loop and initially directed to the capture column and detectors. The capture column mobile phase consisted of 1% acetonitrile aqueous solution at 2 mL/min. After 1 min of elution, analytical mobile phase was applied at a flow rate of 2 mL/min. The analytical mobile phase contained 65% acetonitrile and 35% aqueous solution of 0.06-M-ammonium formate. The time-profile percentages of the parent ligand and radiometabolites were then generated from the HPLC results and applied to the total plasma time activity curve (TAC) to generate a metabolite-corrected plasma TAC of the parent as well as a plasma radiometabolites TAC.
[11C]MDTC brain PET kinetic modeling and test-retest analyses
[11C]MDTC brain PET image processing and kinetic analysis were performed using the software package PMOD (v3.7, PMOD Technologies Ltd., Zurich, Switzerland). When necessary, post-reconstruction inter-frame motion correction was applied using PMOD using a static reference frame from the average of frames corresponding to 30–60 min p.i., with rigid, frame-by-frame matching of dynamic data to the reference image. PET-MRI co-registration was completed using PMOD as previously described [11]. The attenuation map was not realigned to the emission data after co-registration since there was no significant motion detected during any scan, as assessed by head counts monitored over the dynamic PET. Segmentation of T1-weighted MR images were performed with the FreeSurfer image analysis suite (http://surfer.nmr.mgh.harvard.edu/). Eleven regions of interest (ROIs) were selected for this study, including cerebellar cortex, brainstem, thalamus, striatum, hippocampus, temporal cortex, occipital cortex, cingulate cortex, frontal cortex, parietal cortex, and the corpus callosum. PET TACs were generated for all subjects using the defined ROIs.
Compartmental model fitting was applied to the regional TACs. A two-tissue compartment model (2TCM) with 4 parameters (K1, k2, k3, k4) was fitted with the metabolite-corrected arterial input function [12, 13]. A three-tissue compartment with metabolite model (3TCM) was also used. This model includes a separate input function and compartment for brain-penetrant metabolites with nonspecific binding [14]. The fitted parameters from this model were K1p, k2p, k3, and k4 of the [11C]MDTC based on the metabolite-corrected input function and K1m and k2m of the metabolites based on the plasma metabolites input function. The blood volume fraction (vB) was first fitted using whole plasma input function without correction and then fixed for fitting other parameters with the metabolite-corrected plasma input function. The whole plasma input was used because whole blood activities were not measured. In addition, a delay of plasma data was also fitted using whole brain uptake and then fixed for all ROIs. The primary outcome was the total distribution volume, VT, which reflects the equilibrium ratio of [11C]MDTC concentration in tissue to the concentration in arterial plasma [12]. The distribution volume in the specific compartment, VS, was also evaluated for the 3TCM [12, 14]. Nonlinear least-squares analysis was performed, with the Marquardt algorithm for parameter estimation.
Within-subject changes in VT between test and retest were calculated as ΔVT = 2·(VT1–VT2)/(VT1 + VT2), with the numeric subscripts distinguishing values from test (1) and retest (2). The mean absolute deviation, 2·|VT1–VT2|/(VT1 + VT2), was also calculated and reported as a metric of reliability. A Bland-Altman plot was also generated to compare regional VT estimates between the test and retest scans. Similar test-retest analysis was also applied to VS.
[11C]MDTC PET/CT whole-body dosimetry
Each whole-body PET/CT was performed on a Biograph mCT PET/CT system (Siemens Healthineers). A low-dose CT was first performed to obtain an attenuation map, followed by a 60-min dynamic whole-body PET scan that began at time of bolus injection. The average injected dose of radioactivity was 614.8 ± 86.0 MBq with injected mass 0.6 ± 0.1 μg. The dynamic PET protocol included 8 repeated whole-body scan frames, each with 7 bed positions to cover each participant from vertex to mid-thigh. The time per bed position and total scan time for each frame were 0.3 min/bed and 2.3 min, 0.5 min/bed and 3.5 min, 0.7 min/bed and 4.7 min, 0.8 min/bed and 5.8 min, 1 min/bed and 7 min, 1.5 min/bed and 10.5 min, 2 min/bed and 14 min, and 2 min/bed and 14 min, respectively. The images were reconstructed using the ordered-subset expectation maximization (OSEM) algorithm with corrections for attenuation, scatter, randoms, and dead time.
The ROIs of 25 organs of interest were delineated on each co-registered CT image by a trained radiologist. The ROIs were then applied to the dynamic PET images to obtain TACs that were generated without decay correction. Time-integrated activity coefficients in hours were calculated by normalizing the TACs by injected dose and standard organ volume estimates (ICRP 89 adult male and female reference computational phantoms) and then integrating the normalized TACs using trapezoidal integration. The area under the curve was extrapolated beyond the end of the scan by computing the integral to infinity, using a bi-exponential fit to the data 15- to 60-min postinjection. Time-integrated activity coefficients were then entered into OLINDA software (Version 2.1, Vanderbilt University, TN, USA) [15] for estimation of radiation dose in individual organs and the whole-body effective dose for each participant. The final results were computed by averaging across all four participants.
Statistical analysis
Statistical analyses were performed using SPSS Statistics (Version 25.0, IBM Corp., Armonk, NY, USA). Data were checked for outliers, and descriptive statistics were obtained. Normality of the data was assessed using the Shapiro-Wilk test. Data are presented as mean ± standard deviation unless otherwise noted. Compartmental model fitting was assessed by visual inspection of the model fit to the TACs and by relative goodness of fit using the Akaike information criterion (AIC) and F-test [16]. Multivariate analysis of variance (MANOVA) was used to evaluate if age, sex, or BMI had effect on VT or VS values.
Results
[11C]MDTC murine radiometabolite study
Mouse plasma RP-HPLC data showed evidence of radiometabolites of [11C]MDTC, although there was no evidence of any radiometabolites that eluted after [11C]MDTC (Supplemental Fig. S2). Complementary RP-HPLC using the murine brain samples indicates that those plasma metabolites that eluted between 3.5 and 6 min are brain penetrant in mice.
Pharmacological effects in human
Injection of [11C]MDTC in humans did not yield adverse effects, and there were no clinical changes in pre- versus postinjection laboratory (blood, electrocardiogram) results.
Arterial plasma measures
Arterial blood plasma from each unique human participant across the [11C]MDTC brain PET studies (n = 10) was evaluated for radiotracer metabolism. HPLC revealed more polar metabolites that separated well from the parent compound, which agrees with HPLC data from mice (Fig. 1A). The parent compound fraction declined relatively quickly over the 90 min, with ~15% remaining at 60 min and less than 10% parent at 90 min. The fraction of radiometabolites rapidly increased and plateaued at ~70% after 30 min (Fig. 1B). The overall plasma measurement of [11C]MDTC peaked within the first two min p.i and declined over the course of the 90-min scan (Fig. 1C). The total activity of radiometabolites in plasma peaked around 15 min and then gradually decreased (Fig. 1D).
Behavior of [11C]MDTC in human blood. A A representative RP-HPLC chromatogram of plasma isolated from a blood sample collected 90 min after [11C]MDTC administration, revealing metabolites that separated well from the parent [11]MDTC. Radiometabolites that may enter the brain are labeled. B Across the healthy population (n = 10), the parent [11C]MDTC fraction declined relatively quickly over the 90 min, with ~15% remaining at 60 min and less than 10% parent at 90 min. The data at 0 min is the radiochemical purity (0.981 ± 0.013). Radiometabolites increased to 70% at 30 min. C Plasma parent activity peaked in the first 2 min after bolus injection of [11C]MDTC (insert) with subsequent decline over 90 min p.i. D Radiometabolite plasma activity peaked at 15 min. Data are shown from a representative participant
Brain activity and modeled binding
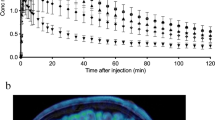
[11C]MDTC showed peak uptake across all brain ROIs within 1 to 2 min, followed by fast washout from brain tissue (Fig. 2A–D). The peak whole brain uptake was 4.88 ± 1.58% of the injected dose. Cortical regions (both cerebral cortex and cerebellar cortex) showed highest uptake, with peak standardized uptake values (SUVs) of 2.0 to 3.0 for cortical regions. The ROI with lowest uptake was the corpus callosum, with a peak SUV of 1.5. Cortical regions showed faster clearance than the white matter regions from 15-min to 60-min postinjection. After 60 min, all ROIs showed similar, relatively low activity.
Average time activity curves (TACs) (N = 10) across 11 brain regions. Top, A cerebellar cortex and subcortical TACs for the full 90 min, and B the first 16 min is shown separately. Bottom, C other cortical and corpus callosal TACs from the full 90 min and D the first 16 min. Regions are listed in each panel (top, bottom) in order of highest to lowest peak activity
Kinetic modeling
The blood volume fraction estimated from the whole plasma data was 3.2 ± 0.7% (coefficient of variation = 21%). The average fitted delay was 2.7 ± 2.6 s. Visually, both the 2TCM and 3TCM fitted the TACs well across all brain ROIs (Fig. 3). The Akaike information criteria (Supplemental Table S1) from both 2TCM and 3TCM were very similar, while 3TCM had smaller AIC values in 60 out of 110 regions (54.5%). The F-test indicated that the 3TCM was a significantly better fit (α = 0.05) compared to 2TCM in 53 out of those 60 regions.
The estimated parameters from 2TCM and from 3TCM are shown in Supplemental Table S2 and Supplemental Table S3, respectively. K1 was estimated well (mean fitting standard error (SE) = 3.8%) across the ROIs (0.12 ± 0.03 mL/min/cm3) using 3TCM (Supplemental Table S3) except corpus callosum that had a SE of 6.6%. The standard error of k2p, k3, k4, VT, and VS was all larger than 5% for most of the regions. The mean SE was 13.9% for VT and 61.2% for VS, respectively, for 3TCM. VT values estimated from 3TCM were highest in the thalamus and lowest in the hippocampus (Fig. 4, Supplemental Table S3). The VS values were highest in corpus callosum and thalamus and lowest in the cingulate cortex. There was no effect of sex, age, or BMI on 3TCM regional VT and VS for the ten young healthy participants in this study.
Test-retest variability
The [11C]MDTC brain PET test-retest data (N = 5) demonstrated good reproducibility of 3TCM-derived VT estimates, with mean test-retest variability in regional VT (ΔVT ) = 6.75 ± 5.15%. The mean absolute deviation in regional VT = 9.91 ± 2.81%, except carpus callosum which had a mean absolute deviation of 21.1%. The Bland-Altman plot showed good agreement between the test and retest 3TCM VT values as evidenced by low absolute bias (−0.052) with small standard deviation (0.063, Supplemental Fig. S1). The mean absolute deviation in regional VS = 40.42 ± 14.54%, indicating poor test-retest variability. The Bland-Altman plot showed a bias of −0.002 with a standard deviation of 0.095 (Supplemental Fig. S1).
[11C]MDTC whole-body dosimetry
The organs with greatest radiation-absorbed doses included the gallbladder wall, liver, urinary bladder wall, kidneys, and adrenals (Table 1). Gallbladder wall was found to be the critical organ (32.48 μSv/MBq). The effective dose was 5.29 ± 1.25 μSv/MBq. This is similar to the other [11C]-based brain tracers such as [11C]PiB that has an effective dose of 4.74 μSv/MBq [17].
Discussion
We present first-in-human neuroimaging data using [11C]MDTC PET for imaging the CB2R in human brain, with complementary whole-body dosimetry. Both [11C]MDTC PET and PET/CT were tolerated well. [11C]MDTC human dosimetry yielded an estimated whole-body effective dose of 5.29 μSv/MBq, which is a dose burden comparable to that of other 11C-labeled radiotracers [18]. [11C]MDTC has fast pharmacokinetics in healthy human brain, with fast uptake to peak regional brain activity around 1 to 2 min, followed by subsequent washout. Cortical regions and thalamus had higher [11C]MDTC uptake (peak SUV 2–3) compared to corpus callosum (peak SUV ~1.5).
The lipophilicity value (clogD7.4 = 3.86) of MDTC is in a conventionally optimal range for good blood-brain barrier permeability [19,20,21,22]. It is also similar to that of another published CB2R PET tracer [23]. The murine radiometabolite studies indicated the presence of plasma radiometabolites that eluted before [11C]MDTC, and one or more brain-penetrant radiometabolites were detected in RP-HPLC of mouse brain homogenate. If similar brain-penetrant radiometabolites also emerge after injection of [11C]MDTC in humans, regular compartmental models such as 1TCM and 2TCM with metabolite-corrected plasma input function cannot be applied well to analyze [11C]MDTC images. Logan graphical analysis is also not applicable to quantify the [11C]MDTC binding outcomes in the presence of brain-penetrant radiometabolites. A 3TCM approach that includes a separate input function and compartment for the radiometabolites entering the human brain was therefore applied here. Results from this human [11C]MDTC brain PET study demonstrated that the 3TCM fitting outperformed the 2TCM in half (54.5%) of the TACs. Since the 2TCM does not separate the contribution of brain-penetrant radiometabolites, it is presently advisable to use the 3TCM.
The 3TCM assumes the brain-penetrant radiometabolite contributes to nonspecific binding alone. The identity of the [11C]MDTC metabolite that enters the brain is currently unknown, and yet we report that radiometabolites of [11C]MDTC are less lipophilic than that of the parent tracer (Fig 1A). While further characterization of the brain-penetrant radiometabolite is needed, the assumption that the [11C]MDTC metabolite does not bind the CB2R is supported by the finding that the binding affinity of cannabinoid ligands is proportional to lipophilicity [24].
The estimated regional VT and VS from 3TCM were very low (Fig 3), consistent with low specific binding. Ex vivo studies have demonstrated very low CB2R distribution in healthy human brain [25]. The average K1p of [11C]MDTC estimated from 3TCM was ~0.11 mL/min/cm3, which is higher than that of [11C]NE40 brain PET that yields mean K1 ~0.07 mL/min/cm3 [26]. Together, the observed low VT and VS values across the brain using [11C]MDTC PET are likely not due to poor radiotracer delivery and instead due to the low availability of the CB2R across the human brain in health. The low availability of CB2R in normal brain also resulted in poor estimates of VS, which had large standard errors (61.2%) and poor test-retest variability.
There were no observed effects of biological sex, age, or BMI on regional VT. However, we acknowledge the limitation of our relatively small sample of young and healthy participants who underwent [11C]MDTC brain PET (4 women, 6 men), and future work should investigate the potential effects of these biological factors on [11C]MDTC binding further.
In summary, these first-in-human data support the safety and potential utility of [11C]MDTC PET for estimating the availability of the CB2R in human brain, particularly in conditions with a putative neuroinflammatory component. However, rigorous assessment of the brain-penetrating radiometabolite is recommended before the movement of [11C]MDTC PET into study of conditions with high CB2R expression.
Data availability
The datasets generated and analyzed in this study are available from the corresponding author on reasonable request.
References
Xin Q, et al. The impact of cannabinoid type 2 receptors (CB2Rs) in neuroprotection against neurological disorders. Acta Pharmacol Sin. 2020;41(12):1507–18.
Basavarajappa BS, et al. Endocannabinoid system in neurodegenerative disorders. J Neurochem. 2017;142(5):624–48.
Solas M, et al. CB2 receptor and amyloid pathology in frontal cortex of Alzheimer's disease patients. Neurobiol Aging. 2013;34(3):805–8.
Gómez-Gálvez Y, et al. Potential of the cannabinoid CB(2) receptor as a pharmacological target against inflammation in Parkinson's disease. Prog Neuropsychopharmacol Biol Psychiatry. 2016;64:200–8.
García MC, et al. Identification of CB2 receptors in human nigral neurons that degenerate in Parkinson's disease. Neurosci Lett. 2015;587:1–4.
Cristino L, Bisogno T, Di Marzo V. Cannabinoids and the expanded endocannabinoid system in neurological disorders. Nat Rev Neurol. 2020;16(1):9–29.
Horti AG, et al. Synthesis and biodistribution of [11C]A-836339, a new potential radioligand for PET imaging of cannabinoid type 2 receptors (CB2). Bioorg Med Chem. 2010;18(14):5202–7.
Yao BB, et al. Characterization of a cannabinoid CB2 receptor-selective agonist, A-836339 [2,2,3,3-tetramethyl-cyclopropanecarboxylic acid [3-(2-methoxy-ethyl)-4,5-dimethyl-3H-thiazol-(2Z)-ylidene]-amide], using in vitro pharmacological assays, in vivo pain models, and pharmacological magnetic resonance imaging. J Pharmacol Exp Ther. 2009;328(1):141–51.
Rahmim A, et al. Statistical dynamic image reconstruction in state-of-the-art high-resolution PET. Phys Med Biol. 2005;50(20):4887–912.
Hilton J, et al. Column-switching HPLC for the analysis of plasma in PET imaging studies. Nucl Med Biol. 2000;27(6):627–30.
Coughlin JM, et al. The distribution of the alpha7 nicotinic acetylcholine receptor in healthy aging: an in vivo positron emission tomography study with [(18)F]ASEM. Neuroimage. 2018;165:118–24.
Innis RB, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27(9):1533–9.
Carson RE. Parameter estimation in positron emission tomography. In: Principles And Applications For The Brain And Heart. New York, N Y, USA: Raven Press; 1986. p. Xiii+690. Illus 347-390.
Fujita M, et al. Kinetic and equilibrium analyses of [(123)I]epidepride binding to striatal and extrastriatal dopamine D(2) receptors. Synapse. 1999;34(4):290–304.
Stabin MG, Sparks RB, Crowe E. OLINDA/EXM: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med. 2005;46(6):1023–7.
Glatting G, et al. Choosing the optimal fit function: comparison of the Akaike information criterion and the F-test. Med Phys. 2007;34(11):4285–92.
Scheinin NM, et al. Biodistribution and radiation dosimetry of the amyloid imaging agent 11C-PIB in humans. J Nucl Med. 2007;48(1):128–33.
van der Aart J, et al. Radiation dose estimates for carbon-11-labelled PET tracers. Nucl Med Biol. 2012;39(2):305–14.
Dishino DD, et al. Relationship between lipophilicity and brain extraction of C-11-labeled radiopharmaceuticals. J Nucl Med. 1983;24(11):1030–8.
Kao CH, et al. Evaluation of [76Br]FBAU 3',5'-dibenzoate as a lipophilic prodrug for brain imaging. Nucl Med Biol. 2002;29(5):527–35.
Kubinyi H. The quantitative analysis of structure-activity relationships. In: Abraham DJ, editor. Burger’s Medicinal Chemistry and Drug Discovery, in Burger’s Medicinal Chemistry and Drug Discovery. New York: Wiley; 1995. p. 497–571.
Welch MJ, et al. Biodistribution of N-alkyl and N-fluoroalkyl derivatives of spiroperidol; radiopharmaceuticals for PET studies of dopamine receptors. Int J Rad Appl Instrum B. 1986;13(5):523–6.
Evens N, et al. Synthesis and biological evaluation of carbon-11- and fluorine-18-labeled 2-oxoquinoline derivatives for type 2 cannabinoid receptor positron emission tomography imaging. Nucl Med Biol. 2009;36(4):455–65.
Katoch-Rouse R, et al. Synthesis, structure-activity relationship, and evaluation of SR141716 analogues: development of central cannabinoid receptor ligands with lower lipophilicity. J Med Chem. 2003;46(4):642–5.
Liu QR, et al. Species differences in cannabinoid receptor 2 (CNR2 gene): identification of novel human and rodent CB2 isoforms, differential tissue expression and regulation by cannabinoid receptor ligands. Genes Brain Behav. 2009;8(5):519–30.
Ahmad R, et al. Decreased in vivo availability of the cannabinoid type 2 receptor in Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2016;43(12):2219–27.
Acknowledgements
This publication was supported by the Johns Hopkins Institute for Clinical and Translational Research (ICTR), which is funded in part by Grant Number UL1 TR 001079 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the Johns Hopkins ICTR, NCATS, or NIH. We also acknowledge EB024495 for support.
Funding
This publication was made possible by the Johns Hopkins Institute for Clinical and Translational Research (ICTR), which is funded in part by Grant Number UL1 TR 001079 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the Johns Hopkins ICTR, NCATS, or NIH. We also acknowledge EB024495 for support.
Author information
Authors and Affiliations
Contributions
YD, JC, ML, AH, and MP contributed to the study conception and design. Material preparation, data collection, and analysis were performed by all authors. The first draft of the manuscript was written by YD and JC, and each author read, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Johns Hopkins Institutional Review Board and was conducted under a FDA-approved US investigational new drug application (IND 131367). All experimental animal protocols were performed in accordance with relevant federal and state laws and institutional guidelines and approved by the Animal Care and Use Committee of the Johns Hopkins Medical Institutions.
Consent to participate
Written, informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Neurology
Supplementary information
ESM 1
(DOCX 271 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Du, Y., Coughlin, J.M., Brosnan, M.K. et al. First-in-human imaging using [11C]MDTC: a radiotracer targeting the cannabinoid receptor type 2. Eur J Nucl Med Mol Imaging 50, 2386–2393 (2023). https://doi.org/10.1007/s00259-023-06170-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-023-06170-y