Abstract
Purpose
Lymphovascular invasion (LVI) impairs surgical outcomes in lung adenocarcinoma (LAC) patients. Preoperative prediction of LVI is challenging by using traditional clinical and imaging parameters. The purpose of this study was to investigate the value of the radiomics nomogram integrating clinical factors, CT features, and maximum standardized uptake value (SUVmax) to predict LVI and outcome in LAC and to evaluate the additional value of the SUVmax to the PET/CT-based radiomics nomogram.
Methods
A total of 272 LAC patients (87 LVI-present LACs and 185 LVI-absent LACs) with PET/CT scans were retrospectively enrolled, and 160 patients with SUVmax ≥ 2.5 of them were used for PET radiomics analysis. Clinical data and CT features were analyzed to select independent LVI predictors. The performance of the independent LVI predictors and SUVmax was evaluated. Two-dimensional (2D) and three-dimensional (3D) CT radiomics signatures (RSs) and PET-RS were constructed with the least absolute shrinkage and selection operator algorithm and radiomics scores (Rad-scores) were calculated. The radiomics nomograms, incorporating Rad-score and independent clinical and CT factors, with SUVmax (RNWS) or without SUVmax (RNWOS) were built. The performance of the models was assessed with respect to calibration, discrimination, and clinical usefulness. All the clinical, PET/CT, pathologic, therapeutic, and radiomics parameters were assessed to identify independent predictors of progression-free survival (PFS).
Results
CT morphology was the independent LVI predictor. SUVmax provided better discrimination capability compared with CT morphology in the training set (P < 0.001) and test set (P = 0.042). A total of 1409 CT and PET radiomics features were extracted and reduced to 8, 8, and 10 features to build the 2D CT-RS, 3D CT-RS, and the PET-RS, respectively. There was no significant difference in AUC between the 2D-RS and 3D-RS (P > 0.05), and 2D CT-RS showed a relatively higher AUC than 3D CT-RS. The CT-RS, the CT-RNWOS, and the CT-RNWS showed good discrimination in the training set (AUC [area under the curve], 0.799, 0.796, and 0.851, respectively) and the test set (AUC, 0.818, 0.822, and 0.838, respectively). There was significant difference in AUC between the CT-RNWS and CT-RNWOS (P = 0.044) in the training set. Decision curve analysis (DCA) demonstrated the CT-RNWS outperformed the CT-RS and the CT-RNWOS in terms of clinical usefulness. Furthermore, DCA showed the PETCT-RNWS provided the highest net benefit compared with the PET-RNWS and CT-RNWS. PFS was significantly different between the pathologic and RNWS-predicted LVI-present and LVI-absent patients (P < 0.001). Carbohydrate antigen 125 (CA125), carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), pathologic LVI, histologic subtype, and SUVmax were independent predictors of PFS in the 244 CT-RNWS-predicted cohort; and CA125, NSE, pathologic LVI, and SUVmax were the independent predictors of PFS in the 141 PETCT-RNWS-predicted cohort.
Conclusions
The radiomics nomogram, incorporating Rad-score, clinical and PET/CT parameters, shows favorable predictive efficacy for LVI status in LAC. Pathologic LVI and SUVmax are associated with LAC prognosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer is one of the leading causes of cancer-related mortality worldwide [1,2,3]. Non-small cell lung cancer (NSCLC) accounts for approximately 80% of lung cancers, while lung adenocarcinoma (LAC) is the major histological subtype of NSCLC [3, 4]. Surgical resection is considered the standard treatment for localized NSCLC. However, the survival rate remains unsatisfactory due to tumor recurrence after surgery. Even for stage I or II NSCLC, the 5-year survival was between 30 and 60%, and at least 30% stage I NSCLC patients experience tumor recurrence after complete resection [2]. Identification of prognostic biomarkers is therefore essential to stratify patients with higher risks for clinical decision of adjuvant therapy [3, 5].
Lymphovascular invasion (LVI), defined as the infiltration of neoplastic cells within arterial, venous, or lymphatic lumens, has been proven to induce the first step of local recurrence and distant diffusion of the tumor. LVI has been recognized as an unfavorable prognostic factor in NSCLC patients [1,2,3,4,5,6,7,8,9,10,11,12]. Some scholars suggested cataloging LVI status into the forthcoming tumor-node-metastasis staging of lung cancer [1, 3, 5, 13]. Preoperative prediction of LVI is still challenging because LVI is a histologic status that can only be postoperatively diagnosed with a surgical specimen.
CT, F-18 fluorodeoxyglucose (FDG) positron emission tomography (PET)/CT and MRI have become important tools for tumor detection, diagnosis, staging, clinical decision-making, follow-up, and prediction of prognosis in patients with lung cancer. However, few imaging parameters have been used in predicting LVI of lung cancer. Noda et al. [14] showed that the maximum standardized uptake value (SUVmax) of LAC was a potential imaging biomarker for predicting tumor LVI. Tsuchiya et al. [15] found that ADC histogram analyses on the basis of the entire tumor volume were able to stratify LVI in NSCLC. However, these imaging parameters for prediction of LVI in lung cancer have not been widely recognized and still need further validation.
Recently, radiomics, which extracts and analyzes quantitative features from images, has been proven to be a favorable biomarker to identify additional characteristics of the tumor heterogeneity and microenvironment that is invisible to human eyes [16,17,18,19]. Radiomics has had success in detection of tumor [20, 21], prediction of histology and mutations [22,23,24,25], prediction of prognosis [26,27,28,29], and assessment of treatment effect [30] of lung cancer. However, there was no previous study focused on the value of metabolic parameters and PET/CT-based radiomics in preoperative prediction of LVI in LAC.
The purpose of this study was to investigate whether the radiomics nomograms integrating clinical factors, CT features, and metabolic parameters could be used to predict LVI and outcome of patients with LAC. In addition, the incremental value of the SUVmax to the PET/CT-based radiomics nomogram was evaluated.
Methods
Patients
Ethical approval was obtained for this retrospective study, and the need of written informed consent was waived.
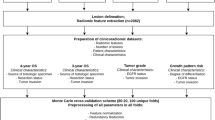
We searched the medical records of two hospitals (The Affiliated Hospital of Qingdao University and the Qingdao Central Hospital) to derive the surgically confirmed LAC from October 2011 to January 2018. A total of 272 LACs (113 males and 159 females; median age, 62 years; age range, 27–83 years) were enrolled according to the following inclusion criteria: (1) patients with a pathologically confirmed LAC with a definite LVI status on surgically resected specimens; LVI was identified as the presence of neoplastic cells within arterial, venous, or lymphatic channels during routine histologic evaluation with hematoxylin and eosin (H&E) stains, D2-40, and CD31 stains; (2) patients underwent PET/CT scan less than 15 days before surgery; (3) PET/CT images were satisfactory for analysis; (4) patients with complete clinical data; (5) the maximum diameter of tumor > 10 mm. The exclusion criteria were as follows: (1) patients received anti-tumor therapy before surgery; (2) patients suffering from other pulmonary diseases that might affect image analysis; (3) patients with other malignancies; (4) cystic lung cancer. According to the TRIPOD statement [31], 140 patients from The Affiliated Hospital of Qingdao University constituted the training set, whereas 132 patients from the Qingdao Central Hospital constituted the test set.
Clinical information were obtained by searching medical records, including age, gender, smoking history, and serum levels of four tumor markers which were routinely measured within 2 weeks before surgery: carcinoembryonic antigen (CEA), carbohydrate antigen 125 (CA125), neuron-specific enolase (NSE), and cytokeratin 19 fragments (CYFRA21-1).
A pathologist (J. W) with 8 years’ experience in lung diseases interpreted the pathology including LVI status, the stage, and the histologic subtype. The NSCLC pathologic TNM staging was performed based on the guidelines from the American Joint Committee on Cancer (AJCC) “Cancer Staging Manual, eighth edition”. In “2015 WHO Classification of Lung Tumour,” the histologic subtypes of LACs are identified according to the morphological patterns of the tumors. The LACs with the lepidic, acinar, or papillary growth patterns are regarded as low-risk subtypes, while the tumors displaying micropapillary (> 5% increments), solid (> 5% increments), invasive mucinous, colloid, fetal, enteric, minimally invasive, or preinvasive growth patterns are more invasive and are considered to be high-risk subtypes. The epidermal growth factor receptor (EGFR) gene status, the surgical approach, and the treatment after surgery were also recorded.
PET/CT image acquisition
Patients were instructed to fast for 6 h prior to F-18 FDG-PET/CT acquisition. Blood glucose levels measured before scanning were lower than 11.0 mmol/l in all patients. All patients underwent PET/CT scanning using a Discovery VCT 64 PET/CT system (GE Healthcare, Milwaukee, USA). A 3.7–5.5 MBq/kg dose of FDG was administered intravenously, and 60 min later, whole-body CT, covering the vertex to the pelvis, was acquired using the following parameters: detector coverage, 40 mm; coverage speed, 29.46 mm/s; gantry rotation time, 0.7 s; tube voltage, 120 kVp; tube current, 110 mA; pitch, 0.516:1; field of view, 70 cm; matrix, 512 × 512; and slice thickness, 3.75 mm. After CT scan, the emission scan was followed by a 1.5–2 min transmission scan per bed position. Images of seven to nine bed positions (70 cm of axial field of view) were acquired. PET images were reconstructed using a 3-dimensional (3D) ordered subset expectation maximization algorithm with 20 subsets and 2 iterations). Patients held their breath for scanning in the supine position. Axial thin-slice chest images were reconstructed with a slice thickness of 1.25 mm and interval of 1.25 mm with a lung algorithm.
PET/CT features analysis
The PET/CT images were analyzed by two radiologists (reader 1, P.N and reader 2, G. Y with 5 and 8 years’ experience in the interpretation of PET/CT images, respectively). Blinded to the clinical and pathologic information, the two readers analyzed the following CT features by consensus: the maximum diameter of the tumor on the axial thin-slice CT image; peripheral location (yes or no, “peripheral location” was defined as subsegmental location of the tumor); lobulation (present or not, “lobulation” was defined as irregular undulation of the nodule margin), spiculation (present or not, “spiculation” was regarded as the presence of 2 mm or thicker strands extending from the nodule margin into the lung parenchyma without reaching the pleural surface), bubble lucency (present or not, “bubble lucency” was defined as small spots of round or ovoid air attenuation within a nodule), air bronchogram (present or not, “air bronchogram” was defined as an air-filled bronchi within a nodule), vessel convergence (present or not, “vessel convergence” was defined as multiple blood vessels gathered toward a nodule), pleural indentation (present or not, “pleural indentation” was referred to a linear or triangular strand originating from the nodule surface and reaching the pleural surface), and CT morphology of the nodule (the CT morphology of the nodule was classified into pure ground-glass nodule [pGGN], mixed ground-glass nodule [mGGN], and solid nodule [sN]. A pGGN was defined as pure ground-glass opacity on lung window images [level, − 600 HU; width, 1500 HU] that had no soft-tissue attenuation within the lesion on mediastinal window images [level, 40 HU; width, 400 HU]. A mGGN was defined as a nodule with the mixture of ground-glass opacity and soft-tissue attenuation. A sN was defined as a nodule with pure soft-tissue attenuation). In addition, the SUVmax was measured by drawing a region-of-interest (ROI) encompassing the entire lesion on the axial PET image.
Analysis of the clinical information, CT features, and SUVmax
Univariate analysis was used to compare the differences of the clinical information and CT features between LVI-present and LVI-absent groups, and a multiple logistic regression analysis was used to identify the independent predictors significantly associated with LVI. Odds ratios (OR) as estimates of relative risk with 95% confidence intervals (CI) were obtained for each predictor. The diagnostic performance of the independent predictors and the SUVmax for predicting LVI was assessed by using the receiver operator characteristic (ROC) curve (AUC) and Delong test.
Tumor segmentation and radiomics feature extraction
Two-dimensional (2D) ROIs were manually segmented in the largest cross-sectional area using an ITK-SNAP software (Version 3.8.0, www.itksnap.org). Contouring was drawn slightly within the borders of the tumor on 1.25 mm thickness CT images, but avoiding covering the adjacent blood vessels, bronchus, chest wall, or mediastinum structures.
Three-dimensional (3D) semi-automatic segmentation of the tumor was performed using a Grow-from-Seeds tool for CT images, and a PET-Tumor-Segmentation tool for PET images implemented in the 3D-Slicer software (Version 4.11.0, www.slicer.org). Observers labeled foreground and background pixels in each view, and the algorithm automatically generates the ROI by using these pixel sets. Then a manual editing of the generated contours was performed to remove the adjacent blood vessels, bronchus, chest wall, or mediastinum from the ROI. CT and PET ROI segmentations are presented in Fig. 1a–d.
Construction of radiomics signatures (RSs). a, b CT ROI segmentation. c, d PET ROI segmentation. e, f, g Two-dimensional (2D) CT, three-dimensional (3D) CT, and PET features selection using the least absolute shrinkage and selection operator (LASSO) regularization. h, i, j The selected 8 2D CT features, 8 3D CT features, 10 PET features and their coefficients. k, l, m The 2D CT radiomics scores (Rad-scores), 3D CT Rad-scores, and PET Rad-scores for each patient in the training and test sets
Features were extracted by using a Radcloud platform (Huiying Medical Technology Co., Ltd., http://radcloud.cn) [32, 33]. A total of 1409 radiomics features were extracted from the ROIs. The quantization method of the radiomics calculation was “fixed bin width” with the bin width of 25. The instructions of the radiomics features are shown in Supplementary Methods. The radiomics features are divided into four groups: (1) intensity statistics features, which consists of 19 features that quantitatively delineate the distribution of voxel intensities within the ROI through commonly used and basic metrics; (2) shape features, including 2D and 3D features, are used to reflect the shape and size of the ROI; (3) texture features, composed of 59 features calculated by gray level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), and gray level size zone matrix (GLSZM), quantify the heterogeneity differences of ROI; and (4) filter and wavelet features, which include the intensity and texture features derived from filter transformation and wavelet transformation of the original image, obtained by applying filters such exponential, logarithm, square, square root, and wavelet using eight frequency band combinations (low-high-low [LHL], low-high-high [LHH], high-low-low [HLL], low-low-high [LLH], high-low-high [HLH], high-high-high [HHH], high-high-low [HHL], and low-low-low [LLL]).
Inter- and intra-class correlation coefficients (ICCs) were calculated for evaluation of the inter-reader reliability and intra-reader reproducibility of feature extraction. Reader 1 and reader 2 drew the ROIs of 40 cases (20 LVI-present LACs and 20 LVI-absent LACs) of CT images and PET images randomly selected from the whole cohort. Reader 1 repeated the segmentations 2 weeks later. An ICC greater than 0.75 indicated good agreement of feature extraction. The ROI segmentation for the remaining cases was performed by reader 1.
Construction of the radiomics signatures
In order to prevent overfitting of the signature, dimension reduction of the features was conducted before signature construction following three steps. First, to avoid subjective differences in segmenting the ROIs, the radiomics features with both inter- and intra-reader ICCs > 0.75 were retained. Second, the significantly different features between LVI-present and LVI-absent were chosen by using one-way analysis of variance (ANOVA). Finally, the least absolute shrinkage and selection operator algorithm (LASSO) was performed to select the optimal radiomics features. The radiomics signature (RS) was developed by combining the selected features, and a radiomics score (Rad-score) was calculated for each patient.
Construction of radiomics nomograms and performance assessment of different models
The radiomics nomogram without SUVmax (RNWOS) was developed by incorporating the independent LVI predictors as well as the Rad-score. The radiomics nomogram with SUVmax (RNWS) was developed by combining the independent LVI predictors, the Rad-score, and the SUVmax.
The Hosmer-Lemeshow test was used to evaluate the goodness-of-fit of the nomograms. The diagnostic performance of the RS, the RNWOS, and the RNWS for prediction of LVI in LAC was assessed with respect to AUC, sensitivity, specificity, and accuracy in the training and test sets. The internal validation of the models was assessed by using a bootstrap sampling method. The AUC between each two of the three models was evaluated by using the Delong test. To estimate the clinical utility of the nomograms, a decision curve analysis (DCA) was performed by calculating the net benefits for a range of threshold probabilities in the whole cohort.
Follow-up and survival analysis
Patients were postoperatively followed up with CT every 6–12 months for the first 2 years and then annually. The end point of this study was progression-free survival (PFS), which was defined as the interval between surgery and radiographic detection of progression (relapse), last negative follow-up, or death without evidence of progression. Patients were censored in case of emigration, or on July 31, 2019, whichever came first.
Survival curves were plotted according to the Kaplan-Meier method and compared using the log-rank test. All the prognosis-associated factors (including basic information, serum tumor markers, pathologic LVI status, stage, histologic subtype, surgical approach, treatment after surgery, and EGFR gene status), CT features, SUVmax, and RNWS-predicted LVI status were assessed by univariate analysis based on the Kaplan-Meier method. Variables which had statistical significance in the univariate analysis were considered for the multivariate Cox forward stepwise regression model to identify independent predictors of progression.
Statistics
Univariate analysis (including chi-square test or Mann-Whitney U test, where appropriate) and one-way ANOVA were performed by using SPSS software (Version 25.0, IBM). ICC, LASSO regression analysis, ROC analysis, calibration plots, Hosmer-Lemeshow test, Delong test, DCA, and survival analysis were performed with R statistical software (Version 3.3.3, https://www.r-project.org). A two-sided P < 0.05 was considered statistically significant.
Results
Clinical information and PET/CT features of the patients
The clinical factors, CT features, and SUV max of the patients in the training and test sets are shown in Table 1. Mixed ground-glass nodule (P = 0.017) was proven as independent LVI predictors using the multiple logistic regression analysis (Supplementary Table S1). Tumors with mixed ground-glass morphology (OR, 0.266; 95% CI 0.090–0.790) were likely to be LVI-absent LACs. The AUCs of CT morphology and SUVmax in prediction of LVI were 0.635 and 0.818, respectively, in the training set; and 0.727 and 0.800, respectively, in the test set. SUVmax provided better discrimination capability compared with CT morphology in the training set (P < 0.001) and test set (P = 0.042).
Construction of the 2D and 3D CT-RSs and PET-RS
Of the 1409 radiomics features extracted from 2D/3D ROIs of CT images and 3D ROIs of PET images, 1274 2D CT features, 971 3D CT features, and 1297 PET features had a good inter- and intra-observer agreement, with ICCs > 0.75. A total of 641 2D CT features, 477 3D CT features, and 32 PET features showing significant differences between LVI-present and LVI-absent patients (P = 0.000–0.050) were enrolled into the LASSO regression model (Fig. 1e–g), and 8 2D features, 8 3D CT features, and 10 PET features were finally selected (Fig. 1h–j). The formulas of the three Rad-scores are shown in Supplementary Results.
Low tumor SUVmax brings a great challenge for the PET radiomics analysis as tumor ROIs on PET images are difficult to delineate. Therefore, we enrolled 160 cases (82 cases in the training set; 78 cases in the test set) with SUVmax ≥ 2.5 to perform PET radiomics analysis.
The Rad-scores showed statistically significant difference between LVI-present and LVI-absent groups in the training and test sets (P < 0.001). The three RSs are shown in Fig. 1k–m.
The AUCs of 2D CT-RS and 3D CT-RS were 0.799 (95% CI 0.723–0.862) and 0.765 (95% CI 0.686–0.832), respectively, in the training set; and 0.818 (95% CI 0.741–0.880) and 0.772 (95% CI 0.691–0.840), respectively, in the test set. There was no significant difference in AUC between the two RSs (P = 0.301 and 0.102, in the training and test sets). 2D CT-RS showed a relatively higher AUC than 3D CT-RS did in both the training and test sets; therefore, 2D CT-RS was selected for further construction of nomogram.
Construction of the RNWOS and the RNWS and performance of different models
The CT morphology and 2D RS were incorporated into the CT-RNWOS construction. The CT-RNWS was constructed by integrating the CT morphology, 2D RS and SUVmax. The Hosmer-Lemeshow tests showed good calibration of the CT-RNWOS (P = 0.800) and of the CT-RNWS (P = 0.733). The diagnostic performance of the three CT models is presented in Table 2. There were no significant differences in AUC between each two of the following models (the CT-RS vs the CT-RNWOS, P = 0.448 and 0.102; the CT-RS vs the CT-RNWS, P = 0.055 and 0.096, respectively, in the training set and the test set). There was significant difference in AUC between the CT-RNWS and CT-RNWOS (P = 0.044) in the training set; and there was no significant difference in AUC between the two models (P = 0.182) in the test set. The CT-RNWOS and CT-RNWS are shown in Fig. 2a, b.
The DCA (Fig. 2c) showed that the CT-RNWS had the highest overall net benefit than the CT-RNWOS and the CT-RS in predicting LVI across the majority of the range of reasonable threshold probabilities.
Combing CT morphology and PET-RS, with or without SUVmax, the PET-RNWS and the PET-RNWOS were constructed. The diagnostic performance of these models is shown in Table 3. PETCT-RNWS was developed by combing PET-RS, CT-RS, CT morphology, and SUVmax. The PET-RNWS and PETCT-RNWS are shown in Fig. 3a, b. There were no significant differences in AUC between each two of the following models (the PET-RS vs the PET-RNWOS, P = 0.670 and 0.292; the PET-RS vs the PET-RNWS, P = 0.764 and 0.387; the PET-RNWS vs the PET-RNWOS, P = 0.580 and 0.544; the PET-RS vs the CT-RS, P = 0.942 and 0.528; the PET-RNWOS vs the CT-RNWOS, P = 0.960 and 0.383; the PET-RNWS vs the CT-RNWS, P = 0.578 and 0.965; the PETCT-RNWS vs the PET-RNWS, P = 0.060 and 0.754; the PETCT-RNWS vs the CT-RNWS, P = 0.204 and 0.620, respectively, in the training set and the test set).
The DCA (Fig. 3c) showed that the three RNWSs (including PETCT-RNWS, PET-RNWS, and CT-RNWS) had higher overall net benefit than SUVmax and non-radiomics model (incorporated with SUVmax and CT morphology) in predicting LVI. Furthermore, the PETCT-RNWS outperformed the PET-RNWS and CT-RNWS in clinical usefulness.
Survival prediction
The pathologic and therapeutic factors associated with prognosis are shown in Table 4. As of July 31, 2019, 244 of 272 patients and 141 of 160 patients had completed the PFS follow-up, respectively, in the CT-RNWS-predicted cohort and the PETCT-RNWS-predicted cohort. The overall progression rate was 20.49% (50/244). The median PFS of the patients was 23 months (range, 1–86 months), particularly 19 months (range, 1–63 months) for the pathologic LVI-present patients and 24 months (range, 2–86 months) for the pathologic LVI-absent patients (log-rank test, P < 0.001).
In the 244 CT-RNWS-predicted cohort, the median PFS was 23 months (range, 1–86 months) for the CT-RNWS-predicted LVI-present patients, and the median PFS was 23 months (range, 1–81 months) for the CT-RNWS-predicted LVI-absent patients (log-rank test, P < 0.001). Survival curves according to pathologic and CT-RNWS-predicted LVI status are shown in Fig. 4a, b. In the 141 PETCT-RNWS-predicted cohort, the median PFS was 24 months (range, 2–86 months) for the PETCT-RNWS-predicted LVI-present patients, and the median PFS was 20.5 months (range, 1–68 months) for the PETCT-RNWS-predicted LVI-absent patients (log-rank test, P < 0.001). Survival curves according to pathologic and PETCT-RNWS-predicted LVI status are shown in Fig. 5a, b.
Progression-free survival (PFS) curves according to pathologic LVI status (a) and CT-RNWS-predicted LVI status (b) in the 244 CT-RNWS-predicted cohort. c Forest plot of independent predictors of PFS with a multivariate Cox regression model. LVI, lymphovascular invasion; RNWS, radiomics nomogram with SUVmax; HR, hazard ratio; CI, confidence interval; CA125, carbohydrate antigen 125; CEA, carcinoembryonic antigen; NSE, neuron-specific enolase
Progression-free survival (PFS) curves according to pathologic LVI status (a) and PETCT-RNWS-predicted LVI status (b) in the 141 PETCT-RNWS-predicted cohort. c Forest plot of independent predictors of PFS with a multivariate Cox regression model. LVI, lymphovascular invasion; RNWS, radiomics nomogram with SUVmax; HR, hazard ratio; CI, confidence interval; CA125, carbohydrate antigen 125; NSE, neuron-specific enolase
The predictors of PFS by means of univariate analysis are shown in Supplementary Table S2. In the 244 CT-RNWS-predicted cohort, the multivariate Cox regression analysis (Fig. 4c) showed that CA125, CEA, NSE, pathologic LVI, histologic subtype, and SUVmax were independent predictors of PFS. In the 141 PETCT-RNWS-predicted cohort, CA125, NSE, pathologic LVI, and SUVmax were found to be independent predictors of PFS (Fig. 5c).
Discussion
The purpose of the present study was to investigate the value of a computer-assisted method derived from massive clinical and PET/CT data to preoperatively predict histologic LVI and clinical outcome in LAC patients. We found that the radiomics nomogram integrating Rad-score, SUVmax, clinical factors, and CT features had favorable predictive value for LVI in LAC. The RNWS achieved relatively higher AUCs than the RNWOS did, indicating the additional value of metabolic parameters to the RNWOS in the prediction of LVI. In addition, the pathologic LVI status and SUVmax were independently associated with PFS, indicating SUVmax can be used as a potential biomarker to assist clinical decision-making in LAC patients.
LVI has been shown to induce the first step of tumor recurrence or diffusion, and is proven to independently affect prognosis in patients with lung cancer. From a clinical point of view, it is crucial to preoperatively predict LVI in LAC patients in order to select the proper treatment strategy. Contrary to macrolymphovascular invasion, which can be identified with imaging, LVI is a histologic status that can only be diagnosed with specimens after surgery, leading the preoperative prediction a great challenge with traditional clinical examinations and imaging techniques. In the present study, the traditional clinical factors (including age, gender, smoking history, and four serum tumor markers) and nine CT features (the maximum diameter, peripheral location, lobulation, spiculation, bubble lucency, air bronchogram, vessel convergence, pleural indentation, and CT morphology) were analyzed and only CT morphology was found as significantly independent variable for prediction of LVI. The CT morphology, as an independent LVI predictor, achieved a relatively lower AUC of 0.635 and 0.727, respectively, in the training set and test set, indicating the factor derived from traditional clinical data and CT features made a limited contribution to LVI prediction in LAC patients.
F-18 FDG PET/CT is a widely used imaging modality for characterization and staging of lung cancers. Metabolic parameters have been proven as potential imaging biomarkers for predicting tumor LVI [14, 34]. Hyun SH et al. [34] analyzed PET/CT images and clinical factors of 158 patients with hepatocellular carcinoma (HCC), and found the tumor-to-normal-liver standardized uptake value ratio (TLR) of the tumor, serum alpha-fetoprotein (AFP) level, and tumor size were significantly associated with the presence of microvascular invasion (MVI). A combined model integrating the three independent predictors achieved an AUC of 0.756 for prediction of MVI in HCC patients. Partly in agreement with previous findings of Noda et al. [14] and Hyun SH et al. [34], we found that SUVmax could be used as a favorable LVI predictor, achieving a relatively higher AUC (0.818 and 0.800, respectively, in the training set and the test set) than the independent predictor derived from traditional clinical and CT factors in predicting LVI in LAC patients.
Recently, radiomics has been proven to be an effective imaging modality to identify histological and biological characteristics of tumors beyond visual assessment on CT, PET/CT, and MRI images. Previous studies have shown that radiomics had favorable diagnostic efficacy in preoperative prediction of MVI or LVI in various malignancies. Xu et al. [35] developed a CT radiomics model integrating large-scale clinical factors and imaging findings to predict MVI in 495 HCC patients, achieving an AUC of 0.889 in the test set. Liu et al. [36] showed that the MRI-based radiomics signature in combination with MRI axillary lymph node status was effective in predicting LVI status of patients with invasive breast cancer before surgery with an AUC of 0.763. Liu et al. [37] found that CT texture parameters, including standard deviation, minimum attenuation, and entropy, held great potential in predicting MVI status of gastric cancers.
To the best of our knowledge, the present study is the first one focused on metabolic parameters and PET/CT-based radiomics in LVI prediction of lung cancer. Radiomics feature extraction is a key procedure in the radiomics studies. There is a trade-off while selecting 2D or 3D ROIs. In theory, whole tumor analysis may appear more indicative of tumor heterogeneity than the largest cross-sectional area. However, whether 3D features are definitely superior to 2D features in the practical application has not been verified. In this study, 8 2D features and 8 3D CT features were finally selected for RS development. However, the promising 2D and 3D CT features were fundamentally different. The possible reasons are as follows. First, LASSO regression model was used for dimension reduction and development of RS. Instead of a single feature, a cluster of features with the optimal distinction efficiency were finally selected to build the RS by using LASSO regularization. Second, the same 2D and 3D features may have different quantitative methods, for example, different directions, pixel distance, or aggregation methods. Thus, they may reflect different tumor heterogeneity. We compared the performance of 2D and 3D CT radiomics and found there was no significant difference in AUC between the 2D and 3D radiomics models. As 2D CT-RS showed a relatively higher AUC than 3D CT-RS did and was easy to achieve, it is recommended to choose 2D features in clinical practice. Our results were consistent with Shen et al. [38] who found that 2D features performed slightly better than 3D features in prediction of prognosis in NSCLC. The combined radiomics nomograms, incorporating Rad-score and independent clinical and CT factors, with or without SUVmax were built, achieved favorable efficacy in predicting LVI. We also evaluated the performance of PET radiomics and found that there were no significant differences in AUC between PET and CT radiomics models, indicating that the PET- and CT-based radiomics shared almost the same value in prediction of LVI in LACs. Additionally, the nomogram with SUVmax achieved a relatively higher AUC and higher clinical net benefit than the nomogram without SUVmax did, indicating the additional value of the metabolic parameter in the contribution to LVI prediction in LAC patients.
Sufficient analysis of clinical and imaging information facilitates correct diagnosis and management of lung cancer. Both PET/CT features and Rad-score are derived from the primary images, but they represent two different methods of image analysis. As macroscopic observations of imaging features, traditional image interpretation is subjective and prone to be influenced by the experience of the radiologists. However, the reproducibility and simplicity make it a basic tool to evaluate the characteristics of LAC and predict its biological behaviors. On the other hand, radiomics, a computer-assisted technique, although limited by its complexity of calculation and lack of standardization, is now a noninvasive and quantifiable modality to reveal the microscopic features associated with tumor heterogeneity. Therefore, integrating the two imaging analytic methods as well as clinical factors within a combined nomogram can allow for a better decision-making for LAC patients.
Previous investigations have shown that radiomics can be used for prediction of survival in patients with lung cancer. Kirienko et al. [27] analyzed clinical parameters and PET/CT-based radiomics features of 295 NSCLC patients, and found that the radiomics signature, combined clinical factors, PET and CT radiomics, had been identified and validated for the prediction of disease-free survival in patients with NSCLC treated by surgery with an AUC of 0.65. Wang et al. [28] found an integrative nomogram incorporated CT radiomic, clinical, and hematological features improved survival prediction in locally advanced NSCLC patients. Being different from the previous studies, we applied LVI as a stratifying factor and investigated the survival prediction value of the LVI-predicted radiomics nomogram. Partly in agreement with the results of previous studies [3, 4, 6, 39], we found that CA125, NSE, pathologic LVI, and SUVmax were independent risk factors of PFS, suggesting their prognostic value for the long-term management of LAC patients.
Several limitations of this study should be mentioned. As a retrospective study performed at two institutes, patient selection and time-trend biases were inevitable. Therefore, prospective multi-institutional studies are required to further validate the result. Second, a fixed tube current was used in PET/CT scans; therefore, the image noise changed across patients, and the intrinsic noise varied across the ROIs. The current modulation technique will be applied to acquire more reliable CT images in our future study.
Conclusions
In conclusion, the radiomics nomogram, incorporating clinical and PET/CT parameters, has favorable predictive efficacy for LVI status in LAC patients. Pathologic LVI and SUVmax can be used to predict outcome of LAC. As a noninvasive and quantitative method, the radiomics nomogram may serve as a potential biomarker to supplement the traditional clinical and imaging modalities for personalized treatment in LAC patients, although this still requires further validation before widespread application in clinical practice.
Data availability
The datasets generated and analyzed during the current study are not publicly available due to patient privacy concerns but are available from the corresponding author on reasonable request.
Change history
28 July 2020
Figures 1 and 4 are incorrect in the original manuscript.
References
Wang S, Zhang B, Qian J, Qiao R, Xu J, Zhang L, et al. Proposal on incorporating lymphovascular invasion as a T-descriptor for stage I lung cancer. Lung Cancer. 2018;125:245–52.
Sung SY, Kwak YK, Lee SW, Jo IY, Park JK, Kim KS, et al. Lymphovascular invasion increases the risk of nodal and distant recurrence in node-negative stage I-IIA non-small-cell lung cancer. Oncology. 2018;95:156–62.
Kinoshita T, Ohtsuka T, Yotsukura M, Asakura K, Goto T, Kamiyama I, et al. Prognostic impact of preoperative tumor marker levels and lymphovascular invasion in pathological stage I adenocarcinoma and squamous cell carcinoma of the lung. J Thorac Oncol. 2015;10:619–28.
Kuo SW, Chen JS, Huang PM, Hsu HH, Lai HS, Lee JM. Prognostic significance of histologic differentiation, carcinoembryonic antigen value, and lymphovascular invasion in stage I non-small cell lung cancer. J Thorac Cardiovasc Surg. 2014;148:1200–7 e3.
Patel AJ, Daniel G, Naidu B, Bishay E. The significance of microvascular invasion after complete resection of early-stage non-small-cell lung cancer. Interact Cardiovasc Thorac Surg. 2016;22:101–5.
Okiror L, Harling L, Toufektzian L, King J, Routledge T, Harrison-Phipps K, et al. Prognostic factors including lymphovascular invasion on survival for resected non-small cell lung cancer. J Thorac Cardiovasc Surg. 2018;156:785–93.
Ramnefjell M, Aamelfot C, Aziz S, Helgeland L, Akslen LA. Microvascular proliferation is associated with aggressive tumour features and reduced survival in lung adenocarcinoma. J Pathol Clin Res. 2017;3:249–57.
Park C, Lee IJ, Jang SH, Lee JW. Factors affecting tumor recurrence after curative surgery for NSCLC: impacts of lymphovascular invasion on early tumor recurrence. J Thorac Dis. 2014;6:1420–8.
Mollberg NM, Bennette C, Howell E, Backhus L, Devine B, Ferguson MK. Lymphovascular invasion as a prognostic indicator in stage I non-small cell lung cancer: a systematic review and meta-analysis. Ann Thorac Surg. 2014;97:965–71.
Al-Alao BS, Gately K, Nicholson S, McGovern E, Young VK, O'Byrne KJ. Prognostic impact of vascular and lymphovascular invasion in early lung cancer. Asian Cardiovasc Thorac Ann. 2014;22:55–64.
Hishida T, Yoshida J, Maeda R, Ishii G, Aokage K, Nishimura M, et al. Prognostic impact of intratumoural microvascular invasion and microlymphatic permeation on node-negative non-small-cell lung cancer: which indicator is the stronger prognostic factor? Eur J Cardiothorac Surg. 2013;43:772–7.
Hanagiri T, Takenaka M, Oka S, Shigematsu Y, Nagata Y, Shimokawa H, et al. Prognostic significance of lymphovascular invasion for patients with stage I non-small cell lung cancer. Eur Surg Res. 2011;47:211–7.
Noma D, Inamura K, Matsuura Y, Hirata Y, Nakajima T, Yamazaki H, et al. Prognostic effect of lymphovascular invasion on TNM staging in stage I non-small-cell lung cancer. Clin Lung Cancer. 2018;19:e109–22.
Noda Y, Goshima S, Kanematsu M, Watanabe H, Kawada H, Kawai N, et al. F-18 FDG uptake on positron emission tomography as a predictor for lymphovascular invasion in patients with lung adenocarcinoma. Ann Nucl Med. 2016;30:11–7.
Tsuchiya N, Doai M, Usuda K, Uramoto H, Tonami H. Non-small cell lung cancer: whole-lesion histogram analysis of the apparent diffusion coefficient for assessment of tumor grade, lymphovascular invasion and pleural invasion. PLoS One. 2017;12:e0172433.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–77.
Hassani C, Varghese BA, Nieva J, Duddalwar V. Radiomics in pulmonary lesion imaging. AJR Am J Roentgenol. 2019;212:497–504.
Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, et al. Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer. 2018;115:34–41.
Sollini M, Antunovic L, Chiti A, Kirienko M. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imaging. 2019. https://doi.org/10.1007/s00259-019-04372-x.
Kirienko M, Cozzi L, Rossi A, Voulaz E, Antunovic L, Fogliata A, et al. Ability of FDG PET and CT radiomics features to differentiate between primary and metastatic lung lesions. Eur J Nucl Med Mol Imaging. 2018;45:1649–60.
Yang X, He J, Wang J, Li W, Liu C, Gao D, et al. CT-based radiomics signature for differentiating solitary granulomatous nodules from solid lung adenocarcinoma. Lung Cancer. 2018;125:109–14.
Jiang M, Sun D, Guo Y, Guo Y, Xiao J, Wang L, et al. Assessing PD-L1 expression level by radiomic features from PET/CT in nonsmall cell lung cancer patients: an initial result. Acad Radiol. 2019. https://doi.org/10.1016/j.acra.2019.04.016.
Kang F, Mu W, Gong J, Wang S, Li G, Li G, et al. Integrating manual diagnosis into radiomics for reducing the false positive rate of (18)F-FDG PET/CT diagnosis in patients with suspected lung cancer. Eur J Nucl Med Mol Imaging. 2019. https://doi.org/10.1007/s00259-019-04418-0.
Tu W, Sun G, Fan L, Wang Y, Xia Y, Guan Y, et al. Radiomics signature: a potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer. 2019;132:28–35.
Yu L, Tao G, Zhu L, Wang G, Li Z, Ye J, et al. Prediction of pathologic stage in non-small cell lung cancer using machine learning algorithm based on CT image feature analysis. BMC Cancer. 2019;19:464.
Ahn HK, Lee H, Kim SG, Hyun SH. Pre-treatment (18)F-FDG PET-based radiomics predict survival in resected non-small cell lung cancer. Clin Radiol. 2019;74:467–73.
Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45:207–17.
Wang L, Dong T, Xin B, Xu C, Guo M, Zhang H, et al. Integrative nomogram of CT imaging, clinical, and hematological features for survival prediction of patients with locally advanced non-small cell lung cancer. Eur Radiol. 2019;29:2958–67.
Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281:947–57.
Xu Y, Hosny A, Zeleznik R, Parmar C, Coroller T, Franco I, et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res. 2019;25:3266–75.
Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594.
Nie P, Yang G, Wang Z, Yan L, Miao W, Hao D, et al. A CT-based radiomics nomogram for differentiation of renal angiomyolipoma without visible fat from homogeneous clear cell renal cell carcinoma. Eur Radiol. 2020;30:1274–84.
Liu J, Sun D, Chen L, Fang Z, Song W, Guo D, et al. Radiomics analysis of dynamic contrast-enhanced magnetic resonance imaging for the prediction of sentinel lymph node metastasis in breast cancer. Front Oncol. 2019;9:980.
Hyun SH, Eo JS, Song BI, Lee JW, Na SJ, Hong IK, et al. Preoperative prediction of microvascular invasion of hepatocellular carcinoma using (18)F-FDG PET/CT: a multicenter retrospective cohort study. Eur J Nucl Med Mol Imaging. 2018;45:720–6.
Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133–44.
Liu Z, Feng B, Li C, Chen Y, Chen Q, Li X, et al. Preoperative prediction of lymphovascular invasion in invasive breast cancer with dynamic contrast-enhanced-MRI-based radiomics. J Magn Reson Imaging. 2019. https://doi.org/10.1002/jmri.26688.
Liu S, Liu S, Ji C, Zheng H, Pan X, Zhang Y, et al. Application of CT texture analysis in predicting histopathological characteristics of gastric cancers. Eur Radiol. 2017;27:4951–9.
Shen C, Liu Z, Guan M, Song J, Lian Y, Wang S, et al. 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung cancer. Transl Oncol. 2017;10:886–94.
Shiono S, Abiko M, Sato T. Positron emission tomography/computed tomography and lymphovascular invasion predict recurrence in stage I lung cancers. J Thorac Oncol. 2011;6:43–7.
Funding
This study was funded by the National Natural Science Foundation of China (81701688 and 81601527); the Science and Technology Project of Southern District of Qingdao City (2020-2-004-YY); the Natural Science Foundation of Shandong Province (ZR2017BH096 and ZR2017MH036); the Key Research and Development Project of Shandong Province (2018GSF118078); and the Postdoctoral Science Foundation of China (2018 M642617).
Author information
Authors and Affiliations
Contributions
Literature search: Pei Nie, Guangjie Yang; Study design: Zhenguang Wang; Data collection: Pei Nie, Guangjie Yang, Yan Lei, Wenjie Miao, Yanli Duan, Yanli Wang, Aidi Gong, Yujun Zhao, Jie Wu, Chuantao Zhang, Maolong Wang, Mingming Yu, Dacheng Li; Data analysis: Guangjie Yang, Pei Nie, Ning Wang, Jingjing Cui, Yanqin Sun, Yangyang Wang; Manuscript writing: Pei Nie, Guangjie Yang; Manuscript review: Zhenguang Wang. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Research involving human participants
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee of The Affiliated Hospital of Qingdao University (Approval No. QYFY WZLL 25580) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was waived for this retrospective study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised. Figures 1 and 4 are updated.
This article is part of the Topical Collection on Oncology - Chest
Electronic supplementary material
ESM 1
(PDF 421 kb)
Rights and permissions
About this article
Cite this article
Nie, P., Yang, G., Wang, N. et al. Additional value of metabolic parameters to PET/CT-based radiomics nomogram in predicting lymphovascular invasion and outcome in lung adenocarcinoma. Eur J Nucl Med Mol Imaging 48, 217–230 (2021). https://doi.org/10.1007/s00259-020-04747-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-020-04747-5