Abstract
This review discusses the basics of cardiovascular gene therapy, the results of recent human clinical trials, and the rapid progress in imaging techniques in cardiology. Improved understanding of the molecular and genetic basis of coronary heart disease has made gene therapy a potential new alternative for the treatment of cardiovascular diseases. Experimental studies have established the proof-of-principle that gene transfer to the cardiovascular system can achieve therapeutic effects. First human clinical trials provided initial evidence of feasibility and safety of cardiovascular gene therapy. However, phase II/III clinical trials have so far been rather disappointing and one of the major problems in cardiovascular gene therapy has been the inability to verify gene expression in the target tissue. New imaging techniques could significantly contribute to the development of better gene therapeutic approaches. Although the exact choice of imaging modality will depend on the biological question asked, further improvement in image resolution and detection sensitivity will be needed for all modalities as we move from imaging of organs and tissues to imaging of cells and genes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Coronary heart disease is the most common cause of mortality and morbidity in industrialized countries. Better understanding of the molecular and genetic basis of these diseases has made gene therapy a potential new alternative for the treatment of cardiovascular diseases [1]. With the use of gene transfer techniques it is possible to modify somatic cells in ischemic myocardium and skeletal muscle to overexpress or inhibit pathologically important proteins and achieve therapeutic effects [2]. The main goal of gene therapy has been therapeutic angiogenesis, with the aim of improving blood flow in ischemic muscles [3]. Blood vessels and myocardium are easily accessible with currently available catheters, making them particularly useful targets for gene therapy. Experimental studies have established the proof-of-principle that gene transfer to the cardiovascular system can achieve therapeutic effects. First human clinical trials provided initial evidence of feasibility and safety of cardiovascular gene therapy. However, phase II/III clinical trials have so far been rather disappointing and one of the major problems in cardiovascular gene therapy has been the inability to verify gene expression in the target tissue. New imaging techniques could significantly contribute to the development of better gene therapeutic approaches. In this review we discuss the basics of cardiovascular gene therapy, the results of recent human clinical trials, and the rapid progress in imaging techniques in cardiology.
Vascular growth factors
Therapeutic angiogenesis can be achieved by using angiogenic growth factors or their genes [2]. Endothelium-specific growth factors include members of the vascular endothelial growth factor (VEGF) family and the angiopoietin (Ang) family. Other factors which promote angiogenesis are members of the fibroblast growth factor (FGF) family, hepatocyte growth factor (HGF), and some cytokines, such as monocyte chemotactic protein (MCP-1) [3].
Vectors for cardiovascular gene therapy
One of the most important determinants of the success of gene therapy is efficient delivery and expression of therapeutic genes in target cells [4]. The ideal gene delivery system will be safe, efficient, cell specific, and regulated. Various methods have been developed to deliver genes into the myocardium. Some of these methods involve the direct use of plasmid DNA with or without carrier molecules, which usually are lipids or cationic polymers. The main disadvantage of the nonviral approach is a low gene transfer efficiency [1]. Therefore, viral vectors have been developed for gene delivery. Viruses have evolved efficient mechanisms to enter cells and they use the host cell machinery to express their own genes [1]. To develop viral vectors, wild-type viruses have been engineered to deliver and express only the gene of interest in target cells. Because most viruses are either pathogens or have a pathogenic potential, viral vectors have been modified to eliminate their ability to replicate in the host. Several different viruses have been used to generate viral vectors with different biological characteristics. In cardiology, adenoviral vectors have been most commonly used for phase II/III clinical applications. Also, plasmid vectors have been used in the clinics [3].
Delivery of vectors to myocardium
Delivery of the vector significantly contributes to the expression of the therapeutic gene in the target tissue (Fig. 1). Vector delivery to ischemic myocardium for coronary heart disease has been achieved using intramyocardial injections via thoracotomy and intracoronary injections [3, 5]. Also, a recently introduced catheter-based electromechanical mapping system (NOGA, Biosense Webster, Inc) seems to be very promising [6]. The regions of infarcted or ischemic myocardium can be distinguished from healthy myocardium by virtue of the reductions in both electrical voltage and mechanical activity assessed by the NOGA system. Additionally, NOGA-guided injection catheters enable gene transfer targeted selectively to ischemic regions of the heart. Intramyocardial injection catheter systems are also available from other manufacturers. As an alternative route, pericardial delivery of the vectors can be used to transfer genes to the myocardium and the outer surface of coronary arteries. Local delivery to small arterioles and capillaries can be achieved by coated stents or coated biodegradable microspheres. Also, perivascular collars or sheaths and biodegradable gels can be used for gene delivery [1].
Gene transfer as a tool to induce therapeutic vascular growth. a Gene delivery through major arteries. b Myocardium can be approached from ventricles, through coronary arteries, through epicardial surface, or during bypass surgery. c Peripheral vascular disease and some lymphatic disorders can be treated through intravascular or intramuscular routes or during surgery. Adapted from Yla-Herttuala S and Alitalo K [3]
Clinical trials
The vast majority of studies in cardiovascular gene therapy have been preclinical. Induction of therapeutic angiogenesis has been the major target for cardiovascular gene therapy. Most of the published vascular gene therapy trials have been phase I studies and there are only a few randomised, double-blinded, placebo-controlled phase II/III studies. Data regarding completed and ongoing gene therapy clinical trials can be found from http://www4.od.nih.gov/oba/rac/clinicaltrial.htm and http://www.wiley.co.uk/wileychi/genmed/clinical/.
Although preclinical results and several phase I trials have provided positive outcomes in the clinics, recently conducted trials with cardiac gene transfer have not fulfilled expectations of significant clinical improvements in randomized, controlled, blinded studies. Intravascular catheter-mediated VEGF-A gene transfer with plasmid/liposome to human coronary arteries in conjunction with PTCA was demonstrated to be safe and feasible in a randomised, placebo-controlled phase I/IIa study [5]. However, no therapeutic efficiency was shown by control angiography in the follow-up after 6 months.
The AGENT (Adenovirus Fibroblast Growth Factor Angiogenic Gene Therapy) Trial [7] tested adenovirus-mediated FGF4 gene transfer to coronary arteries. Although phase II results were positive using an exercise tolerance test as an endpoint [7] (Table 1), a phase III trial with this compound has recently been stopped because of the lack of clinical efficacy in interim analysis. The KAT (Kuopio Angiogenesis Trial) [8] tested intracoronary adenovirus-VEGF-A gene transfer in coronary heart disease patients. Gene transfer was done during PTCA and stenting operation. Although the adenovirus group had significantly increased myocardial perfusion as measured with SPECT 6 months after the treatment, no significant difference was found between the treatment and control groups on an exercise test. Similarly, the Euroinject One Trial did not meet its primary endpoint in patients treated with intracardiac naked VEGF-A plasmid injection using the NOGA catheter system [9]. Also, recombinant protein trials have not met their endpoints. VIVA (Vascular Endothelial Growth Factor in Ischemia for Vascular Angiogenesis) [10] and FIRST (FGF-Initiating Revascularization Trial) [11] trials did not show clear clinical benefit whereas the REVASC Trial has shown a positive outcome as measured by 1-mm ST segment depression on exercise tolerance test 26 weeks after intramyocardial delivery of adenovirus-VEGF-A121 [12].
One of the major problems in the conducted phase II/III clinical trials has been the verification of the transduction efficiency and gene expression in the transduced myocardium. One of the most likely reasons for inconsistent clinical benefit is the low transduction efficiency in myocardium [4]. Thus, in future clinical trials, it would be extremely important to visualize transgene expression in the myocardium. Also, verification of the vector delivery and retention in the target tissue would provide important information for the future design of clinical trials. Since biopsy samples and analysis of circulating blood can only provide circumstantial evidence about the transduction level and transgene expression in the myocardium, imaging techniques applicable to clinical cardiology should be developed to evaluate these critical parameters in addition to safety, feasibility, and efficacy [4].
Background of cardiac molecular imaging
Traditionally, investigators monitor cardiac gene expression by using conventional reporter genes such as β-galactosidase (β-gal) [13] and chloramphenicol-acetyl transferase (CAT) [14]. These techniques, however, require invasive biopsy or postmortem tissue sampling for analysis. In contrast, the development of molecular imaging now allows noninvasive, quantitative, and repetitive imaging of targeted macromolecules and biological processes in living organisms [15]. In general, two fundamental elements are required: (a) molecular probes that can indicate gene expression and (b) methods to monitor these events [16]. For the former, the two most widely used strategies are direct and indirect imaging. For the latter, a vast array of imaging techniques such as bioluminescence imaging (BLI), computed tomography (CT), magnetic resonance imaging (MRI), single-photon emission computed tomography (SPECT), positron emission tomography (PET), and ultrasound are available, as outlined in another section (“Primer on molecular imaging technology” by C. Levin).
Direct molecular imaging involves direct probe–target interaction. The target can be a receptor, enzyme, or mRNA. For probe–receptor imaging, a recent example of cardiac application involves imaging αvβ3 integrin receptor expressed in angiogenic vessels after myocardial infarction by using 111In-RP748, a radiolabeled quinolone targeted at αvβ3 [17]. For probe–enzyme imaging, the most well-known cardiac application is 18F-fluorodeoxyguclose ([18F]FDG) to assess for tissue viability after myocardial infarction. The [18F]-labeled glucose analog is transported across the intact cell membrane, undergoes phosphorylation by hexokinase, and is retained in the cell in proportion to the rate of cellular glycolysis [18]. The radioactive [18F] undergoes positron annihilation into two high-energy γ rays (511 keV) which can be detected as coincidence signals by PET. For probe–mRNA imaging, radiolabeled antisense oligonucleotide (RASON) probes can be used [19, 20]. These radiolabeled probes are typically 12–35 nucleotides long and are complementary to a small segment of the target mRNA. However, the RASON approach is less practical at this point because of (a) a low number of target mRNA (∼1,000 copies) per cell compared with proteins (>10,000 copies), (b) limited tracer penetration across the cell membrane, (c) poor intracellular stability, (d) slow washout of unbound oligonucleotide probes, and (e) low target/background ratios [21]. For similar reasons, probe–DNA imaging within the nuclear membrane will be extremely difficult and is not yet feasible. Overall, the main disadvantage of direct molecular imaging is that it requires synthesizing a customized probe for every product of interest (e.g., receptor, enzyme, or mRNA). This can be time consuming and lack the generalizability that is needed for most applications.
Indirect molecular imaging using reporter genes and reporter probes has only recently been validated. Using molecular biology techniques, the reporter gene is first introduced into target tissues by viral or nonviral vectors. The promoter or enhancer elements can be cloned into these vectors to drive the transcription of a reporter gene. The promoter activity can be “constitutive” (always on), “inducible” (turned on or off), or “tissue-specific” (expressed only in the heart, liver, or other organs). Next, translation of the mRNA leads to a reporter protein that can interact with the reporter probe. The signals can then be detected by various imaging modalities such as BLI, PET, or MRI. The main advantage of indirect molecular imaging is that by altering various components, the reporter gene can provide information about the regulation of DNA by upstream promoter [22], the fate of intracellular protein trafficking [23], and the efficiency of vector transduction into cells [24]. Likewise, the reporter probe itself does not have to be changed if one wishes to study a new biological process, which saves valuable time needed to synthesize, test, and validate new radiotracer agents [24]. However, the main disadvantage is that it is a surrogate marker of the physiologic process of interest rather than a direct measurement of the receptor density, intracellular enzyme, or mRNA copies as discussed above for direct molecular imaging, which are likely to be more clinically relevant. Other issues such as the safety of chromosomal integration in the cellular system, the potential for host immune response against reporter genes, and the possibility of reporter gene silencing [25, 26] will need to be investigated.
Cardiac reporter gene imaging studies
Most molecular imaging studies have focused on cancer biology [27, 28], and only recently has reporter gene imaging of cardiac transgene expression been investigated. In the following section, a summary of some of the most relevant studies published over the past years will be discussed.
Bioluminescence imaging
In the first proof-of-principle study involving cardiac optical bioluminescence imaging, adenovirus with a constitutive cytomegalovirus (CMV) promoter driving firefly luciferase reporter gene [Ad-CMV-Fluc; 1×109 particle forming unit (pfu)] was injected into the rat myocardium via aseptic lateral thoracotomy [29]. The reporter probe d-luciferin (125 mg/kg body weight) was injected intraperitoneally before imaging. Interaction of Fluc and d-luciferin generates light photons (2–3 eV) that can be detected by an ultra-sensitive charged coupled device (CCD) camera. Figure 2 shows noninvasive assessment of cardiac transgene expression over 2 weeks. The cardiac Fluc activity peaked within the first 3–5 days but declined rapidly thereafter owing to host cellular immune response against the adenoviral vector [30]. Leakage into the systemic circulation allowed the adenovirus to bind to Coxsackie-adenovirus receptors (CAR) on hepatocytes, leading to expression at an unintended target site [31]. The in vivo imaging results correlated well with in vitro enzyme assays (r2=0.92), indicating that BLI can be used in parallel or in lieu of traditional biochemical assays. Interestingly, the kinetics of cardiac transgene expression varied significantly among different animals, suggesting that the efficiency of gene transfer in human trials may also vary due to inter-individual response. The main advantage of BLI is low cost setup, which allows for high throughput studies in small animals. The main drawback is that the exact location of cardiac transgene location could not be determined exactly owing to photon attenuation and scattering [24].
Optical imaging of cardiac reporter gene expression. Control rat transduced with Ad-CMV-HSV1-sr39tk (1×109 pfu) shows background signal. Study rat transduced with Ad-CMV-Fluc (1×109 pfu) emits significant cardiac firefly luciferase (FL) activity at days 2, 5, 8, and 14 (P<0.05 vs control). Significant hepatic FL activity is seen, starting at day 8. The heart from the same rat is shown, explanted and sliced into three sections at day 14. FL activity is localized at the anterolateral wall of the left ventricle along the site of virus injection. Note the bioluminescence scales are different for control rat, study rat days 2–5, and study rat days 8–14 to account for the wide range of cardiac FL activity observed. RA right atrium, RV right ventricle, LA left atrium, LV left ventricle, RLU/min relative light units per minute. Reproduced from Wu JC, et al. with permission [29]
Positron emission tomography
For these reasons, imaging of human cardiac gene therapy would most likely require PET-based reporter genes and reporter probes. The cellular concept of detecting reporter gene and reporter probe interaction within the myocardium was demonstrated initially by using postmortem autoradiography [32]. In explanted heart tissues, the transgene expression within sites injected with adenovirus carrying the herpes simplex virus type 1 thymidine kinase (HSV1-tk) reporter gene showed a 3.4±2.2-fold higher level of [124I]-2′-fluoro-2′-deoxy-5-iodo-1-β-d-arabino-furanosyluracil ([125I]FIAU) reporter probe radioactivity compared with uninjected sites. Subsequently, in the first proof-of-principle study using in vivo cardiac PET imaging, adenovirus carrying a mutant version of the HSV1-tk reporter gene (Ad-CMV-HSV1-sr39tk) was injected intramyocardially [33]. Similar to the BLI study, adenoviral transduction led to robust but transient (∼2 weeks) gene expression in the myocardium as assessed by the PET reporter probe 9-(4-[18F]fluoro-3 hydroxymethylbutyl) guanine ([18F]FHBG). With PET imaging, the transgene expression could be localized tomographically at the anterolateral wall as shown on short, vertical, and horizontal axes (Fig. 3). In a follow-up study, Inubushi et al. examined the quantitative aspects of the HSV1-sr39tk and [18F]FHBG system [34]. Myocardial [18F]FHBG accumulation was visualized with viral titers down to 1×107 pfu but not with 1×106 pfu of HSV1-sr39tk. For comparison, cardiac clinic trials typically use higher dosages of adenovirus at 2×1010−1×1011 pfu [7, 8], suggesting that the detection sensitivity for clinical imaging should be possible in the future. More recently, gamma camera imaging of cardiac transgene expression using adenoviral-mediated expression of human sodium iodide symporter (Ad-CMV-hNIS) and 123I or 99mTc as reporter probes was shown to be a practical and effective alternative to HSV1-tk and HSV1-sr39tk imaging [35]. The main advantage of small animal PET imaging is that it can be scaled upward for human PET imaging in the future. On the other hand, the main drawback is higher cost per study session and the need for a cyclotron facility, which many research institutions do not have.
PET imaging of cardiac reporter gene expression. a At day 4, whole-body small animal PET image of a rat shows focal cardiac [18F]FHBG activity at the site of intramyocardial Ad-CMV-HSV1-sr39tk injection. Liver [18F]FHBG activity is also seen owing to systemic adenoviral leakage with transduction of hepatocytes. A control rat injected with Ad-CMV-Fluc shows no [18F]FHBG activity in the cardiac and hepatic regions. Significant gut and bladder activities are seen for both study and control rats owing to the route of [18F]FHBG clearance. b Tomographic views of cardiac small animal PET images. The [13N]NH3 (gray scale) images of perfusion are superimposed on [18F]FHBG images (color scale) demonstrating HSV1-sr39tk reporter gene expression. [18F]FHBG activity is seen in the anterolateral wall for the study rat compared with background signal in the control rat. Note: perpendicular lines represent the axes for vertical and horizontal cuts. Color scale is expressed as %ID/g. Reproduced from Wu JC, et al. with permission [33]
Tissue-specific imaging
In previous imaging studies [29, 33, 34], both transient cardiac gene expression and unintended hepatic expression were observed. These observations suggest that ischemic heart patients who underwent intracoronary infusion of adenovirus carrying either fibroblast growth factor (FGF) [7] or vascular endothelial growth factor (VEGF) [8] in clinical trials would likely have encountered these two issues. In future studies, the design of the delivery vector will need to be vastly improved. An ideal vector system will demonstrate high infectivity of cardiac muscle cells, minimal infectivity of unintended target site, and no elicitation of host immune response [36]. The issue of transient gene expression may be addressed by substituting the adenovirus with less immunogenic vectors such as the adeno-associated virus [37] or gutless adenovirus [38]. The issue of unintended target site expression may be addressed by using cardiac tissue-specific promoters such as myosin light chain kinase (MLC) that drive the reporter genes instead of the constitutive CMV promoter [39]. A recent imaging study also demonstrated that rats injected with Ad-MLC-Fluc in the myocardium had more restricted gene expression in the myocardium compared with rats injected with Ad-CMV-Fluc [22]. However, tissue-specific promoters are inherently less robust in driving gene expression compared with constitutive promoters. Currently efforts are underway to use a two-step transcriptional activation (TSTA) system for amplifying gene expression from a weak tissue specific promoter [40, 41]. Despite these potential options, it remains a formidable challenge to translate results from small animals to large animals and eventually to human gene therapy trials.
Large animal imaging
The feasibility of transferring imaging results from a small animal PET scanner to a clinical PET scanner (ECAT EXACT scanner, Siemens/CTI) has been demonstrated recently in a porcine model [42]. Bengel et al. showed that myocardial tissues infected with adenovirus expressing HSV1-tk had significantly higher [124I]FIAU retention and the in vivo images correlated with postmortem autoradiography and immunohistochemistry. However, the signal to background ratio at the site of HSV1-tk injection was only ∼1.25 within the first 30 min post injection. Afterwards, there was further significant washout from 45 to 120 min post injection as the [124I]FIAU retention became similar to that in control myocardial regions. Thus, the combination of HSV1-tk/[124I]FIAU may be less ideal than the HSV1-sr39tk/[18F]FHBG combination for imaging cardiac transgene expression [43], consistent with data from the oncology literature [44].
Imaging myocardial angiogenesis
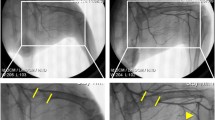
While a great deal of effort has been focused on the development of strategies to enhance neoangiogenesis in the heart, there remains a need for effective measurement in patients. Indeed, most trials have relied on indirect evidence of myocardial angiogenesis, such as changes in clinical performance (i.e., increased treadmill exercise times or decreased anginal symptoms) or physiological consequences (i.e., improved myocardial perfusion on SPECT or contractility on echocardiography) [36]. In contrast, the neovasculature offers multiple targets for imaging which may not be expressed by the established vasculature. For instance, the αvß3 integrin is expressed in angiogenic vessels and represents a novel target for direct molecular imaging of the natural angiogenesis [45]. In a proof of principle study involving rats and dogs that underwent left anterior descending artery (LAD) or left circumflex artery (LCX) ligation, serial in vivo dual-isotope SPECT imaging with the 111In-labeled αvß3-targeted agent (111In-RP748) demonstrated focal radiotracer uptake in hypoperfused regions where angiogenesis was stimulated (Fig. 4) [17]. There was a fourfold increase in myocardial radiotracer uptake in the infarct region, which was also confirmed histologically. Thus, targeted imaging of the angiogenic process in the heart is feasible and may allow sensitive assessment of patients following myocardial infarction or following angiogenic gene therapies.
Molecular imaging of the myocardial angiogenic process. a Serial in vivo 111In-RP748 SPECT short axis, vertical long axis (VLA), and horizontal long axis (HLA) images at 20 min and 75 min after tracer injection in a dog 3 weeks after LAD infarction. 111In-RP748 SPECT images were registered with 99mTc-MIBI perfusion images (third row). The 75-min 111In-RP748 SPECT images were colored red and fused with 99mTc-MIBI images (green) to better demonstrate localization of 111In-RP748 activity within the heart (color fusion, bottom row). Right ventricular (RV) and left ventricular (LV) blood pool activity is seen at 20 min. White arrows indicate region of increased 111In-RP748 uptake in the anterior wall. This corresponds to the anteroapical 99mTc-sestamibi perfusion defect (yellow arrows). b Sequential 99mTc-sestamibi (top row) and 111In-RP748 in vivo SPECT HLA images at 90 min after injection (middle row) from a dog at 8 h (acute), 1 week, and 3 weeks after LAD infarction. Increased myocardial 111In-RP748 uptake is seen in the anteroapical wall at all three time points. Color fusion 99mTc-MIBI (green) and 111In-RP748 (red) images (bottom row) demonstrate 111In-RP748 uptake within 99mTc-MIBI perfusion defect. c Ex vivo 99mTc-sestamibi (left) and 111In-RP748 (center) images of myocardial slices from a dog 3 weeks after LAD occlusion, with color fusion image on the right. Short axis slices are in the standard orientation. Yellow arrows indicate anterior location of nontransmural perfusion defect region; white arrows indicate the corresponding area of increased 111In-RP748 uptake. Although this study was not performed after the administration of neoangiogenic gene delivery, it demonstrates the promise of such a technique for demonstration of the efficacy of gene therapy. Reproduced from Meoli DF, et al. with permission [17]
Imaging bicistronic reporter genes
Similarly, indirect molecular imaging can also be used to track cardiac gene therapy. Conceptually, this can be achieved by linking a PET reporter gene to a therapeutic gene of interest. To first demonstrate that the expression of two linked genes would have high fidelity, Chen et al. constructed a bicistronic adenoviral vector whereby a CMV-driven receptor-based PET receptor gene (a mutant dopamine 2 receptor, or D2R80a) was coupled to an enzyme-based PET reporter gene (HSV1-sr39tk) using an internal ribosomal entry site (Ad-CMV-D2R80a-IRES-HSV1-sr39tk) [46]. After injection into the rat myocardium, longitudinal imaging with PET reporter probes 3-(2-[18F]fluoroethyl)spiperone ([18F]FESP) and [18F]FHBG, respectively, revealed a good correlation between the two linked PET reporter genes (r2=0.73; P<0.001). These results suggest that if one of the PET reporter genes is replaced with a therapeutic gene, a correlated expression would likely exist and allow for indirect monitoring of the therapeutic gene. Besides the IRES system, other techniques of linking the therapeutic gene to the reporter gene are also available, including using two separate delivery vectors [47], fusion of reporter genes [48], bi-directional transcription [49], and the dual promoter approach, as described below.
Imaging VEGF therapeutic and PET reporter genes
In the first proof of principle study using PET imaging to track therapeutic gene expression, Wu et al. constructed an adenovirus with a CMV promoter driving a VEGF121 therapeutic gene and a second CMV promoter driving the HSV1-sr39tk reporter gene [50]. The two expression cassettes are separated by poly adenine sequences (Ad-CMV-VEGF121-polyA-CMV-HSV1-sr39tk-polyA). The adenovirus was injected into the myocardium of adult rats with myocardial infarction by ligation of the LAD artery. Control animals received adenovirus without an expression cassette (Ad-null) instead. Reporter gene expression persisted for only 2 weeks owing to host cellular immune immune repsponse. Repeat adenoviral injection into the same animals at 2 months did not induce any reporter gene expression owing to host humoral immune response (Fig. 5) [30]. At 10 weeks, the mean densities of capillaries (747±104 versus 450±101 per mm2) and small blood vessels (8.1±0.8 versus 5.1±1.2 per mm2) were significantly higher in the VEGF-treated study group compared with the control group as assessed by histological staining for anti-CD31 and anti-smooth muscle actin antibodies (P<0.05 for both). For functional studies, left ventricular ejection fraction (LVEF) showed mild improvement in the VEGF-treated study group (from 43.4±8.1% at baseline to 47.3±12.5% at week 10) compared with the control group (47.5±9.3% to 45.2±8.4%), but this did not reach statistical significance. The VEGF-treated study animals also showed an encouraging trend toward lower [13N]NH3 perfusion defects (15.2±3.1% to 13.8±2.6%) and [18F]FDG metabolism deficits (12.7±4.3% to 11.5±4.6%), but the changes were not statistically significant. As expected, the control group did not show any significant changes in perfusion (14.0±4.0% to 15.3±4.1%) or metabolism (13.4±2.3% to 15.1±3.0%) scores (P=NS) (Fig. 6). Taken together, these results suggest that the microscopic level of neovasculature induced by VEGF did not translate into significant changes in clinically relevant physiological parameters such as myocardial contractility, perfusion, and metabolism under the study conditions tested.
PET imaging of cardiac angiogenic gene therapy. a Schematic of Ad-CMV-VEGF121-CMV-HSV1-sr39tk-mediated gene expression. Two separate gene cassettes with CMV promoters driving the expression of a VEGF121 therapeutic gene and an HSV1-sr39tk reporter gene separated by polyA tails. The translated product of VEGF121 is soluble and excreted extracellularly, whereas the translated product of HSV1-sr39tk (HSV1-sr39TK) traps [18F]FHBG intracellularly by phosphorylation. b Noninvasive imaging of the kinetics of cardiac transgene expression. Gene expression peaked at day 1 and rapidly decreased thereafter. A second injection (arrow) of Ad-CMV-VEGF121-CMV-HSV-sr39tk at day 60 yielded no detectable signal on days 62 and 64. Error bars represent mean±SEM. c A representative rat scanned longitudinally with transaxial [18F]FHBG PET images shown at similar slice levels of the chest cavity. The gray scale is normalized to the individual peak activity of each image. In this rat, myocardial [18F]FHBG accumulation was visualized at the anterolateral wall (arrow) from day 1 to day 14 but not day 17, 62, or 64. d In vivo gene, perfusion, and metabolism imaging with PET. At day 2, representative images showing normal perfusion ([13N]NH3) and metabolism ([18F]FDG) in a sham rat, anterolateral infarction in a control rat (Ad-null), and anterolateral infarction in a VEGF-treated study rat (Ad-CMV-VEGF121-CMV-HSV1-sr39tk) in short, vertical, and horizontal axes (gray scale). The color scale is expressed as % ID/g for [18F]FHBG uptake. As expected, the sham and control animals both had background [18F]FHBG signal only (blue color) that outlined the shape of the chest cavity. In contrast, the study rat showed robust HSV1-sr39tk reporter gene activity near the site of injection. Reproduced from Wu JC, et al. with permission [50]
Evaluation of myocardial functional improvement after VEGF121 gene therapy. a Ten weeks after myocardial infarction, echocardiography was performed to evaluate cardiac function by M-mode (top row) and short-axis views during systole (middle row) and diastole (bottom row). The left ventricular ejection fraction (LVEF) and fractional shortening (FS) were compared for all three groups. As expected, sham animals had normal LVEF and FS that remained unchanged. Control animals (Ad-null) showed slightly decreased LVEF and FS from baseline to week 10 while study animals (Ad-CMV-VEGF121-CMV-HSV1-sr39tk) showed slightly increased LVEF and FS from baseline to week 10. However, neither trend was statistically significant (n=10; P=NS). b Evaluation of changes in myocardial perfusion and metabolism in relation to VEGF121 gene therapy. Ten weeks after myocardial infarction, [13N]NH3 perfusion and [18F]FDG metabolism imaging were performed again for sham, control (Ad-null), and study (Ad-CMV-VEGF121-CMV-HSV1-sr39tk) animals. A representative sham animal showed normal myocardial perfusion and metabolism while control and VEGF study animals had anterior and anterolateral wall defects on polar maps. Compared with baseline, the extent of perfusion and metabolism improved slightly for VEGF study animals but did not reach statistical significance (n=10; P=NS)
Summary
These findings may shed light on the interpretation of recent cardiac gene therapy trials. Historically, the field of cardiac angiogenesis attracted much attention from the cardiovascular community in the late 1990s as most animal studies and phase 1 nonrandomized trials uniformly showed positive results [3]. However, recent phase 2 randomized trials such as the VIVA [10], FIRST [11], AGENT [7], and KAT [8] have demonstrated neither consistent nor substantial efficacy. In retrospect, several lessons can be learned from these trials. They showed that angiogenesis is a complex process regulated by the interaction of various growth factors and likely cannot be duplicated by use of a single protein or gene injection for a brief period of time [51]. The ideal injection method, delivery vector, and patient population remain to be determined. Because there was no available method of assessing gene expression in vivo, the investigators were blinded as to whether the lack of symptomatic improvement was a result of transient gene expression, poor delivery technique, or host inflammatory response. Thus, biological issues related to pharmacokinetics, functional, and physiological effects of gene expression will need to be fully understood before clinical efficacy of cardiac gene therapy can be expected [36]. This is exactly this reason that molecular imaging can and should be used for monitoring gene transfer.
Conclusion
Despite recent inconsistent clinical results in phase II/III clinical trials for coronary heart disease, intramyocardial gene transfer still has great potential to become an efficient treatment option for severely affected patients. However, many practical questions remain in routine clinical assessment of gene therapy, such as: (1) Has the vector reached its target site? (2) Do other nontarget tissues also show gene expression? (3) How long does the gene expression last? (4) What are the optimal route, timing, and dosage of gene delivery? (5) Is the level of gene expression sufficient to induce a therapeutic effect? (6) Does the kinetics of gene expression correlate with functional improvement? In these regards, cardiac imaging methods can provide significant new information about the functionality and efficacy of current vector and gene delivery systems. This information is likely to be vital for the better design of local gene transfer methods and selection of the best treatment genes [4].
Cardiac molecular imaging combines the disciplines of cell biology, molecular biology, synthetic chemistry, medical physics, and translational sciences into a new research paradigm. As discussed here and elsewhere, optical bioluminescence and fluorescence imaging have excellent detection sensitivity but limited depth penetration. MRI provides spectacular image resolution but lacks detection sensitivity. PET has both great detection sensitivity and spatial resolution but requires a nearby cyclotron facility. Although the exact choice will depend on the biological question asked, further improvement in image resolution and detection sensitivity will be needed for all modalities as we move from imaging of organs and tissues to imaging of cells and genes. Likewise, key research areas that remain to be addressed in molecular imaging include: (1) Is there a host immune response against reporter genes? (2) Do the reporter probes adversely affect cellular metabolism? (3) Can we image gene expression quantitatively and accurately? Undoubtedly, active collaboration among basic researchers and imaging experts will be needed as the field moves forward from animal studies to human trials with targeted human imaging.
References
Yla-Herttuala S, Martin JF. Cardiovascular gene therapy. Lancet 2000;355:213–22
Markkanen JE, Rissanen TT, Kivela A, Yla-Herttuala S. Growth factor-induced therapeutic angiogenesis and arteriogenesis in the heart-gene therapy. Cardiovasc Res 2005;65:656–64
Yla-Herttuala S, Alitalo K. Gene transfer as a tool to induce therapeutic vascular growth. Nat Med 2003;9:694–701
Yla-Herttuala S, Markkanen JE, Rissanen TT. Gene therapy for ischemic cardiovascular diseases: some lessons learned from the first clinical trials. Trends Cardiovasc Med 2004;14:295–300
Laitinen M, Hartikainen J, Hiltunen MO, Eranen J, Kiviniemi M, Narvanen O, et al. Catheter-mediated vascular endothelial growth factor gene transfer to human coronary arteries after angioplasty. Hum Gene Ther 2000;11:263–70
Rutanen J, Rissanen TT, Markkanen JE, Gruchala M, Silvennoinen P, Kivela A, et al. Adenoviral catheter-mediated intramyocardial gene transfer using the mature form of vascular endothelial growth factor-D induces transmural angiogenesis in porcine heart. Circulation 2004;109:1029–35
Grines CL, Watkins MW, Helmer G, Penny W, Brinker J, Marmur JD, et al. Angiogenic Gene Therapy (AGENT) trial in patients with stable angina pectoris. Circulation 2002;105:1291–7
Hedman M, Hartikainen J, Syvanne M, Stjernvall J, Hedman A, Kivela A, et al. Safety and feasibility of catheter-based local intracoronary vascular endothelial growth factor gene transfer in the prevention of postangioplasty and in-stent restenosis and in the treatment of chronic myocardial ischemia: phase II results of the Kuopio Angiogenesis Trial (KAT). Circulation 2003;107:2677–83
Kastrup J, Jorgensen E, Ruck A, Tagil K, Glogar D, Ruzyllo W, et al. Direct intramyocardial plasmid vascular endothelial growth factor-A165 gene therapy in patients with stable severe angina pectoris A randomized double-blind placebo-controlled study: the Euroinject One trial. J Am Coll Cardiol 2005;45:982–8
Henry TD, Annex BH, McKendall GR, Azrin MA, Lopez JJ, Giordano FJ, et al. The VIVA trial: vascular endothelial growth factor in ischemia for vascular angiogenesis. Circulation 2003;107:1359–65
Simons M, Annex BH, Laham RJ, Kleiman N, Henry T, Dauerman H, et al. Pharmacological treatment of coronary artery disease with recombinant fibroblast growth factor-2: double-blind, randomized, controlled clinical trial. Circulation 2002;105:788–93
Stewart DJ. A phase 2, randomized, multicenter, 26-week study to assess the efficacy and safety of BIOBYPASS (AdgvVEGF121.10) delivered through minimally invasive surgery versus maximum medical treatment in patients with severe angina, advanced coronary artery disease, and no options for revascularization. Circulation 2002:1 (Abstract)
Schröder G, Risch K, Nizze H, Kolls J, Reinke P, Brock J, et al. Immune response after adenoviral gene transfer in syngeneic heart transplants: effects of anti-CD4 monoclonal antibody therapy. Transplantation 2000;70:191–8
Kass-Eisler A, Falck-Pedersen E, Alvira M, Rivera J, Buttrick PM, Wittenberg BA, et al. Quantitative determination of adenovirus-mediated gene delivery to rat cardiac myocytes in vitro and in vivo. Proc Natl Acad Sci U S A 1993;90:11498–502
Blasberg RG, Tjuvajev JG. Molecular-genetic imaging: current and future perspectives. J Clin Invest 2003;111:1620–9
Herschman HR. Molecular imaging: looking at problems, seeing solutions. Science 2003;302:605–8
Meoli DF, Sadeghi MM, Krassilnikova S, Bourke BN, Giordano FJ, Dione DP, et al. Noninvasive imaging of myocardial angiogenesis following experimental myocardial infarction. J Clin Invest 2004;113:1684–91
Schelbert HR. 18F-deoxyglucose and the assessment of myocardial viability. Semin Nucl Med 2002;32:60–9
Shi N, Boado RJ, Pardridge WM. Antisense imaging of gene expression in the brain in vivo. Proc Natl Acad Sci U S A 2000;97:14709–14
Tavitian B, Terrazzino S, Kuhnast B, Marzabal S, Stettler O, Dolle F, et al. In vivo imaging of oligonucleotides with positron emission tomography. Nat Med 1998;4:467–71
Gambhir SS, Barrio JR, Herschman HR, Phelps ME. Imaging gene expression: principles and assays. J Nucl Cardiol 1999;6:219–33
Boecker W, Bernecker OY, Wu JC, Zhu X, Sawa T, Grazette L, et al. Cardiac-specific gene expression facilitated by an enhanced myosin light chain promoter. Mol Imaging 2004;3:69–75
Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci U S A 2002;99:15608–13
Wu JC, Sundaresan G, Iyer M, Gambhir SS. Noninvasive optical imaging of firefly luciferase reporter gene expression in skeletal muscles of living mice. Molec Ther 2001;4:297–306
Kim YH, Lee DS, Kang JH, Lee YJ, Chung JK, Roh JK, et al. Reversing the silencing of reporter sodium/iodide symporter transgene for stem cell tracking. J Nucl Med 2005;46:305–11
Wu JC, Wang D, Chen IY, Park JM, Patel MR, Gheysens O, et al. Molecular mechanisms of reporter gene silencing and reversal in cell transplant imaging. Mol Imaging Biol 2005;7:107 (Abstract)
Gambhir SS, Barrio JR, Wu L, Iyer M, Namavari M, Satyamurthy N, et al. Imaging of adenoviral-directed herpes simplex virus type 1 thymidine kinase reporter gene expression in mice with radiolabeled ganciclovir. J Nucl Med 1998;39:2003–11
Tjuvajev JG, Finn R, Watanabe K, Joshi R, Oku T, Kennedy J, et al. Noninvasive imaging of herpes virus thymidine kinase gene transfer and expression: a potential method for monitoring clinical gene therapy. Cancer Res 1996;56:4087–95
Wu JC, Inubushi M, Sundaresan G, Schelbert HR, Gambhir SS. Optical imaging of cardiac reporter gene expression in living rats. Circulation 2002;105:1631–4
Yang Y, Li Q, Ertl HC, Wilson JM. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J Virol 1995;69:2004–15
Guzman RJ, Lemarchand P, Crystal RG, Epstein SE, Finkel T. Efficient gene transfer into myocardium by direct injection of adenovirus vectors. Circ Res 1993;73:1202–7
Bengel FM, Anton M, Avril N, Brill T, Nguyen N, Haubner R, et al. Uptake of radiolabeled 2′-fluoro-2′-deoxy-5-iodo-1-beta-D-arabinofuranosyluracil in cardiac cells after adenoviral transfer of the herpesvirus thymidine kinase gene: the cellular basis for cardiac gene imaging. Circulation 2000;102:948–50
Wu JC, Inubushi M, Sundaresan G, Schelbert HR, Gambhir SS. Positron emission tomography imaging of cardiac reporter gene expression in living rats. Circulation 2002;106:180–3
Inubushi M, Wu JC, Gambhir SS, Sundaresan G, Satyamurthy N, Namavari M, et al. Positron-emission tomography reporter gene expression imaging in rat myocardium. Circulation 2003;107:326–32
Miyagawa M, Beyer M, Wagner B, Anton M, Spitzweg C, Gansbacher B, et al. Cardiac reporter gene imaging using the human sodium/iodide symporter gene. Cardiovasc Res 2005;65:195–202
Pislaru S, Janssens SP, Gersh BJ, Simari RD. Defining gene transfer before expecting gene therapy: putting the horse before the cart. Circulation 2002;106:631–6
Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol 2005;23:321–8
Fleury S, Driscoll R, Simeoni E, Dudler J, von Segesser LK, Kappenberger L, et al. Helper-dependent adenovirus vectors devoid of all viral genes cause less myocardial inflammation compared with first-generation adenovirus vectors. Basic Res Cardiol 2004;99:247–56
Franz WM, Rothmann T, Frey N, Katus HA. Analysis of tissue-specific gene delivery by recombinant adenoviruses containing cardiac-specific promoters. Cardiovasc Res 1997;35:560–6
Iyer M, Wu L, Carey M, Wang Y, Smallwood A, Gambhir SS. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci U S A 2001;98:14595–600
Chen IY, Ray S, Padmanabhan P, Gheysens O, Deroose C, Caffarelli A, et al. A generalizable bi-directional vector for non-invasive imaging and amplification of transgene expression mediated by a weak cardiac-specific promoter. Mol Imaging Biol 2005;7:104 (Abstract)
Bengel FM, Anton M, Richter T, Simoes MV, Haubner R, Henke J, et al. Noninvasive imaging of transgene expression by use of positron emission tomography in a pig model of myocardial gene transfer. Circulation 2003;108:2127–33
Miyagawa M, Anton M, Haubner R, Simoes MV, Stadele C, Erhardt W, et al. PET of cardiac transgene expression: comparison of 2 approaches based on herpesviral thymidine kinase reporter gene. J Nucl Med 2004;45:1917–23
Min JJ, Iyer M, Gambhir SS. Comparison of [18F]FHBG and [14C]FIAU for imaging of HSV1-tk reporter gene expression: adenoviral infection vs stable transfection. Eur J Nucl Med Mol Imaging 2003;30:1547–60
Sipkins DA, Cheresh DA, Kazemi MR, Nevin LM, Bednarski MD, Li KC. Detection of tumor angiogenesis in vivo by alphaVbeta3-targeted magnetic resonance imaging. Nat Med 1998;4:623–6
Chen IY, Wu JC, Min JJ, Sundaresan G, Lewis X, Liang Q, et al. Micro-positron emission tomography imaging of cardiac gene expression in rats using bicistronic adenoviral vector-mediated gene delivery. Circulation 2004;109:1415–20
Yaghoubi SS, Wu L, Liang Q, Toyokuni T, Barrio JR, Namavari M, et al. Direct correlation between positron emission tomographic images of two reporter genes delivered by two distinct adenoviral vectors. Gene Ther 2001;8:1072–80
Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res 2004;64:1323–30
Sun X, Annala AJ, Yaghoubi SS, Barrio JR, Nguyen KN, Toyokuni T, et al. Quantitative imaging of gene induction in living animals. Gene Ther 2001;8:1572–9
Wu JC, Chen IY, Wang Y, Tseng JR, Chhabra A, Salek M, et al. Molecular imaging of the kinetics of vascular endothelial growth factor gene expression in ischemic myocardium. Circulation 2004;110:685–91
Dor Y, Djonov V, Abramovitch R, Itin A, Fishman GI, Carmeliet P, et al. Conditional switching of VEGF provides new insights into adult neovascularization and pro-angiogenic therapy. Embo J 2002;21:1939–47
Acknowledgements
The authors have indicated they have no financial conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, J.C., Yla-Herttuala, S. Human gene therapy and imaging: cardiology. Eur J Nucl Med Mol Imaging 32 (Suppl 2), S346–S357 (2005). https://doi.org/10.1007/s00259-005-1897-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-005-1897-6