Abstract
Promoter choice is an important step in recombinant protein production, which directly determines the expression manner as constitutive or inducible and the expression level of the recombinant protein. This study aims to investigate the applicability of heterologous yeast promoters (Kluyveromyces marxianus TPI, Hansenula polymorpha PMA, Candida tropicalis ICL, and Saccharomyces cerevisiae CUP) in Pichia pastoris. The regulation mode of the CtICL and ScCUP promoters in P. pastoris was found to be inducible and that of the KmTPI and HpPMA was constitutive. The carbon sources in which the promoters exhibited the highest activity were determined as glycerol for PMA and TPI, glucose for CUP, and ethanol for ICL. The DNA region showing the highest activity was determined as 1000 bp for all promoters by promoter deletion analysis. Results from the study demonstrate the potential of inducible and constitutive heterologous promoters allowing expression under different conditions in the P. pastoris expression system and offers alternatives to frequently used promoters.
Key points
• Heterologous promoters exhibited similar expression pattern in P. pastoris with its native host.
• HpPMA has the highest promoter activity among the heterologous promoters tested.
• Reporter gene expression with ScCUP is responsive to elevating Cu2+in P. pastoris.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Promoters are molecular tools that allow the control of recombinant protein production at the level of transcription. The optimum expression of a foreign protein in the selected expression system is inherently related to the success of recombinant protein production (Vogl and Glieder 2013). This led to attempts to establish molecular tool libraries for controlling gene expression (Redden and Alper 2015; Yan et al. 2022a). To achieve different expression levels of the gene of interest, the construction of promoter libraries consisting of different strength promoters are among these studies. Recently, promoter libraries have been introduced for the most frequently used Pichia pastoris promoters (Hartner et al. 2008; Qin et al. 2011).
Promoter libraries can be created using different approaches, such as random mutation (Bakke et al. 2009; Qin et al. 2011) and deletion and duplication of putative transcription factor binding sites (Hartner et al. 2008; Erden-Karaoğlan et al. 2022). It is also possible to obtain novel hybrid synthetic promoters by combining the regulatory regions from different promoters (Blazeck et al. 2012; Zhao et al. 2021). In order for these synthetic promoters to be broad-spectrum capable of transcription in different host systems, they require an inter-species combination of all essential and efficient promoter elements (Tang et al. 2020). Therefore, studies on the characterization of native promoters to reveal the effects of promoter structure on promoter strength and performance constitute the first step as the starting point of synthetic biology studies (Redden and Alper 2015; Feng and Marchisio 2021). To shed light on future synthetic biology studies, and especially for multi-host applications, the characterization of the native promoters isolated from other yeasts was considered in the present study, as they provide higher potential than endogenous promoters. Also, characterized alternative promoters have been presented for the P. pastoris expression system.
In this study, the feasibility of heterologous yeast promoters in the P. pastoris expression system was investigated. Within the scope of the study, constitutive Kluyveromyces marxianus TPI and Hansenula polymorpha PMA promoters and inducible Candida tropicalis ICL and Saccharomyces cerevisiae CUP1 promoters were tested. In addition, relative strengths were determined by comparison with the most commonly used P. pastoris promoters, AOX1 and GAP. As a result, new promoters with different expression modes whose regulation is supported by the P. pastoris expression system have been introduced.
Materials and methods
Bioinformatics analysis
In this study, C. tropicalis ICL, H. polymorpha PMA, K. marxianus TPI, and S. cerevisiae CUP promoters were tested to drive recombinant protein expression in P. pastoris. NCBI accession numbers of genes regulated by the respective promoters; XM_002550358.1 for C. tropicalis ICL, F109913.2 for H. polymorpha PMA, AJ577476.2 for K. marxianus TPI, and 856450 for S. cerevisiae CUP. The promoters to be tested in recombinant protein production were determined as 1500 bp DNA region at the 5′ end of the gene, starting from the translation initiation codon and covering the nucleotides -1500 to -1.
Strains, vectors, and culture conditions
Escherichia coli XL1-Blue (Promega, Madison, WI, USA) was used for subcloning studies and P. pastoris GS115 (his4−) (Invitrogen, Waltham, MA, USA) was used as the host system for protein expression. Expression cassettes were inserted into the P. pastoris genome through the his4 locus. To construct the expression cassettes containing heterologous promoters, the vectors pPICZαA/HIS4-XylB and pGAPZαA/HIS4-XylB obtained from the vectors produced in our previous study (Karaoğlan et al. 2016) were used (Fig. 1). Expression vectors containing the heterologous promoters were generated by excising the AOX1 promoter with restriction enzymes and ligation of the promoters to be tested to the same restriction enzyme recognition regions.
E. coli cells used in the cloning and plasmid amplification studies were cultured in LB Miller Broth (1% tryptone, 0.5% yeast extract and 1.0% NaCI), LB Miller Agar (1% tryptone, 0.5% yeast extract, 1.0% NaCI, and 1.5% Agar), LB Lennox Broth (1% tryptone, 0.5% yeast extract, and 0.5% NaCI), and LB Lennox Agar (1% tryptone, 0.5% yeast extract, 0.5% NaCI and 1.5% Agar).
P. pastoris GS115 cells cultured in YPD Broth (1% yeast extract, 2% peptone, 2% dextrose), YPD Agar (1% yeast extract, 2% peptone, 2% dextrose and 1.5% Agar), MD broth (2% glucose, 1.34% YNB and 4 × 10–5% biotin), and MD Agar (2% glucose, 1.34% YNB, 4 × 10–5% biotin and 1.5% agar) media for growth and/or selection of the cells. Minimal media for recombinant protein production contained 1.34% YNB, 4 × 10–5% biotin, 100 mM potassium phosphate pH 6 buffer, and 1% carbon source. Rich media for recombinant protein production contained 2% peptone, 1% yeast extract, 1.34% YNB, 4 × 10–5% biotin, and 100 mM phosphate buffer pH 6.0 and 1% carbon source. Glycerol was used as a carbon source for the cell accumulation phase and the carbon sources determined for each promoter were used in the protein production phase.
Construction and cloning of the expression vectors containing heterologous promoters
In the first stage, the promoter regions tested were 1500 bp DNA originating from the region at the 5′ end of the respective gene. ScCUP, HpPMA, and KmTPI promoters were amplified from chromosomal DNA samples of the relevant yeasts using the primers (Table 1) designed based on the genome sequences given in the NCBI (National Center of Biotechnology Information) database. Since C. tropicalis yeast is pathogenic (neurotoxic), CtICL promoter sequence has been synthesized commercially.
Heterologous yeast promoters obtained by PCR were digested with BglII and AsuII and ligated into the AOX1 promoter-excised pPICZαA/HIS4-XylB plasmid. The general representation of these expression vectors is given in Fig. 1. The resulting expression vectors were linearized with SspI through the marker HIS4 gene and transformed into the electrocompetent P. pastoris GS115 cells by electroporation. The selection of transformant cells recovering the HIS4 gene was performed on minimal media containing glucose.
The expression cassettes were integrated into the P. pastoris GS115 genome at the his4 locus. Southern blot analysis was performed to confirm the integration of the expression cassettes and determine the number of integrated expression cassette copy numbers according to the method applied by Karaoğlan et al. (2016). The clones containing a single copy were selected for each promoter to eliminate the gene copy number effect in comparison analysis and the studies were continued with shake flask experiments.
Shake flask experiments with heterologous promoters
The protein expression under the regulation of tested heterologous promoters was analyzed using different carbon sources. The carbon sources tested for each promoter are given in Table 2. The best carbon source that provides the highest production was determined.
Clones containing a single copy expression cassette determined by Southern blot analysis were first inoculated from frozen culture into test tubes containing 5 mL of YPD medium and incubated at 28 °C and 250 rpm for 24 h. These cultures were used to inoculate 50 mL of BMGY medium with an initial density of OD600nm = 0.1. Cultures reached an optical density at 600 nm (OD600) of 8–10 were centrifuged, the cell pellets were washed, and resuspended in 50 mL of induction media containing different carbon sources. Incubation was carried out at 28 °C and 250 rpm for 72 h. During fermentation, samples were taken at regular intervals (every 12 h). Samples taken were centrifuged 10,000 × g for 5 min, and the collected supernatant samples were kept at + 4 ℃ until activity analysis. Enzyme activity and growth of the cells were followed during fermentation. As a result of the data obtained, the carbon source in which the highest activity was obtained under shake flask conditions was determined for each promoter and further analyses were carried out with these carbon sources.
5′-End deletion analysis of the heterologous promoters
The first trials of the study were carried out with 1500 bp DNA regions of the heterologous promoters considering the general promoter lengths reported for yeast promoters in the literature between 1000 and 1500 bp (Kristiansson et al. 2009; Xu et al. 2021). Then, 5′-end deletion analysis was performed to identify the DNA regions that mediate the highest expression level of the promoters, and promoter activities of 1500, 1000, and 500 bp-DNA regions of the promoters were compared. Results also provide a preliminary result for the detection of the exact promoter regions of heterologous promoters that function in the P. pastoris expression system. Cloning studies of the 1000 and 500 bp promoter variants were carried out with the same strategy described for 1500 bp DNA regions.
Promoters were tested for protein production under the conditions determined and using the carbon sources selected in the previous step. Enzyme activity and cell growths (optical density) were analyzed in samples taken at the 12th hour of fermentation.
While evaluating the results, different lengths of each promoter were compared within itself. Promoter activities were calculated relative to the activity of promoter variant with the highest activity. For each heterologous promoter, the DNA region that exhibited the highest activity was identified and used in comparative analysis with the commonly used P. pastoris promoters AOX1 and GAP.
Comparison of heterologous promoters with P. pastoris AOX1 and GAP promoters
It was aimed to reveal the relative activities of the heterologous promoters compared to well-known P. pastoris promoters. For this purpose, heterologous promoters with the highest activity under optimum conditions were compared with P. pastoris AOX1 and GAP promoters.
Cloning and verification analyses were performed as detailed under the construction and cloning of the expression vectors containing the heterologous promoters section. The expression vectors containing P. pastoris AOX1 and GAP promoters, pPICZαA/HIS4-XylB, and pGAPZαA/HIS4-XylB, were digested with SspI for the integration into the his4 locus and transferred into competent P. pastoris GS115 cells. Clones containing a single copy expression cassette were chosen by Southern blot to eliminate the effect of gene copy number effect in comparison analysis as described in previous steps.
Recombinant xylanase production was carried out under the regulation of the promoters tested with the selected clones. Cell growth and enzyme activities were monitored during the fermentation. Promoter strengths were determined indirectly by measuring xylanase enzyme activity and were calculated relative to the AOX1 promoter activity, which was accepted as 100%.
Determination of xylanase enzyme activity
Xylanase enzyme activities were measured to determine the promoter’s strengths. Enzyme activities were analyzed with the dinitrosalicylic (DNS) method (Karaoğlan et al. 2016). The samples were diluted to the appropriate ratio determined by preliminary tests, and 100 µL of the diluted samples were added to the 900 µL of the reaction solution containing 1% (w/v) beechwood xylan in 0.05 M sodium citric acid buffer (pH 5). The reaction mixture was incubated at 50 °C for 5 min. At the end of the incubation, 100 µL of the reaction mixture was added to 900 µL of DNS solution to stop the reaction. The mixture was incubated at 100 °C for 5 min and, after cooling on ice, the values were read spectrophotometrically at 540 nm wavelength. A standard curve was drawn with 1–10 µmol of xylose. One unit of the xylanase activity was defined as the amount of enzyme required to produce 1 µmol of reducing sugar under the assay conditions described.
Analysis of the proteins by SDS-PAGE
Proteins in supernatant samples taken during protein production under the regulation of heterologous promoters were analyzed by SDS-PAGE. The analysis was performed using 5% polyacrylamide stacking gel and 10% polyacrylamide separating gel. Supernatant samples were prepared with 4 × sodium dodecyl sulfate (SDS) gel loading buffer (200 mM Tris base, pH 6.8, 8% SDS, 0.4% bromophenol blue, 40% glycerol, 100 mM DTT (1,4-dithiothreitol)) in a total volume of 25 µL and denatured at 70 °C for 10 min. Electrophoresis of the samples was carried out in 1 × TGS (0.025 M Tris base, 0.192 M glycine, 0.1% SDS, pH 8.3) buffer at 150 V for 1 h.
The gel was agitated in the staining solution (0.1% Coomassie brilliant blue R-250, 50% methanol, and 10% acetic acid) on the orbital shaker for 1 h. The stained gel was then washed by agitation in the destaining solution (40% methanol and 10% acetic acid) until the protein bands appeared. Finally, the gel was hydrated with ultrapure water and the gel image was recorded.
Analysis of the proteins by western blot
Western blot analysis was performed to specifically detect histidine-labeled protein in supernatant samples. The proteins separated on polyacrylamide gel was transferred to the PVDF membrane using an Owl HEP-1 Electroblotter (Thermo Fisher Scientific, Waltham, MA, USA) by applying 20 V for 1 h. The proteins transferred to the membrane were detected using a SuperSignal West HisProbe Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
Results
Construction and cloning of the expression vectors containing 1500 bp DNA regions of the heterologous promoters
In this study, the applicability of the inducible C. tropicalis ICL and S. cerevisiae CUP promoters and the constitutive K. marxianus TPI and H. polymorpha PMA promoters in the P. pastoris expression system was investigated. Considering that yeast promoter lengths are generally between 1000 and 1500 bp, the first 1500 bp regions were tested to analyze the function of the promoters (Kristiansson et al. 2009; Xu et al. 2021).
Analysis of the heterologous promoters on different carbon sources
Heterologous promoter activities were analyzed using different carbon sources. Recombinant protein productions with the heterologous promoters were performed in both minimal and rich media containing the carbon sources to be tested. Clones containing a single copy expression cassette were first cultured in glycerol-containing media for cell accumulation to an OD600 of 8–10. The cells were then harvested and resuspended in protein production media containing different carbon sources. Fermentation was continued for 72 h with an addition of a carbon source to a final concentration of 1% every 12 h. Xylanase activity values measured in samples taken during the fermentation showed that an additional induction caused a fluctuation in specific productivities depending on the variation in cell growth under the shake flask conditions. Considering the cell densities and enzyme activities, we determined the sampling time as the 12th hour of the protein production phase for comparative analysis. Samples taken from the 12th hour of the fermentation with minimal and rich defined media were compared. As the difference between the promoter activities was observed more clearly with higher activity values depending on higher biomass densities, the study continued with rich defined media. Enzyme activities produced on different carbon sources with the clones harboring heterologous promoters are given in Fig. 2.
Heterologous promoter activities tested on different carbon sources with rich media. Samples were taken from the 12th hour of the fermentation. Experiments were done in triplicates. Each carbon source was added at 1% concentration. Copper (Cu2+) addition for the CUP promoter was 100 µM. Glu, glucose; MeOH, methanol; Gly, glycerol; EtOH, ethanol; Act, acetate. A Xylanase activity (U/mL), B xylanase activity per optical density (U/mL/OD)
Results obtained with the ScCUP promoter showed that the glucose medium containing 100 µM Cu2+ provided 34.8% higher enzyme activity than the methanol medium containing 100 µM Cu2+ and 35% higher enzyme activity than the glycerol medium containing 100 µM Cu2+. In accordance with the results, glucose was chosen as the carbon source to be used for the ScCUP promoter in subsequent analyses. Then, the effect of different Cu2+ concentrations on promoter activity was examined. Promoter activities were determined in 12-h samples taken from the cultures containing 0, 10, 50, 100, and 200 µM Cu2+ in glucose media (Table 3). This showed that the ScCUP promoter was regulated as inducible with Cu2+ in P. pastoris, as in S. cerevisiae.
The results were calculated relative to the value obtained at the Cu2+ concentration providing the highest activity. As seen in Table 3, there was not a significant increase in promoter activities above the 50 µM Cu2+ concentration and the highest activity was achieved with 100 µM. Further analysis was continued with 100 µM Cu2+ concentration.
The HpPMA promoter has been reported as a constitutive promoter in the literature (Cox et al. 2000). The HpPMA promoter activity was tested in glucose, glycerol, and also methanol media for the P. pastoris expression system. Results showed that the promoter activity detected in glycerol media was approximately 35% higher than in the glucose and methanol media, and glycerol was chosen as the carbon source for further analysis. In this study, the highest activity among heterologous promoters was achieved with this promoter.
The K. marxianus TPI promoter has been reported as a constitutive promoter in the literature (Dohner et al. 2004). Therefore, recombinant protein production trials have been tested with glucose and glycerol carbon sources. The performance of the promoter was also tested in methanol media. No significant difference was observed for the TPI promoter in the promoter activity in glucose, glycerol, and methanol carbon sources. In further analysis, we continued with glycerol, a non-fermentable carbon source (with no by-product production) as a carbon source (Zepeda et al. 2018; Tomàs-Gamisans et al. 2019).
In previous studies, the CtICL promoter was reported to be induced in different yeasts with acetate and/or ethanol (Umemura et al. 1995; Atomi et al. 1995). Therefore, the CtICL promoter was tested in ethanol and acetate carbon sources. The results obtained showed that protein expression was recorded in both carbon sources and 30% higher activity was determined with ethanol carbon source compared to acetate. Glucose and methanol carbon sources were also tested with and without ethanol induction. The activities recorded in glucose and methanol media were 20% and 4.5% of the activity determined in the ethanol medium, which was the highest activity observed, while the levels were 38% and 75% in the ethanol-induced glucose and methanol media, respectively. Response to ethanol was also observed in these media, and its expression was observed to be higher in methanol media than in glucose media. Further analysis continued with ethanol as the carbon source providing the highest activity for the ICL promoter.
5′-End deletion analysis of the heterologous promoters
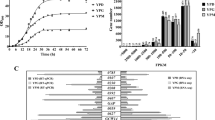
Deletion analysis was performed at the 5′ end of heterologous promoters to determine the DNA region exhibiting the highest activity, and the promoter variants of 1000 and 500 bp lengths were obtained. All clones were analyzed by Southern blot, and clones containing a single copy expression cassette were selected to eliminate the differences that may arise due to gene copy number differences in the comparison of productions. Recombinant xylanase production was carried out with the obtained clones. The results were evaluated according to the xylanase enzyme activity measured in the supernatant samples. Thus, the activity of the promoter variant with the highest activity was accepted as 100% for each promoter. The others were calculated relative to these results (Fig. 3A). As seen in the activity graph, statistical analysis of the results showed that the shortest DNA regions with the highest activity for all four promoters were 1000 bp variants. The activities of the heterologous promoters relative to the PMA promoter with the highest activity are given in Fig. 3B. Activity values were 5.1 U/mL for CUP (1000 bp), 4.4 U/mL for ICL (1000 bp), 0.8 U/mL for TPI (1000 bp), and 25.4 U/mL for PMA (1000 bp).
A Relative xylanase activity values (%) obtained with different DNA region lengths of the heterologous promoters. B Relative xylanase activity values (%) of the heterologous promoters obtained with DNA regions exhibiting the highest promoter activities. Fermentations were carried out with optimum carbon sources determined for each promoter. Samples were taken from the 12th hour of the fermentation. Experiments were done in triplicates. The results were statistically analyzed for each promoter by one-way analysis of variance (ANOVA) using SigmaPlot 11 (Systat Software, Inc., San Jose, CA, USA) program, and the differences between means were determined using the Duncan’s multiple range test at a 95% confidence interval (p < 0.05). Activity values with different lowercase letters define the significant difference (p < 0.05) between the promoter strengths
Supernatant samples taken from xylanase productions with 1500, 1000, and 500 bp DNA regions of heterologous promoters were analyzed by SDS-PAGE (Fig. 4). In the SDS-PAGE gel images, a protein band of approximately 26 kDa was observed representing the xylanase enzyme. Although the protein concentrations did not differ significantly within the different length variants of each promoter, it was observed that the protein bands representing the PMA promoter exhibiting the highest promoter activity had a higher density than that of the other promoters. Conversely, the lowest protein concentration was seen in samples of the TPI promoter with the weakest promoter activity.
Comparison of recombinant protein production levels of heterologous promoters and commonly used P. pastoris promoters AOX1 and GAP
The heterologous promoters examined in the study were compared with inducible AOX1 and constitutive GAP promoters, which are the most used promoters in the P. pastoris expression system (Cereghino and Cregg 2000). The reporter protein xylanase was produced under the regulation of promoters to be compared and promoter activities were determined by xylanase activity. Production studies were carried out using the DNA region and optimum carbon source providing the highest activity for each promoter, and the enzyme activity was analyzed in the supernatant samples taken at the 12th hour of fermentation. In the evaluation of the results, the enzyme activity values obtained with the AOX1 were accepted as 100% and the relative promoter strengths of the heterologous promoters were calculated (Fig. 5).
Comparison of heterologous promoters with the P. pastoris AOX1 and GAP promoters. Fermentations were carried out with optimum carbon sources determined for each promoter. Samples were taken from the 12th hour of the fermentation. Experiments were done in triplicates. Fermentations of the AOX1 and GAP promoters were carried out using methanol and glucose carbon sources, respectively
Results showed no significant difference between the activities of the heterologous CUP and ICL promoters, which were shown to function inducible in P. pastoris. The relative activity values of the heterologous promoters were approximately 10% of the AOX1 promoter and 21% of the GAP promoter. The PMA promoter, which was found to function constitutively in the P. pastoris expression system with the highest activity among the heterologous promoters, exhibited activity at the level of 42% of the AOX1 promoter and 81% of the GAP promoter. The TPI promoter had the lowest activity among the heterologous promoters, and the promoter activity was determined to be 2% of the AOX1 promoter and 4% of the GAP promoter. Proteins in supernatant samples were analyzed by SDS-PAGE and Western blot. SDS-PAGE gel image and Western blot X-ray film image of the samples are given in Fig. 6. SDS-PAGE and Western blot results specifically showed a protein band of approximately 26 kDa representing the xylanase enzyme.
Discussion
In this study, the applicability of heterologous yeast promoters (KmTPI, HpPMA, CtICL, and ScCUP) was tested in a P. pastoris expression system for recombinant protein production. The results showed that the HpPMA promoter provided constitutive expression and had the highest activity among the promoters analyzed. Several studies have been conducted to investigate the compatibility of the molecular tools of the H. polymorpha and P. pastoris expression systems with each other. A H. polymorpha–derived promoter, the HpMOX (promoter region of methanol oxidase gene), was applied for recombinant protein production in P. pastoris and shown to be effectively induced under tested conditions (Mombeni et al. 2020). Also, the HpFMD promoter (promoter region of formate dehydrogenase) was tested in P. pastoris and was observed to have up to 3.5 times higher expression compared to the strongest endogenous P. pastoris methanol utilization pathway promoters (Vogl et al. 2020). Besides, P. pastoris AOX1 and GCW14 promoters were also transferred to H. polymorpha and have been observed to be functional in directing protein expression (Raschke et al. 1996; Rodriguez et al. 1996; Yan et al. 2022b). These studies show that the machinery of these two related organisms successfully supports each other’s molecular tools. However, the constitutive KmTPI promoter which was reported to be strong in the literature by Dohner et al. (2004), exhibited a weak activity in the P. pastoris expression system. The authors reported that seven possible transcription starting points were identified in the KmTPI promoter and the efficiency of the promoter could be adjusted depending on the transcription start site selected for the microorganism in which the expression takes place (Dohner et al. 2004). Also, transcription factors provided by the expression system act as activators and repressors to further increase or decrease the transcription level (McMillan et al. 2019). In the P. pastoris expression system, KmTPI may not have reached the activity level reported in K. marxianus due to these reasons. In addition, the relative promoter activity of the P. pastoris TPI promoter on glucose media has been reported to be approximately 10–20% of the GAP promoter (Stadlmayr et al. 2010; Cai et al. 2021). These results suggest that TPI regulation is not strongly supported in the P. pastoris expression system.
The CtICL and ScCUP promoters have been reported to exhibit inducible expression in previous studies (Atomi et al. 1995; Umemura et al. 1995; Gross et al. 2000). In S. cerevisiae, the CtICL promoter was analyzed in acetate, glycerol, lactate, ethanol, and oleate media. Recombinant protein production results demonstrated that it could be induced in these media (Umemura et al. 1995). In the E. coli expression system, the CtICL promoter has been tested in acetate medium and has been shown to be functional and drive inducible expression (Atomi et al. 1995). In our study, the CtICL activity was investigated in ethanol, acetate, and also glucose and methanol carbon sources with and without ethanol induction. We observed varying promoter activity levels in all media (between 4.5 and 75% compared to the activity recorded in ethanol, which provided the highest activity). It was also determined that the CtICL responded to ethanol induction in glucose and methanol. As a consequence, it is seen that this promoter is inducible in different expression systems.
Sometimes the expression manner of promoters might be different in heterologous expression systems from that in its native system. For instance, the ScCUP promoter, which directs inducible protein expression in its native system, S. cerevisiae (Gross et al. 2000), was determined to exhibit constitutive expression in Kluyveromyces lactis (Macreadie et al. 1991). In our study, we also detected the expression from ScCUP promoter under the uninduced conditions in rich media. However, reporter gene expression was also responsive to Cu2+ and increased significantly with elevating Cu2+ concentrations. In a previous study using the ScCUP promoter in the P. pastoris expression system, intracellular β-galactosidase production was performed, and the results showed that the promoter responds to increased levels of Cu2+ (Koller et al. 2000). It is thought that the difference in induction levels may be related to the difference in production manner and the different proteins produced. Furthermore, even when no Cu2+ was added, Koller et al. (2000) also observed a small amount of promoter activity (3.8% of the activity recorded with a 100 µM concentration of Cu2+). Our results obtained with minimal media conditions showed no significant reporter protein expression without copper induction. These results suggest that media conditions influence recorded basal promoter activity (Gorman et al. 1986).
The heterologous promoters examined in the study were compared with inducible AOX1 and constitutive GAP promoters. The activity results relative to the most used promoters in P. pastoris may guide the selection for applications where the revealed heterologous promoters have the potential for further studies. For example, the HpPMA promoter, which is strong compared to the other heterologous promoters, can be used as an alternative in studies where the constitutive promoter is preferred for recombinant protein production. The strengths of CtICL and ScCUP promoters were found to be 10% compared to the AOX1 promoter, and the KmTPI promoter was at the level of 4% of the GAP promoter. These promoters have the potential to be used in protein productions where relatively weak promoters give more successful results, such as for toxic protein production (Duzenli and Okay 2020). Another example is that Shen et al. (2021) tested a weak promoter, PDog2p300, in P. pastoris to regulate antibiotic-resistant gene expression to reduce the metabolic burden and decrease the cost of the antibiotics for the selection of multi-copy strains and have achieved successful results.
In future studies, it is possible to increase the promoter activities by analyzing the promoter regions in more detail and identifying cis-acting and trans-acting factors. Furthermore, the identified cis-acting regions can be used in synthetic biology studies to develop new synthetic and hybrid promoters, and the advantages of the regulatory mechanisms of these promoters can be exploited in new promoters.
Data availability
The authors confirm that the datasets supporting the findings and conclusions of this study are available within the article.
References
Atomi H, Umemura K, Higashijima T, Kanai T, Yotsumoto Y, Teranishi Y, Ueda M, Tanaka A (1995) The upstram region of the isocitrate lyase gene (UPR-ICL) of Candida tropicalis induces gene expression in both Saccharomyces cerevisiae and Escherichia coli by acetate via two distinct promoters. Arch Microbiol 163:322–328. https://doi.org/10.1007/BF00404204
Bakke I, Berg L, Aune TE, Brautaset T, Sletta H, Tøndervik A, Valla S (2009) Random mutagenesis of the Pm promoter as a powerful strategy for improvement of recombinant-gene expression. Appl Environ Microbiol 75(7):2002–2011. https://doi.org/10.1128/aem.02315-08
Blazeck J, Garg R, Reed B, Alper HS (2012) Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol Bioeng 109:2884–2895. https://doi.org/10.1002/bit.24552
Cai P, Duan X, Wu X, Gao L, Ye M, Zhou YJ (2021) Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res 49:7791–7805. https://doi.org/10.1093/nar/gkab535
Cereghino JL, Cregg JM (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev 24(1):45–66. https://doi.org/10.1111/j.1574-6976.2000.tb00532.x
Cox H, Mead D, Sudbery P, Eland M, Mannazzu I, Evans L (2000) Constitutive expression of recombinant proteins in the methylotrophic yeast Hansenula polymorpha using the PMA1 promoter. Yeast 16:1191–1203. https://doi.org/10.1002/1097-0061(20000930)16:13%3C1191::aid-yea589%3E3.0.co;2-2
Duzenli OF, Okay S (2020) Promoter engineering for the recombinant protein production in prokaryotic systems. AIMS Bioeng 7(2):62–81. https://doi.org/10.3934/bioeng.2020007
Dohner L, Becher D, Salim S, Siekstele R, Sasnauskas K (2004) DNA sequence comprising regulatory regions for expressing proteins. U.S. 20040161841, Google Patents, https://patents.google.com/patent/US20040161841. Accessed 13 May 2022.
Erden-Karaoğlan F, Karaoğlan M, Yılmaz G, Yılmaz S, Inan M (2022) Deletion analysis of Pichia pastoris alcohol dehydrogenase 2 (ADH2) promoter and development of synthetic promoters. Biotechnol J 17:e2100332. https://doi.org/10.1002/biot.202100332
Feng X, Marchisio MA (2021) Saccharomyces cerevisiae promoter engineering before and during the Synthetic Biology Era. Biology 10(6):504. https://doi.org/10.3390/biology10060504
Gorman JA, Clark PE, Lee MC, Debouck C, Rosenberg M (1986) Regulation of the yeast metallothionein gene. Gene 48(1):13–22. https://doi.org/10.1016/0378-1119(86)90347-1
Gross C, Kelleher M, Iyer VR, Brown PO, Winge DR (2000) Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J Biol Chem 275:32310–32316. https://doi.org/10.1074/jbc.m005946200
Hartner FS, Ruth C, Langenegger D, Johnson SN, Hyka P, Lin-Cereghino GP, Lin-Cereghino J, Kovar K, Cregg JM, Glieder A (2008) Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res 36(12):e76. https://doi.org/10.1093/nar/gkn369
Karaoğlan M, Erden-Karaoğlan F, Inan M (2016) Comparison of ADH3 promoter with commonly used promoters for recombinant protein production in Pichia pastoris. Protein Expr Purif 121:112–117. https://doi.org/10.1016/j.pep.2016.01.017
Koller A, Valesco J, Subramani S (2000) The CUP1 promoter of Saccharomyces cerevisiae is inducible by copper in Pichia pastoris. Yeast 16:651–656. https://doi.org/10.1002/(sici)1097-0061(200005)16:7%3C651::aid-yea580%3E3.0.co;2-f
Kristiansson E, Thorsen M, Tamás MJ, Nerman O (2009) Evolutionary forces act on promoter length: Identification of enriched cis-regulatory elements. Mol Biol Evol 26(6):1299–1307. https://doi.org/10.1093/molbev/msp040
Macreadie IG, Horaitis O, Vaughan PR, Clark-Walker GD (1991) Constitutive expression of the Saccharomyces cerevisiae CUP1 gene in Kluyveromyces lactis. Yeast 7(2):127–135. https://doi.org/10.1002/yea.320070206
McMillan J, Lu Z, Rodriguez JS, Ahn TH, Lin Z (2019) YeasTSS: an integrative web database of yeast transcription start sites. Database 2019:baz048 (https://dx.doi.org/10.1093%2Fdatabase%2Fbaz048)
Mombeni M, Arjmand S, Siadat SOR, Alizadeh H, Abbasi A (2020) PMOX: A new powerful promoter for recombinant protein production in yeast Pichiapastoris. Enzym Microb Technol 139:109582. https://doi.org/10.1016/j.enzmictec.2020.109582
Qin X, Qian J, Yao G, Zhuang Y, Zhang S, Chu J (2011) GAP promoter library for fine-tuning of gene expressionin Pichia pastoris. Appl Environ Microbiol 77(11):3600–3608. https://doi.org/10.1128/aem.02843-10
Raschke WC, Neiditch BR, Hendricks M, Cregg JM (1996) Inducible expression of a heterologous protein in Hansenula polymorpha using the alcohol oxidase 1 promoter of Pichia pastoris. Gene 177:163–167. https://doi.org/10.1016/0378-1119(96)00293-4
Redden H, Alper H (2015) The development and characterization of synthetic minimal yeast promoters. Nat Commun 6:7810. https://doi.org/10.1038/ncomms8810
Rodriguez L, Narciandi RE, Roca H, Cremata J, Montesinos R, Rodriguez E, Grillo JM, Muzio V, Herrera LS, Delgado JM (1996) Invertase secretion in Hansenula polymorpha under the AOX1 promoter from Pichia pastoris. Yeast 12:815–822. https://doi.org/10.1002/(sici)1097-0061(199607)12:9%3c815::aid-yea916%3e3.0.co;2-h
Shen Q, Yu Z, Zhou XT, Zhang SJ, Zou SP, Xiong N, Xue YP, Liu ZQ, Zhen YG (2021) Identification of a novel promoter for driving antibiotic-resistant genes to reduce the metabolic burden during protein expression and effectively select multiple integrations in Pichiapastoris. Appl Microbiol Biotechnol 105:3211–3223. https://doi.org/10.1007/s00253-021-11195-0
Stadlmayr G, Mecklenbräuker A, Rothmüller M, Maurer M, Sauer M, Mattanovich D, Gasser B (2010) Identification and characterisation of novel Pichia pastoris promoters for heterologous protein production. J Biotechnol 150(4):519–529. https://doi.org/10.1016/j.jbiotec.2010.09.957
Tang H, Wu Y, Deng J, Chen N, Zheng Z, Wei Y, Luo X, Keasling JD (2020) Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites 6–10(8):320. https://doi.org/10.3390/metabo10080320
Tomàs-Gamisans M, Ødum ASR, Workman M, Ferrer P, Albiol J (2019) Glycerol metabolism of Pichia pastoris (Komagataella spp.) characterised by 13C-based metabolic flux analysis. N Biotechnol 50:52–59. https://doi.org/10.1016/j.nbt.2019.01.005
Umemura K, Atomi H, Kanai T, Teranishi Y, Ueda M, Tanaka A (1995) A novel promoter, derived from the isocitrate lyase gene of Candida tropicalis, inducible with acetate in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 43(3):489–492. https://doi.org/10.1007/BF00218454
Vogl T, Glieder A (2013) Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnol 30(4):385–404. https://doi.org/10.1016/j.nbt.2012.11.010
Vogl T, Fischer JE, Hyden P, Wasmayer R, Sturmberger L, Glieder A (2020) Orthologous promoters from related methylotrophic yeasts surpass expression of endogenous promoters of Pichia pastoris. AMB Express 10(1):38. https://doi.org/10.1186/s13568-020-00972-1
Xu L, Liu P, Dai Z, Fan F, Zhang X (2021) Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants. Microb Cell Fact 20:148. https://doi.org/10.1186/s12934-021-01641-z
Yan C, Yu W, Yao L, Guo X, Zhou YJ, Gao J (2022) Expanding the promoter toolbox for metabolic engineering of methylotrophic yeasts. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-022-11948-5
Yan C, Yu W, Zhai X, Yao L, Guo X, Gao J, Zhou YJ (2022) Characterizing and engineering promoters for metabolic engineering of Ogataea polymorpha. Synth Syst Biotechnol 7(1):498–505. https://doi.org/10.1016/j.synbio.2021.12.005
Zepeda AB, Pessoa A, Farías JG (2018) Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Braz J Microbiol 49:119–127. https://doi.org/10.1016/j.bjm.2018.03.010
Zhao Y, Liu S, Lu Z, Zhao B, Wang S, Zhang C, Xiao D, Foo JL, Yu A (2021) Hybrid promoter engineering strategies in Yarrowia lipolytica: isoamyl alcohol production as a test study. Biotechnol Biofuels 14:149. https://doi.org/10.1186/s13068-021-02002-z
Funding
This study was funded by the Scientific and Technological Research Council of Turkey (TÜBİTAK) (Project Number117Z580).
Author information
Authors and Affiliations
Contributions
FEK and MK designed the study and conducted experiments, analyzed data, and wrote the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Erden-Karaoğlan, F., Karaoğlan, M. Applicability of the heterologous yeast promoters for recombinant protein production in Pichia pastoris. Appl Microbiol Biotechnol 106, 7073–7083 (2022). https://doi.org/10.1007/s00253-022-12183-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12183-8