Abstract
Extracellular glycosyl hydrolases are uncommon in lactobacilli and include amylases and fructosidases mediating starch and fructan utilization, respectively. Extracellular arabinanases have not been described in lactobacilli. This study is aimed at identifying the function of an arabinan utilization operon in Lactobacillus crispatus DSM29598 and at characterizing two putative extracellular arabinanases that are located on that operon. The arabinan utilization operon of L. crispatus DSM29598 encodes enzymes for degradation of arabinan, α-galactosidases, β-galactosidases, and enzymes and for utilization of arabinose including phosphoketolase. The two putative extracellular arabinanases, AbnA and AbnB, are homologous to family GH43 endo-arabinanases. In Lactobacillaceae, homologs of these enzymes were identified exclusively in vertebrate-adapted species of the genus Lactobacillus. L. crispatus grew with arabinan from sugar beet pectin as sole carbon source, indicating extracellular arabinanase activity, and produced lactate and acetate, indicating metabolism via the phosphoketolase pathway. The two arabinanases AbnA and AbnB were heterologously expressed and purified by affinity chromatography. AbnA hydrolyzed linear and branched arabinan, while AbnB hydrolyzed only linear arabinan. The optimum pH for AbnA and AbnB was 6 and 7.5, respectively; 40 °C was the optimum temperature for both enzymes. The application of arabinan degrading L. crispatus as probiotic or as synbiotic with pectins may improve the production of short-chain fatty acids from pectin to benefit host health.
Key points
• An arabinan utilization operon in L. crispatus encodes two extracellular arabinanases.
• The same operon also encodes metabolic genes for arabinose conversion.
• In Lactobacillaceae, extracellular arabinanases are exclusive to Lactobacillus species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lactobacilli have adapted to nutrient-rich environments including ecological niches in plants, humans, and animals but also are associated with food and feed fermentations (Zheng et al. 2020). Their competitiveness in nutrient-rich environments is based on rapid conversion of abundant carbon sources (Gänzle 2015; Zheng et al. 2015; Duar et al. 2017). Carbohydrate metabolism of lactobacilli relies mainly on intracellular conversion of mono-, and di-, and tri-saccharides. Extracellular glycosyl hydrolases are rarely present in lactobacilli, and the utilization of oligosaccharides with a degree of polymerization four or higher is limited by transport to the cytoplasm (Gänzle and Follador 2012). Extracellular amylases of lactobacilli hydrolyze amylose, amylopectin, or pullulan and were mostly identified in host-adapted Lactobacillus species (Gänzle and Follador 2012; Zheng et al. 2015). Other extracellular glycosyl hydrolases in lactobacilli include the fructosidases FosE and FruA in Lacticaseibacillus paracasei and Lactobacillus crispatus, respectively (Yong et al. 2007; Loponen and Gänzle 2018), and glucansucrases and fructansucrases, which are frequent in Liquorilactobacillus and Limosilactobacillus species (van Hijum et al. 2006; Zheng et al. 2020).
Monocotyledonous and dicotyledonous plants contain arabinoxylans and pectins, respectively, as major components of the plant cell wall (Gigli-Bisceglia et al. 2020). Pectins are composed of homogalacturonan, rhamnogalactan I, and rhamnogalactan II; rhamnogalactan I is substituted with branched α-(1 → 3), α-(1 → 5) and α-(1 → 3,5) linked arabinan side chains; side chains in rhamnogalactan I and rhamnogalactan II also include α- and β-linked galactose or galactans (Willats et al. 2001; Ridley et al. 2001). Plant foods rich in pectin include vegetables, e.g., tomatoes and carrots, and fruits, e.g., apples or citrus fruits. Arabinoxylans are composed of a xylose backbone which is substituted with arabinose, branched arabino-oligosaccharides, or other substituents (Saulnier et al. 2007; Rumpagaporn et al. 2015). Monocotyledonous plants include cereals such as corn, wheat, rice, and sorghum, which are staple foods of the diet in agricultural societies. Accordingly, cereal arabinoxylans contribute substantially to the human fiber intake (Rumpagaporn et al. 2015). Metabolism of arabinoxylans by colonic microbiota generates short-chain fatty acids as main mediators of health benefits (Bindels et al. 2015; Koh et al. 2016). Arabinoxylan hydrolysis is mediated by extracellular glycosidase hydrolases and polysaccharide lyases of intestinal bacteria including Bacteroides thetaiotaomicron, Bacteroides ovatus, and Roseburia intestinalis (Flint et al. 2008; Koropatkin et al. 2012). Species in the genera Lactobacillus, Lactilactobacillus, and Limosilactobacillus are stable components of intestinal microbiota of animals; however, lactobacilli are now known to utilize arabinoxylans or pectins. Furfurilactobacillus rossiae metabolizes xylo-oligosacchrides (Pontonio et al. 2016). Two pentosanases were identified in the genome of F. rossiae; however, these proteins are intracellular oligosaccharide hydrolases as their sequences do not include a predicted signal peptide (De Angelis et al. 2014; Pontonio et al. 2016).
The genome sequence of L. crispatus DSM29598 indicates the presence of an arabinan utilization operon. This gene cluster is unusual in two respects. First, most Lactobacillus species, including L. crispatus, do not ferment pentoses and thus lack the capability to utilize products of arabinan hydrolysis (Zheng et al. 2020). Second, extracellular glycosyl hydrolases that enable growth with pentosans or pectins as sole substrate have not been described in lactobacilli. Therefore, the aim of this study was to explore the structure and function of the arabinan utilization operon, as well as the presence of related arabinanases in lactic acid bacteria. Two extracellular arabinanases of L. crispatus DSM29598 were characterized by cloning and expression of their catalytic domain and testing their activities and functional properties.
Materials and methods
Strain and growth conditions
Strains and plasmids used in this study are shown in Table 1. The sourdough isolate L. crispatus DSM29598 (Li et al. 2020) was cultivated in modified deMan-Rogosa-Sharpe (Gänzle and Vogel 2003) (mMRS) medium at 37 °C. Escherichia coli BL21 was cultured in Luria-Bertani (LB) medium at 37 °C. E. coli DH5α with pET28a+ was grown in LB medium with kanamycin (50 mg L−1), which were also used for antibiotic-resistant E. coli selection. The frozen stock culture was inoculated on agar plates; single colonies were inoculated in 1 mL broth, subcultured with 1% inoculum in broth.
Identification of arabinan utilization operon in L. crispatus DSM29598 and analysis of the domain organization of putative arabinanases
The putative function of predicted open reading frames in the arabinan utilization operon in the genome sequences of L. crispatus DSM29598 (accession number: JAATOH000000000) was analyzed by using the blastp tool of NCBI, searching against the UniProtKB/Swiss-Prot and the non-redundant protein sequence collections on the NCBI database. Signal peptides of the putative extracellular endo-arabinanases AbnA and AbnB were predicted by SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/data.php) based on the amino acid sequences of AbnA and AbnB. The prediction of domains in AbnA and AbnB was conducted by comparison of amino acid (AA) sequences using the Conserved Domain Database on the NCBI website.
Phylogenetic analysis of arabinanases in bacteria
Bacterial protein sequences of pentosanases and arabinanases were retrieved from the National Center for Biotechnology Information database using protein BLAST and AbnA and AbnB of L. crispatus DSM29598 as query sequences. Representative sequences from every species of lactic acid bacteria and sequences from representative species of other genera were selected with a cut-off value of 70% coverage and 40% amino acid identity to AbnA or AbnB. A phylogenetic tree was constructed using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al. 1992), and the bootstrap support values were calculated from 100 replicates by MEGAX (Stecher et al. 2020).
Growth of L. crispatus with different carbon sources
L. crispatus was subcultured twice in mMRS basal broth containing 1% linear arabinan from sugar beet pectin (arabinose:galactose:rhamnose:galacturonic acid = 85.2:7.6:1.5:5.7; Megazyme Inc., Chicago, USA), 1% polygalacturonic acid from citrus pectin (galacturonic acid:galactose:arabinose:rhamnose:xylose:glucose = 94:1.0:0.2:1.0:1.0:0.3, Megazyme), 0.5% starch (Megazyme), or mMRS, respectively and grown overnight. The cultures were subsequently used to inoculate (1–2% inoculum) mMRS broth containing the sugars at the same concentrations. The optical density at 600 nm (OD600) and pH of the culture were measured after overnight growth. Lactic acid and acetic acid production was quantified by HPLC as described (Tang et al. 2017). All experiments were done in triplicate.
Hydrolysis of linear and branched arabinan by L. crispatus
L. crispatus was grown in mMRS broth containing 1% linear arabinan and cells were harvested by centrifugation. Cells were fractionated at 4 °C to obtain concentrated culture supernatant, cell wall fraction, and concentrated cytoplasmic extract as described (Yong et al. 2007). Cell fractions were incubated with the same volume of 1% linear arabinan (Megazyme) or branched arabinan from sugar beets (arabinose:galactose:rhamnose:galacturonic acid:other sugars = 69:18.7:1.4:10.2:0.7, Megazyme) in 50 mM citrate phosphate buffer (pH 6.0) at 40 °C for 20 min. Protein concentrations were determined with the Bradford reagent (Sigma), using the manufacturer’s instructions. The enzyme activities are expressed as the amount of reducing sugar released from linear-arabinan or arabinan per minute per mg of protein. The experiments were done in triplicate.
Cloning, expression, and purification of arabinanases
The sequences of AbnA and AbnB without the signal peptide and the SLAP domain were amplified by PCR with AbnA-F and AbnA-R, and AbnB-F and AbnB-R (Table 2). The products of PCR were purified and ligated into pET28a+ containing a C-terminal His-tag for purification of AbnA and AbnB. The recombinant plasmid was transformed into E. coli BL21; transformants were selected on LB agar with 50 mg L−1 of kanamycin and verified by DNA sequencing with primers shown in Table 2. Recombinant E. coli were cultivated in LB broth with kanamycin at 37 °C until the OD600 reached 0.5–0.8. Then, IPTG was added to the medium to a concentration of 0.5 mM to induce enzyme expression, and the culture was further incubated at 20 °C for 20 h.
The proteins were purified by using His-pure Ni-NTA column and His-Pur Cobalt Resin (Thermo Scientific, Waltham, MA, USA) following the suppliers’ manuals. In brief, AbnA was purified with washing buffer with 25 mM and 50 mM of imidazole for both columns. AbnA was eluted from the Ni-NTA column with 250 mM imidazole or eluted from the cobalt column with a gradient from 100 to 250 mM imidazole. For the purification of AbnB, buffers were prepared with 50 mM sodium phosphate, 1 M sodium chloride, 0.1% Triton-X100, and 3 mM β-mercaptoethanol. The concentrations of imidazole in buffers were equilibration buffer, 25 mM; washing buffer, 25 mM or 35 mM; and elution buffers 150 mM, 200 mM, or 250 mM. All buffers were adjusted to pH 8.0. The concentrated active fractions were stored in PBS buffer (pH 7.4) at 4 °C or − 80 °C. The degree of purification was determined by SDS-PAGE gels (Supplemental Fig. S1). The protein concentration was determined as described above. The protein purified by Ni-NTA columns was used for the biochemical characterization, and protein purified by cobalt columns was used for the qualitative and quantitative analysis of substrate specificity.
Substrate specificity of AbnA and AbnB
Purified AbnA and AbnB were mixed with the same volume of 0.5% linear arabinan from sugar beet pectins, branched arabinan from sugar beet pectins, rye arabinoxylan (all from Megazyme), corn arabinoxylan (Agrifiber Holdings LLC, Illinois, USA), xylan from birchwood (Megazyme), or arabinogalactan from larch wood (Megazyme) in 50 mM citrate phosphate buffer (pH 6.0) and incubated at 37 °C for 20 min. Qualitative determination of polysaccharide hydrolysis by purified AbnA and AbnB was performed with high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). In brief, reaction mixtures were separated on a Carbopac PA20 column coupled to an ED40 chemical detector (Dionex, Oakville, Canada). Water (A), 0.2 M NaOH (B), and 1 M Na acetate (NaOAc) (C) were used as mobile phase with the following gradient: 0 min, 68.3% A, 30.4% B and 1.3% C; 30 min, 54.6% A, 30.4% B and 15.0% C; 50 min, 46.6% A, 30.4% B and 23% C; 95 min, 33.3% A, 30.4% B and 36.3% C; 95.1 min, 63.7% A and 36.3% C; 100 min, 50% A and 50% C; 105 min, 10% A, 73% B and 17% C; 105.1 min, 33.3% A, 30.4% B and 36.3% C; 111 min, 10% A, 73% B and 17% C, followed by re-equilibration. The enzyme activity was quantified by determination of reducing sugars with the 3,5-dinitrosalicylic acid (DNS) method (Farro et al. 2018). One unit of enzyme activity was defined as 1 μmol of reducing sugar as arabinose produced per minute.
Effect of pH and temperature on activity of AbnA and AbnB
The optimum pH for arabinanase activity was determined by incubating purified AbnA or AbnB with 0.5% linear arabinan in the pH range from 2 to 10 at 37 °C for 20 min. The following buffers were used: 50 mM citrate phosphate buffer, pH 2.0 to 7.5; 50 mM potassium phosphate, pH 8.0; 50 mM Tris–HCl, pH 8.5 to 9.0; 50 mM glycine–NaOH, pH 10.0. The maximum temperature for enzymatic activity was determined at the optimum pH by using temperatures ranging from 20 to 80 °C. The results were expressed as percentages of the activity obtained at either the optimum pH or the optimum temperature.
The thermal stability of the purified enzyme was determined by incubating purified AbnA or AbnB at 40 °C, 50 °C, and 60 °C for 5 min, 10 min, and 30 min. Samples were withdrawn at the indicated times, cooled on ice bath, and assayed for the residual enzyme activities at optimum pH and temperature.
Effects of additives on the activity of AbnA and AbnB
The effects of several metals on the enzyme activity were determined at optimum pH and temperature as described in above. Several metals (MnCl2, MgCl2, FeCl2, FeCl3, ZnCl2, CaCl2, KCl2, BaCl2, each at 1 mM) and chelating agent (EDTA, 1 mM) were added into the reaction mixture. The results were expressed as percentages of the activity obtained in the reaction with 18 MΩ water in place of the compounds.
Substrate specificity of AbnA and AbnB
The substrate specificity of enzymes was tested using 0.5% linear arabinan, branched arabinan, rye arabinoxylan, corn arabinoxylan, xylan, and arabinogalactan from larch wood at optimum pH and temperature as described above. Enzyme activity was expressed as unit per milligram protein.
Statistical analysis
Data analysis was performed with IBM SPSS statistics 23 (IBM Corp, Armonk, NY, USA) using one-way analysis of variance (ANOVA). A p value of ≤ 0.05 was considered statistically significant.
Results
Arabinan utilization operon of L. crispatus DSM29598
The annotation of the L. crispatus DSM29598 genome sequence identified an arabinan utilization operon (Fig. 1). The operon starts with open reading frames with homology to transposases or mobile element proteins, suggesting that the operon was acquired by lateral gene transfer. The operon also includes enzymes with activity on arabinans, enzymes related to transport and metabolism of arabinan, arabinose, and galactose, several hypothetical proteins, one putative transcriptional regulator, and several glycosyl hydrolases with predicted activity on arabinans or α- and β-linked galactans (Table 3). The extracellular enzymes AbnA and AbnB include a GH43 domain and are 35% and 36%, respectively, identical to an endo-arabinanase in Thermotoga petrophila (Table 3)(Squina et al. 2010). AbfF is a predicted exo-active enzyme that hydrolyzes non-reducing α-L-arabinofuranoside residues in α-L-arabinosides. AraNPQ and MsmK are putative ABC-type transporters with predicted affinity to arabinose-oligosaccharides and α-galacto-oligosaccharides, respectively (Sá-Nogueira et al. 1997). AbfA and AbfB are predicted intracellular and exo-active arabinan hydrolases. AbfH putatively hydrolyzes β-arabinodisaccharides (Fujita et al. 2014). AraA, XylB, AraD, and XpkA are metabolic enzymes for conversion of arabinose to acetyl-CoA and glyceraldehyde-phosphate via ribulose, L-ribulose-5-phosphate, and D-xylulose-5-phosphate (Sá-Nogueira et al. 1997). Additional glycosyl hydrolases include an intracellular LacLM-type β-galactosidase and the α-galactosidase Aga. GalM converts α-aldoses to the β-anomers, and it is active on D-glucose, L-arabinose, D-xylose, D-galactose, maltose, and lactose. The operon is exceptional in two respects, first, because it encodes for several putative extracellular arabinofuranosidases or arabinanases, which have not been characterized in Lactobacillaceae, second, because it encodes all genes for metabolism of arabinose, which is exceptional in Lactobacillus spp. and has not been described for L. crispatus.
Structure of the arabinan utilization operon in L. crispatus DSM 29598 that includes the genes coding for AbnA and AbnB. The putative function of ORFs was analyzed by blastP (Table 3) and different colors refer to corresponding different protein functions: orange, glycocyl hydrolase; green, transport protein; gray, regulatory protein; yellow, metabolic proteins; blue, mobile element proteins; white, hypothetical proteins. The names of ORFs are from left to right: 1. traA, 2.iep, 3. hypA, 4. abnA, 5. xylB, 6. araD, 7. araR 8. lacM, 9. lacL, 10. araA, 11. abnB, 12. abfH, 13. abfA, 14. hypB, 15. araN, 16. araP, 17. araQ, 18. hypC, 19. abfF, 20. abfB, 21. galM, 22. aga, 23. msmK, 24.yxkF, 25. xpkA
Protein domains of AbnA and AbnB
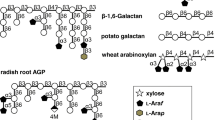
Analysis of the protein sequence of AbnA and AbnB predicted an N-terminal sec-dependent signal peptide mediating protein export with a cleavage site between positions 42 and 43 (VMA-DT) of AbnA and between positions 29 and 30 (LTS-CS) of AbnB (von Heijne 1990). The presence of a signal peptide indicates that AbnA and AbnB are extracellular enzymes. The lack of a signal peptide in AbfF indicates that it is an intracellular enzyme while the homologous enzyme Araf43A in Streptomyces avermitilis is extracellular and includes a signal peptide (Ichinose et al. 2008). Further analyses focused on the extracellular AbnA and AbnB. A predicted GH family 43 domain with 50% amino acid identity to an extracellular arabinanase in Geobacillus stearothermophilus (Shulami et al. 2011) is located between 159 and 539 of AbnA; the same domain with 52% amino acid identity to the extracellular arabinanase in G. stearothermophilus is located between positions 51 and 431 of AbnB (Table 3). The sequences of AbnA and AbnB in positions 548 to 662 and 440 and 549, respectively, are homologous to the C-terminal domain of GH43 enzymes. AbnA additionally includes a C-terminal surface layer protein (SLAP) domain at positions 764 to 817 (Fig. 2a). The presence of an SLAP domain connected to a glycosyl hydrolase domain may indicate that the enzyme is attached to surface layer proteins of L. crispatus (Sillanpää et al. 2000; Li et al. 2020).
Domain prediction of putative arabinanases in L. crispatus DSM29598. a Prediction of domains in AbnA and AbnB based on amino acid sequences. Amino acid sequences of both putative arabinanases were compared with conserved domains. b Phylogenetic analysis of AbnA and AbnB. Amino acid sequences of AbnA and AbnB were used as query sequence to retrieve homologs by protein BLAST on NCBI with cut-off values of 45% protein identity and 80% coverage. The phylogenetic tree shows representative homologs from each species of lactic acid bacteria and homologs from representative species of other genera
Phylogenetic analysis determined the distribution and frequency of arabinanases in bacteria
The amino acid sequences of AbnA and AbnB are 47% identical. Homologous sequences in Lactobacillaceae are exclusively present in strains of the vertebrate host–adapted genus Lactobacillus, i.e., Lactobacillus gallinarum, Lactobacillus xujianguonis, and L. crispatus (Fig. 2b) (Fujisawa et al. 1992; Duar et al. 2017; Meng et al. 2020; Zheng et al. 2020). Homologous sequences were also identified in other lactic acid bacteria (Lactobacillales), predominantly Enterococcus species that are considered to be of intestinal origin (Lebreton et al. 2014) and species of Streptococcus that are also associated with animal or human hosts.
Growth of L. crispatus with different carbon sources
The presence of extracellular arabinanases was confirmed by incubation of L. crispatus in media with linear arabinan from sugar beet pectin and starch and homogalacturonan from citrus pectin as sole carbon sources, as well as mMRS containing glucose, maltose, and fructose (Fig. 3). Extracellular enzymes are required for utilization of polysaccharides to degrade these to oligosaccharides that can be transported to the cytoplasm (Gänzle and Follador 2012). L. crispatus grown in media with linear arabinan had a comparable OD600 and total production of lactic acid and acetic acid with that in mMRS, which indicates that linear arabinan was degraded extracellularly by AbnA or AbnB and utilized via AraA, XylB, AraD, and XpkA. L. crispatus does not harbor extracellular enzymes to utilize starch and homogalacturonan as carbon sources, which is consistent with higher pH, lower OD600, and a lower production of metabolites in media containing starch and citrus pectin as sole carbon sources.
Growth of L. crispatus DSM29598 with different carbon sources. a OD600 and pH of overnight culture grown in mMRS with linear arabinan (black bar), starch (white bar), pectin (gray bar), mMRS (hatched white bar). b Production of lactic acid (black bar) and acetic acid (white bar) of L. crispatus after overnight growth in mMRS with linear-arabinan, starch, pectin, and mMRS. Values for the same analyte differ significantly (p < 0.05) unless the bars share a common superscript. Values ± standard deviation were calculated from three independent experiments
Cellular location of AbnA and AbnB in L. crispatus
The cellular location of AbnA and AbnB was assessed by determination of the arabinanase activity of the supernatant, the cell wall fraction, and the cytoplasmic fraction of L. crispatus grown with LAR (Supplemental Fig. S2). The degradation of linear arabinan by the cell wall fraction and cytoplasmic fraction was higher than the degradation by enzymes present in the supernatant, which indicates the presence of cell wall–associated arabinanase in L. crispatus. However, the degradation of branched arabinan by different cell fractions was comparable.
Cloning, expression, and purification of AbnA and AbnB in E. coli
Catalytic domains of AbnA and AbnB were amplified and ligated in pET 28a+ with a His-tag at the C-terminus. The recombined plasmids, pET-abnA and pET-abnB, were transformed and expressed in E. coli BL21 (Table 1). Overexpressed AbnA and AbnB in the soluble fraction of the cell lysate were purified by His-pure columns. The predicted molecular weights of AbnA and AbnB were 72.11 kDa and 58.56 kDa, respectively, in good accordance with the molecular weight of purified AbnA and AbnB after purification on His-tag columns (Supplemental Fig. S1). Purified AbnB also contained a minor band with slightly lower molecular weight; as this band was absent in purified AbnA (Supplemental Fig. S1A) or uninduced E. coli BL1 pET28a+ (data not shown), it likely represents a hydrolysis product of AbnB.
Arabinan hydrolysis and enzymatic properties of AbnA and AbnB
The degradation of different substrates by purified AbnA and AbnB was assessed using HPAEC-PAD in comparison to of the same substrates incubated with PBS or cell lysate of E. coli BL21 with the pET28a+ empty plasmid as control (Fig. 4). The peaks between 20 and 90 min of retention time were present after reaction with AbnA and AbnB with linear arabinan but not in the controls and thus represent oligosaccharides produced from linear arabinan. AbnA but not AbnB also produced oligosaccharides with branched arabinan as substrate. Both AbnA and AbnB were inactive on any other substrate, including rye arabinoxylan, corn arabinoxylan, xylan, and arabinogalactan.
Separation of oligosaccharides that were released by hydrolysis of different substrates by purified AbnA or AbnB. a Degradation of linear arabinan. b The degradation of branched arabinan. Control shows the reaction of polysaccharides with cell lysis of E. coli BL21 with pET28a+ empty plasmid. The parallel reactions were also conducted with rye arabinoxylan, corn arabinoxylan, xylan, and arabinogalactan (data not shown), in which the differences between control and proteins were not detected
Purified AbnA and AbnB were incubated with linear arabinan in buffers with pH range of 2 to 10 at 37 °C. The activities of AbnA and AbnB were optimal at pH 6 and pH 7.5, respectively (Fig. 5). The optimum temperature of both enzymes was 40 °C (Fig. 5). Both enzymes maintained about 80% of their activity after incubation at 40 °C for 30 min but were inactivated after 5 min at 50 °C or 60 °C (Supplemental Fig. S3).
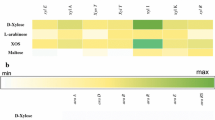
Multiple additives were added into the reaction of AbnA or AbnB with linear arabinan to assess their effect on the activities of two enzymes (Fig. 6). The activity of AbnA was improved by Mn2+, Mg2+, Fe2+, and Fe3+ and reduced by addition of EDTA. The activity of AbnB was not affected by any of the additives.
Effect of different additives on the relative activities of AbnA (black bar) and AbnB (gray bar) with linear arabinan as substrate. Enzyme activities were calculated relative to the reaction without additive. Values obtained for the same protein are significantly different (p < 0.05) unless the bars share a common superscript. Values represent mean ± standard deviation from three independent replicates
The activities of AbnA and AbnB on linear and branched arabinan, rye arabinoxylan, corn arabinoxylan, xylan, and arabinogalactgan were quantified at optimum pH and temperature (Table 4). AbnA degraded linear and branched arabinan with higher activity with the linear substrate while AbnB did not hydrolyze the branched arabinan.
Discussion
This study analyzed an arabinan utilization operon of L. crispatus DSM29598. The operon encodes extracellular arabinan utilization enzymes as well as enzymes for metabolism of arabinose, which are exceptional in Lactobacillus species. Characterization of AbnA and AbnB demonstrated that these enzymes are extracellular endo-arabinanases. Homologous sequences in Lactobacillaceae species are exclusively present in the vertebrate host–adapted genus Lactobacillus.
Until 2015, the fermentation of pentoses by lactobacilli has been used as a tool for their taxonomic identification (Orla-Jensen 1919; Pot et al. 2014; Gänzle 2015); however, fermentation of pentoses in many homofermentative Lactobacillaceae is variable at a species- or strain-level (Zheng et al. 2020). Lactobacillus species do not encode pyruvate formate lyase, or enzymes for homofermentative metabolism of pentoses (Gänzle 2015; Zheng et al. 2015, 2020). Phosphoketolase is present in all Lactobacillaeae but only a few Lactobacillus species including L. hamsteri, L. pasteurii, and L. xujianguonis ferment pentoses (Meng et al. 2020; Zheng et al. 2020). To date, utilization of pentoses by L. crispatus has not been described. The genomic island encoding arabinan utilization in L. crispatus DSM29598 also encodes for AraA, an L-arabinose isomerase, AraD, a L-ribulose-5-phosphate 4-epimerase, XylB, a xylulose or ribulose kinase, and XpkA, a pentose phosphoketolase, and the regulatory proteins AraR. The operon thus provides the full complement of enzymes to allow arabinose utilization via the phosphoketolase pathway (Zhao and Gänzle 2018). The presence of an IS4 transposase gene implies that the arabinan genomic island was obtained by lateral gene transfer. The GC content of arabinan utilization operon is 34.67%, while GC content of genome sequence of L. crispatus DSM29598 is 37.6%. Arabinan metabolism via the phosphoketolase pathway was verified by metabolite analysis (this study), demonstrating that lateral gene transfer bestows pentose utilization to homofermentative lactobacilli, or, when using obsolete terminology, converts an “obligate homofermentative” to a “facultative heterofermentative” organism (Pot et al. 2014; Zheng et al. 2020).
The arabinan utilization operon of L. crispatus also encodes for enzymes that catalyze arabinan hydrolysis, including endo-active extracellular AbnA and AbnB, exo-active AbfA, AbfH, AbfB, and AbfF. AraNPQ transports arabinose or arabino-oligosaccharides into the cells and XylB, AraD, and AraA utilize arabinose as carbon source. These functional enzymes are also encoded by an arabinan operon in Bacillus subtilis (Inácio et al. 2008). The transcriptional regulater AraR was not identified in B. subtilis but found in the arabinan utilization system of G. stearothermophilus (Shulami et al. 2011).
The N-terminal export signal of AbnA and AbnB indicates their extracellular location, which was verified biochemically. AbnA additionally includes a C-terminal surface layer–associated protein (SLAP) domain, which is also found in an extracellular fructosidase (FruA) of L. crispatus DSM29598 (Li et al. 2020). FruA has two SLAP domains with identity of 48–55% with SLAP domain of AbnA. SLAP domains are found in multiple surface layer proteins or surface-associated proteins of lactobacilli (Smit et al. 2001; Antikainen et al. 2002; Åvall-Jääskeläinen et al. 2008; Dohm et al. 2011) and likely mediates association of the enzyme with the cell wall (Li et al. 2020). The combination of GH32 or GH43 domains with SLAP domains is exclusive to vertebrate host–adapted Lactobacillus species (this study, Li et al. 2020). In addition to degrading extracellular polysaccharides, the cell surface–associated enzymes of L. crispatus may contribute to the adhesion of cells to the biofilm matrix or the host epithelium (Frese et al. 2011; Johnson et al. 2013).
AbnA was characterized as endo-arabinanase with activity on linear and branched arabinan, while AbnB only hydrolyzed linear arabinan. Consistent with the activity of extracellular endo-arabinanase in T. petrophila, AbnA showed much lower activity on branched arabinan when compared to linear arabinan (Squina et al. 2010). AbnB is selective for linear arabinan, as was observed for an endo-arabinanase from Bacillus licheniformis (Farro et al. 2018). AbnA activity is partially dependent on metal ions and its activity was reduced to 60% after addition of EDTA, while AbnB activity is metal-independent, consistent with Abn2 in B. subtilis (Inácio and De Sá-Nogueira 2008). Mn2+ stimulated the activity of AbnA in accordance with that of Thermotoga thermarum (Shi et al. 2014). Fe2+ also improved arabinanase activity in Caldicellulorsiruptor saccharolyticus (Hong et al. 2009).
Arabinoxylans and pectins in food are recognized as dietary fiber and improve human health through multiple mechanisms, which include its influence on gut transit and fermentation in the large intestine to yield short-chain fatty acids (SCFA) (Duncan et al. 2016). The utilization of both cell wall polysaccharides by human intestinal microbiota requires dedicated and multiple extracellular and intracellular glycoside hydrolases, lyases, phenolic acid esterases, and transport system (Flint et al. 2008; Koropatkin et al. 2012; Leth et al. 2018). Lactobacilli are stable components of animal intestinal microbiota but L. crispatus DSM29598 is the first strain equipped with an arabinan utilization operon. The lack of enzymes targeting xylans and the substrate specificity of AbnA and AbnB indicate that L. crispatus is unable to degrade cereal arabinoxylans and that the presence of the operon is unrelated to the isolation of the strain from rye sourdough (Li et al. 2020). AbnA and AbnB of L. crispatus hydrolyze the arabinan side chains of rhamnogalactan I in dicotyledonous plants to provide access to a carbon source that is inaccessible to other lactobacilli which inhabit the same ecological niche (Walter 2008, this study). Other glycosyl hydrolases encoded by the arabinan utilization operon of L. crispatus are intracellular and thus not active on polysaccharides; however, they may support intracellular metabolism of pectic oligosaccharides that are generated by enzymes from other gut bacteria.
In conclusion, the arabinan utilization operon of L. crispatus DSM29598 encodes enzymes for hydrolysis and metabolism of extracellular arabinan, arabino-oligosaccharides, and arabinose. These arabinan and arabinose metabolic enzymes of Lactobacillus species were likely acquired by lateral gene transfer. Two extracellular arabinanases, AbnA and AbnB, were characterized as endo-arabinanases which, within the Lactobacillaceae, are exclusive to vertebrate host–adapted Lactobacillus species. AbnA acted on linear and branched arabinan from sugar beet pectin, while AbnB only degraded linear arabinan. The arabinan operon of L. crispatus together with other glycosyl hydrolases utilizes linear and branched arabinans that are present in the cell walls of dicotyledonous plants. Intestinal microbial metabolism of plant cell wall polysaccharides generates short-chain fatty acids that improve the glucose and lipid metabolism to reduce the risk of developing cardiovascular disease, type 2 diabetes, and obesity and also enhance the gastrointestinal barrier function and exert immunoregulatory effects (Deehan et al. 2017; Yan et al. 2018). The application of arabinan degrading L. crispatus as probiotics or as synbiotics with pectins may provide energy for microbial growth and produce short-chain fatty acids to increase host health (Koropatkin et al. 2012).
References
Antikainen J, Anton L, Sillanpää J, Korhonen TK (2002) Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol Microbiol 46:381–394. https://doi.org/10.1046/j.1365-2958.2002.03180.x
Åvall-Jääskeläinen S, Hynönen U, Ilk N, Pum D, Sleytr UB, Palva A (2008) Identification and characterization of domains responsible for self-assembly and cell wall binding of the surface layer protein of Lactobacillus brevis ATCC 8287. BMC Microbiol 8:165. https://doi.org/10.1186/1471-2180-8-165
Bindels LB, Delzenne NM, Cani PD, Walter J (2015) Towards a more comprehensive concept for prebiotics. Nat Rev Gastroenterol Hepatol 12:303–310. https://doi.org/10.1038/nrgastro.2015.47
Bouffard GG, Rudd KE, Adhya SL (1994) Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J Mol Biol 244:269–278
De Angelis M, Bottacini F, Fosso B, Kelleher P, Calasso M, Di Cagno R, Ventura M, Picardi E, Van Sinderen D, Gobbetti M (2014) Lactobacillus rossiae, a vitamin B12 producer, represents a metabolically versatile species within the genus Lactobacillus. PLoS One 9:e207232. https://doi.org/10.1371/journal.pone.0107232
Deehan EC, Duar RM, Armet AM, Perez-Muñoz ME, Jin M, Walter J (2017) Modulation of the gastrointestinal microbiome with nondigestible fermentable carbohydrates to improve human health. Microbiol Spectr 5:BAD-0019-2017. https://doi.org/10.1128/microbiolspec.bad-0019-2017
Dohm N, Petri A, Schlander M, Schlott B, König H, Claus H (2011) Molecular and biochemical properties of the S-layer protein from the wine bacterium Lactobacillus hilgardii B706. Arch Microbiol 193:251–261. https://doi.org/10.1007/s00203-010-0670-9
Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J (2017) Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41:S27–S48. https://doi.org/10.1093/femsre/fux030
Duncan SH, Russell WR, Quartieri A, Rossi M, Parkhill J, Walker AW, Flint HJ (2016) Wheat bran promotes enrichment within the human colonic microbiota of butyrate-producing bacteria that release ferulic acid. Environ Microbiol 18:2214–2225. https://doi.org/10.1111/1462-2920.13158
Farro EGS, Leite AET, Silva IA, Filgueiras JG, de Azevedo ER, Polikarpov I, Nascimento AS (2018) GH43 endo-arabinanase from Bacillus licheniformis: structure, activity and unexpected synergistic effect on cellulose enzymatic hydrolysis. Int J Biol Macromol 117:7–16. https://doi.org/10.1016/j.ijbiomac.2018.05.157
Feldmann SD, Sahm H, Sprenger GA (1992) Cloning and expression of the genes for xylose isomerase and xylulokinase from Klebsiella pneumoniae 1033 in Escherichia coli K12. MGG Mol Gen Genet 234:201–210. https://doi.org/10.1007/BF00283840
Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA (2008) Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. https://doi.org/10.1038/nrmicro1817
Fortina MG, Ricci G, Mora D, Guglielmetti S, Manachini PL (2003) Unusual organization for lactose and galactose gene clusters in Lactobacillus helveticus. Appl Environ Microbiol 69:3238–3243. https://doi.org/10.1128/AEM.69.6.3238-3243.2003
Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, Oh PL, Heng NCK, Patil PB, Juge N, MacKenzie DA, Pearson BM, Lapidus A, Dalin E, Tice H, Goltsman E, Land M, Hauser L, Ivanova N, Kyrpides NC, Walter J (2011) The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet 7:e1001314. https://doi.org/10.1371/journal.pgen.1001314
Fujisawa T, Benno Y, Yaeshima T, Mitsuoka T (1992) Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981). Int J Syst Bacteriol 42:487–491. https://doi.org/10.1099/00207713-42-3-487
Fujita K, Takashi Y, Obuchi E, Kitahara K, Suganuma T (2014) Characterization of a novel β-l-arabinofuranosidase in Bifidobacterium longum: functional elucidation of a duf1680 protein family member. J Biol Chem 289:5240–5249. https://doi.org/10.1074/jbc.M113.528711
Gänzle MG (2015) Lactic metabolism revisited: metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr Opin Food Sci 2:106–117. https://doi.org/10.1016/j.cofs.2015.03.001
Gänzle MG, Follador R (2012) Metabolism of oligosaccharides and starch in lactobacilli: a review. Front Microbiol 3:e340. https://doi.org/10.3389/fmicb.2012.00340
Gänzle MG, Vogel RF (2003) Contribution of reutericyclin production to the stable persistence of Lactobacillus reuteri in an industrial sourdough fermentation. Int J Food Microbiol 80:31–45. https://doi.org/10.1016/S0168-1605(02)00146-0
Gigli-Bisceglia N, Engelsdorf T, Hamann T (2020) Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell Mol Life Sci 77:2049–2077
Hong MR, Park CS, Oh DK (2009) Characterization of a thermostable endo-1,5-α-L-arabinanase from Caldicellulorsiruptor saccharolyticus. Biotechnol Lett 31:1439–1443. https://doi.org/10.1007/s10529-009-0019-0
Ichinose H, Yoshida M, Fujimoto Z, Kaneko S (2008) Characterization of a modular enzyme of exo-1,5-α-L-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl Microbiol Biotechnol 80:399–408. https://doi.org/10.1007/s00253-008-1551-x
Inácio JM, De Sá-Nogueira I (2008) Characterization of abn2 (yxiA), encoding a Bacillus subtilis GH43 arabinanase, Abn2, and its role in arabino-polysaccharide degradation. J Bacteriol 190:4272–4280. https://doi.org/10.1128/JB.00162-08
Inácio JM, Lopes Correia I, de Sá-Nogueira I (2008) Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 154:2719–2729. https://doi.org/10.1099/mic.0.2008/018978-0
Johnson B, Selle K, O’Flaherty S, Goh YJ, Klaenhammer T (2013) Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiol (United Kingdom) 159:2269–2282. https://doi.org/10.1099/mic.0.070755-0
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F (2016) From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. https://doi.org/10.1016/j.cell.2016.05.041
Koropatkin NM, Cameron EA, Martens EC (2012) How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. https://doi.org/10.1038/nrmicro2746
Lebreton F, Willems RJL, Gilmore MS (2014) Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shankar N (eds) Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, pp 1–59
Lee YJ, Lee SJ, Kim SB, Lee SJ, Lee SH, Lee DW (2014) Structural insights into conserved L-arabinose metabolic enzymes reveal the substrate binding site of a thermophilic L-arabinose isomerase. FEBS Lett 588:1064–1070. https://doi.org/10.1016/j.febslet.2014.02.023
Leth ML, Ejby M, Workman C, Ewald DA, Pedersen SS, Sternberg C, Bahl MI, Licht TR, Aachmann FL, Westereng B, Hachem MA (2018) Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat Microbiol 3:570–580. https://doi.org/10.1038/s41564-018-0132-8
Li Q, Loponen J, Gänzle MG (2020) Characterization of the extracellular fructanase FruA in Lactobacillus crispatus and its contribution to fructan hydrolysis in breadmaking. J Agric Food Chem in press 68:8637–8647. https://doi.org/10.1021/acs.jafc.0c02313
Loponen J, Gänzle MG (2018) Use of sourdough in low FODMAP baking. Foods 7:96. https://doi.org/10.3390/foods7070096
Loponen J, Mikola M, Sibakov J (2017) An enzyme exhibiting fructan hydrolase activity. WO 2017/220864 A1
Meng J, Jin D, Yang J, Lai X-H, Pu J, Zhu W, Huang Y, Liang H, Lu S (2020) Lactobacillus xujianguonis sp. nov., isolated from faeces of Marmota himalayana. Int J Syst Evol Microbiol 70:11–15. https://doi.org/10.1099/ijsem.0.003598
Nguyen TH, Splechtna B, Krasteva S, Kneifel W, Kulbe KD, Divne C, Haltrich D (2007) Characterization and molecular cloning of a heterodimeric β-galactosidase from the probiotic strain Lactobacillus acidophilus R22. FEMS Microbiol Lett 269:136–144. https://doi.org/10.1111/j.1574-6968.2006.00614.x
Orla-Jensen S (1919) The lactic acid bacteria. Andr Fred Høst and Son, Copenhagen
Overbeeke N, Fellinger AJ, Toonen MY, van Wassenaar D, Verrips CT (1989) Cloning and nucleotide sequence of the α-galactosidase cDNA from Cyamopsis tetragonoloba (guar). Plant Mol Biol 13:541–550. https://doi.org/10.1007/BF00027314
Pontonio E, Mahony J, Di Cagno R, O’Connell Motherway M, Lugli GA, O’Callaghan A, De Angelis M, Ventura M, Gobbetti M, van Sinderen D (2016) Cloning, expression and characterization of a β-d-xylosidase from Lactobacillus rossiae DSM 15814T. Microb Cell Factories 15:1–13. https://doi.org/10.1186/s12934-016-0473-z
Posthuma CC, Bader R, Engelmann R, Postma PW, Hengstenberg W, Pouwels PH (2002) Expression of the xylulose 5-phosphate phosphoketolase gene, xpkA, from Lactobacillus pentosus MD363 is induced by sugars that are fermented via the phosphoketolase pathway and is repressed by glucose mediated by CcpA and the mannose phosphoenolpyrvate phosphotransferase system. Appl Environ Microbiol 68:831–837. https://doi.org/10.1128/AEM.68.2.831
Pot B, Felis G, De Bruyne K, Tsakalidou E, Papadimitriou K, Leisner J, Vandamme P (2014) The genus Lactobacillus. In: Holzapfel W, Wood B (eds) Lactic acid bacteria: biodiversity and taxonomy. John Wiley & Sons, Inc, Hoboken, pp 249–353
Rhimi M, Ilhammami R, Bajic G, Boudebbouze S, Maguin E, Haser R, Aghajari N (2010) The acid tolerant L-arabinose isomerase from the food grade Lactobacillus sakei 23K is an attractive D-tagatose producer. Bioresour Technol 101:9171–9177. https://doi.org/10.1016/j.biortech.2010.07.036
Ridley BL, O’Neill MA, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57:929–967. https://doi.org/10.1016/S0031-9422(01)00113-3
Rumpagaporn P, Reuhs BL, Kaur A, Patterson JA, Keshavarzian A, Hamaker BR (2015) Structural features of soluble cereal arabinoxylan fibers associated with a slow rate of in vitro fermentation by human fecal microbiota. Carbohydr Polym 130:191–197. https://doi.org/10.1016/j.carbpol.2015.04.041
Russell RRB, Opoku JA, Sutcliffe IC, Tao L, Ferretti JJ (1992) A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem 267:4631–4637
Sá-Nogueira I, Nogueira TV, Soares S, De Lencastre H (1997) The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology 143:957–969. https://doi.org/10.1099/00221287-143-3-957
Saulnier L, Sado PE, Branlard G, Charmet G, Guillon F (2007) Wheat arabinoxylans: exploiting variation in amount and composition to develop enhanced varieties. J Cereal Sci 46:261–281. https://doi.org/10.1016/j.jcs.2007.06.014
Shallom D, Belakhov V, Solomon D, Gilead-Gropper S, Baasov T, Shoham G, Shoham Y (2002) The identification of the acid-base catalyst of α-arabinofuranosidase from Geobacillus stearothermophilus T-6, a family 51 glycoside hydrolase. FEBS Lett 514:163–167. https://doi.org/10.1016/S0014-5793(02)02343-8
Shi H, Ding H, Huang Y, Wang L, Zhang Y, Li X, Wang F (2014) Expression and characterization of a GH43 endo-arabinanase from Thermotoga thermarum. BMC Biotechnol 14:1–9. https://doi.org/10.1186/1472-6750-14-35
Shulami S, Raz-Pasteur A, Tabachnikov O, Gilead-Gropper S, Shner I, Shoham Y (2011) The L-arabinan utilization system of Geobacillus stearothermophilus. J Bacteriol 193:2838–2850. https://doi.org/10.1128/JB.00222-11
Sillanpää J, Martinez B, Antikainen J, Toba T, Kalkkinen N, Tankka S, Lounatmaa K, Keränen J, Höök M, Westerlund-Wikström B, Pouwels PH, Korhonen TK (2000) Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J Bacteriol 182:6440–6450
Smit E, Oling F, Demel R, Martinez B, Pouwels PH (2001) The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J Mol Biol 305:245–257. https://doi.org/10.1006/jmbi.2000.4258
Squina FM, Santos CR, Ribeiro DA, Cota J, de Oliveira RR, Ruller R, Mort A, Murakami MT, Prade RA (2010) Substrate cleavage pattern, biophysical characterization and low-resolution structure of a novel hyperthermostable arabinanase from Thermotoga petrophila. Biochem Biophys Res Commun 399:505–511. https://doi.org/10.1016/j.bbrc.2010.07.097
Stecher G, Tamura K, Kumar S (2020) Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol 37:1237–1239. https://doi.org/10.1093/molbev/msz312/5697095
Tang KX, Zhao CJ, Gänzle MG (2017) Effect of glutathione on the taste and texture of type I sourdough bread. J Agric Food Chem 65:4321–4328. https://doi.org/10.1021/acs.jafc.7b00897
van Hijum SAFT, Kralj S, Ozimek LK, Dijkhuizen L, van Geel-Schutten IGH (2006) Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol Mol Biol Rev 70:157–176. https://doi.org/10.1128/mmbr.70.1.157-176.2006
von Heijne G (1990) The signal peptide. J Membr Biol 115:195–201
Walter J (2008) Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 74:4985–4996. https://doi.org/10.1128/AEM.00753-08
Willats WGT, Mccartney L, Mackie W, Knox JP (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47:9–27. https://doi.org/10.1023/A:1010662911148
Yan YL, Hu Y, Gänzle MG (2018) Prebiotics, FODMAPs and dietary fiber — conflicting concepts in development of functional food products? Curr Opin Food Sci 20:30–37. https://doi.org/10.1016/j.cofs.2018.02.009
Yong JG, Lee JH, Hutkins RW (2007) Functional analysis of the fructooligosaccharide utilization operon in Lactobacillus paracasei 1195. Appl Environ Microbiol 73:5716–5724. https://doi.org/10.1128/AEM.00805-07
Yoshida K, Shindo K, Sano H, Seki S, Fujimura M, Yanai N, Miwa Y, Fujita Y (1996) Sequencing of a 65 kb region of the Bacillus subtilis genome containing the lic and cel loci, and creation of a 177 kb contig covering the gnt-sacXY region. Microbiology 142:3113–3123
Zhao C, Pyle AM (2017) The group II intron maturase: a reverse transcriptase and splicing factor go hand in hand. Curr Opin Struct Biol 47:30–39. https://doi.org/10.1016/j.gde.2016.03.011
Zhao X, Gänzle MG (2018) Genetic and phenotypic analysis of carbohydrate metabolism and transport in Lactobacillus reuteri. Int J Food Microbiol 272:12–21. https://doi.org/10.1016/j.ijfoodmicro.2018.02.021
Zheng J, Ruan L, Sun M, Gänzle MG (2015) A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol 81:7233–7243. https://doi.org/10.1128/AEM.02116-15
Zheng J, Wittouck S, Salvetti E, Franz CMAB, Harris HMB, Mattarelli P, O’Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Gänzle MG, Lebeer S (2020) A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70:2782–2858. https://doi.org/10.1099/ijsem.0.004107
Acknowledgments
Reviewer #2 is acknowledged for critical comments that improved the interpretation of experimental results.
Funding
Funding was provided by the Alberta Wheat Commission, the Saskatchewan Wheat Development Commission, the Minnesota Wheat Research and Promotion Council (grant no. 2018F031R), and the Natural Sciences and Engineering Research Council of Canada (NSERC, grant no. CRDPJ542616-19). Q.L. acknowledges stipend support from the China Scholarship Council, and M.G. acknowledges the Canada Research Chairs program.
Author information
Authors and Affiliations
Contributions
QL and MGG conceived and designed research. QL conducted experiments; QL and MGG wrote the manuscript and read and approved the final manuscript version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 607 kb)
Rights and permissions
About this article
Cite this article
Li, Q., Gänzle, M.G. Characterization of two extracellular arabinanases in Lactobacillus crispatus. Appl Microbiol Biotechnol 104, 10091–10103 (2020). https://doi.org/10.1007/s00253-020-10979-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10979-0