Abstract
Pseudomonas protegens H78 produces multiple secondary metabolites, including antibiotics and iron carriers. The guanosine pentaphosphate or tetraphosphate ((p)ppGpp)-mediated stringent response is utilized by bacteria to survive during nutritional starvation and other stresses. RelA/SpoT homologues are responsible for the biosynthesis and degradation of the alarmone (p)ppGpp. Here, we investigated the global effect of relA/spoT dual deletion on the transcriptomic profiles, physiology, and metabolism of P. protegens H78 grown to mid- to late log phase. Transcriptomic profiling revealed that relA/spoT deletion globally upregulated the expression of genes involved in DNA replication, transcription, and translation; amino acid metabolism; carbohydrate and energy metabolism; ion transport and metabolism; and secretion systems. Bacterial growth was partially increased, while the cell survival rate was significantly reduced by relA/spoT deletion in H78. The utilization of some nutritional elements (C, P, S, and N) was downregulated due to relA/spoT deletion. In contrast, relA/spoT mutation globally inhibited the expression of secondary metabolic gene clusters (plt, phl, prn, ofa, fit, pch, pvd, and has). Correspondingly, antibiotic and iron carrier biosynthesis, iron utilization, and antibiotic resistance were significantly downregulated by the relA/spoT mutation. This work highlights that the (p)ppGpp-mediated stringent response regulatory system plays an important role in inhibiting primary metabolism and activating secondary metabolism in P. protegens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudomonas protegens H78, which was isolated from the rape rhizosphere, can produce numerous antibiotics and siderophores, including pyoluteorin (Plt), 2,4-diacetylphloroglucinol (DAPG), pyrrolnitrin (Prn), orfamide, Pseudomonas fluorescens insecticidal toxin (Fit), hydrogen cyanide (HCN), pyoverdine, and pyochelin (Huang et al. 2017; Liu et al. 2018; Wang et al. 2017). Genomic analysis suggested that P. protegens H78 (GenBank No. CP013184) is highly homologous with P. protegens Pf-5 (Paulsen et al. 2005), CHA0 (Jousset et al. 2014), and Cab57 (Takeuchi et al. 2014). P. protegens species can produce a specific profile of antibiotics, including Plt, DAPG, and Prn (Huang et al. 2017; Jousset et al. 2014; Paulsen et al. 2005; Takeuchi et al. 2014). The Plt biosynthetic gene cluster has two types of evolutionary origins: the P. protegens type (Huang et al. 2017; Jousset et al. 2014; Paulsen et al. 2005; Takeuchi et al. 2014) and Pseudomonas aeruginosa type (M18 and LESB) (Winstanley et al. 2009; Wu et al. 2011). These two origins of plt clusters are identical in gene organization but moderately different in nucleotide and amino acid sequences. However, they are distinct from each other in the pltR-pltL noncoding sequences, aside from the lys box (Paulsen et al. 2005; Wang et al. 2017; Wu et al. 2011). Orfamide is a Pseudomonas-produced cyclic lipopeptide with biosurfactant, antimicrobial, and insecticidal activities (Gross et al. 2007; Loper et al. 2016). Fit is a large protein toxin biosynthesized by the fitABCDEFGH gene cluster in P. protegens CHA0 (Pechy-Tarr et al. 2008). In nature, these secondary metabolites contribute to the ecological and nutritional competition abilities of Pseudomonas spp.
Secondary metabolism, including antibiotic biosynthesis, is subject to the regulation of multiple global and pathway-specific regulatory systems or factors. Classically, the Gac/Rsm signal transduction regulatory system, which is sequentially composed of the two-component system (TCS) GacS/GacA, the RsmY family sRNAs, and the RsmA (Rsm, regulator of secondary metabolism) family RNA-binding proteins, plays a crucial role in the global activation of secondary metabolism in Pseudomonas spp. (Haas and Keel 2003). Gac/Rsm-driven global regulation is generally mediated by pathway-specific regulators, such as the LysR-type transcriptional activator PltR of the Plt biosynthetic operon (Huang et al. 2008). However, the environmental signal that stimulates the phosphorylation of the sensor kinase GacS remains unidentified. Such signalling factors or systems constitute another important array of regulators involved in the biocontrol or virulence of Pseudomonas spp. (Jimenez et al. 2012). A single mutation of any one of the three quorum sensing (QS) systems, including LasI/LasR, RhlI/RhlR, and Pqs/PqsR, causes an enormous increase in Plt biosynthesis in the rhizobacterium P. aeruginosa M18 (Chen et al. 2008; Lu et al. 2009; Yan et al. 2007). Las and Rhl belong to the LuxI/LuxR family QS system (Chen et al. 2008; Yan et al. 2007). Another Lux-type regulatory system, PhzI/PhzR, activates phz operon expression and phenazine biosynthesis in P. chlororaphis (Chancey et al. 1999). Moreover, many nucleotide second messengers, including c-di-GMP, c-di-AMP, cGMP, cAMP, and guanosine pentaphosphate or tetraphosphate ((p)ppGpp), are involved in the global regulation of bacterial cell biology (Jimenez et al. 2012).
(p)ppGpp is the modulator of the stringent response of bacteria to cope with nutritional starvation and other stresses. The RelA/SpoT homologue (RSH) enzymes are responsible for (p)ppGpp biosynthesis and hydrolysis. Two RSH enzymes, RelA and SpoT, exist in Escherichia coli and other β- or γ-proteobacteria (Hauryliuk et al. 2015; Steinchen and Bange 2016). The RelA protein has only synthetase activity and responds to amino acid and fatty acid starvation. SpoT is a bifunctional enzyme with both synthetase and hydrolase activities that is crucial for (p)ppGpp homeostasis. SpoT poorly responds to various stresses, such as fatty acids, carbon sources, phosphate or iron starvation, diauxic shifts, oxidative stress, and hyperosmotic shock (Hauryliuk et al. 2015; Sinha et al. 2019; Steinchen and Bange 2016).
An increasing number of studies have shown that (p)ppGpp globally regulates various aspects of bacterial physiology and metabolism: DNA replication, cell division, DNA transcription, growth, survival, adaption, competence, secondary metabolism, cell motility, biofilm formation, and pathogenicity. Bacterial RNA polymerase (RNAP) has been confirmed to be the first direct target of (p)ppGpp. rRNA transcription and ribosome biogenesis in starving bacteria are thus shut down to conserve energy and survive in adverse conditions (Atkinson et al. 2011; Dalebroux and Swanson 2012; Hauryliuk et al. 2015; Potrykus and Cashel 2008; Steinchen and Bange 2016).
As in E. coli, (p)ppGpp biosynthesis is also catalysed by RelA and SpoT in Pseudomonas spp. The (p)ppGpp-mediated stringent response is involved in regulating virulence or biocontrol in Pseudomonas spp. (Chatnaparat et al. 2015; Manuel et al. 2011, 2012). (p)ppGpp functions as a global signal for activating the virulence of Pseudomonas syringae (Chatnaparat et al. 2015). Moreover, in P. protegens CHA0, (p)ppGpp is required for sustaining biocontrol ability (Takeuchi et al. 2012). The biosynthesis of antibiotics, including DAPG and Plt, is diminished in the relA/spoT double mutant, in which (p)ppGpp biosynthesis is entirely abolished. The relA/spoT double mutation significantly decreases the expression levels of RsmXYZ sRNAs and the gacS/gacA TCS in CHA0; this effect correlates the (p)ppGpp-mediated stringent response with the Gac/Rsm cascade (Takeuchi et al. 2012). However, in two other Pseudomonas strains, the (p)ppGpp-mediated stringent response represses the antifungal activity mediated by the biosynthesis of antibiotics and extracellular enzymes (Manuel et al. 2011, 2012). The relA/spoT mutant of P. chlororaphis PA23 shows increased production of Prn, lipase, and protease, but phenazine production is unchanged (Manuel et al. 2012). Similarly, in Pseudomonas sp. strain DF41, the relA/spoT mutation significantly enhances lipopeptide production and protease activity, thereby contributing to the biocontrol ability of the DF41 strain (Manuel et al. 2011). Therefore, (p)ppGpp-mediated regulation shows a certain level of specificity among various strains. Moreover, no genome-wide transcriptomic or proteomic profile has been reported for stringent response mutants in Pseudomonas spp.
In this study, we compared the transcriptomic profile of the relA/spoT mutant with that of the wild-type strain H78 using RNA sequencing (RNA-seq) after growing the strains to mid- to late log phase (optical density (OD600) = 5.0–6.0) in King’s medium B (KMB) at 28 °C. During this period, P. protegens H78 was still at the stage of rapid growth in the KMB medium with glycerol as carbon source and then it can grow to a maximum OD600 of 14 or above. In addition, systematic phenotype analysis was conducted to assess the diversity and intensity of relA/spoT-mediated regulatory effects on the physiology and metabolism of P. protegens H78. The (p)ppGpp-mediated stringent response extensively inhibited primary metabolism, including DNA replication; bacterial growth; core transcriptional and translational components; amino acid transport and metabolism; carbohydrate and energy metabolism; and transport and utilization of phosphate, sulfate, and nitrogen. However, the stringent response plays a crucial role in maintaining cell survival and globally activating secondary metabolism, including the biosynthesis of antibiotics, iron carriers, and polysaccharides.
Materials and methods
Bacterial strains, plasmids, primers, and culture conditions
Bacterial strains, plasmids, and primers are listed in Supplemental Tables S1 and S2. E. coli was conventionally cultivated in Luria-Bertani (LB) medium at 37 °C. The wild-type strain of P. protegens H78 has been deposited in the China General Microbiological Culture Collection Center (CGMCC 15755). P. protegens H78 and its derivative strains were grown at 28 °C in King’s medium B (KMB) (King et al. 1954), which is composed of 20 g tryptone, 15 ml glycerol, 0.514 g K2HPO4·3H2O, and 0.732 g MgSO4 per litre. Kanamycin and tetracycline were used at working concentrations of 50 and 30 μg ml−1 for P. protegens H78 and 50 and 15 μg ml−1 for E. coli, respectively. In addition, 4 mg l−1 ortho-nitrophenyl-β-d-galactopyranoside (ONPG) in phosphate buffer (pH 7.0) was used to quantify β-galactosidase activity.
DNA manipulation
DNA cloning and recombination were performed following standard methods (Sambrook and Russell 2001). KOD (Toyobo, Osaka, Japan), LA Taq, Taq DNA polymerases (TaKaRa, Kusatsu, Japan), DNA restriction enzymes (NEB, Ipswich, MA), DNA ligase (TaKaRa, Kusatsu, Japan), DNA markers (MBI Fermentas affiliated to Thermo Scientific, Waltham, MA), and other related enzymes were used according to the instructions for the reagents. Genomic DNA from P. protegens H78, plasmid DNA from Pseudomonas or E. coli, and DNA fragments from the gel were purified using the corresponding kits from TaKaRa (Kusatsu, Japan). DNA synthesis and sequencing were performed by Sangon (Shanghai, China) or HuaDa Corporation (Shenzhen, China).
In-frame deletion and heterogeneous complementation of relA and spoT in P. protegens H78
In vitro in-frame knockout and in vivo homologous recombination were utilized to delete relA and spoT in the P. protegens H78 genome according to the construct map displayed in Supplemental Fig. S1. Two flanking fragments (553 and 519 bp) of the relA ORF were individually amplified using the KOD Plus DNA polymerase with two pairs of primers, relA-F1/relA-R1 and relA-F2/relA-R2 (Supplemental Table S2). The respective 3′ and 5′ ends of upstream and downstream fragments share a 15-bp complementary region. The upstream and downstream fragments were mixed as templates in the subsequent fusion PCR with the primers relA-F1 and relA-R2. The fusion PCR product was inserted into the KpnI-HindIII sites of the suicide vector pK18mobsacB. Afterward, the resultant vector, pK18-relA, was transferred from E. coli S17 into P. protegens H78 via biparental hybridization. The single-crossover mutant was first screened on an LB plate with 50 μg ml−1 kanamycin (Km). Then, the double-crossover mutant, which is sensitive to Km and resistant to sucrose, was screened in parallel between an antibiotic-free plate containing 15% sucrose and a kanamycin-containing plate. The resultant relA-deleted mutant was further confirmed using PCR and sequencing. Similarly, the spoT gene was deleted in the relA mutant background (Supplemental Fig. S1; Supplemental Table S1). The transcriptomic results further confirmed that relA and spoT have been successfully deleted in the relA/spoT mutant. The transcripts of these two genes were not detected in the relA/spoT mutant. However, the transcripts of relA and spoT have mean FPKM values of 1980.0 and 3249.6, respectively, in wild-type H78 (Supplemental Table S3).
relA (H78_04755) and spoT (H78_06273) encode the (p)ppGpp synthetase and the bifunctional (p)ppGpp synthetase/hydrolase, respectively. A fragment (2244 bp) that carries the entire ORF and upstream promoter/operator region of the relA gene was amplified and cloned into the KpnI/SacI-digested pME6032 vector. The resulting vector, p6032-relA, was used to complement the relA/spoT mutant of P. protegens H78.
RNA isolation, RNA-seq, and data analysis
P. protegens H78 and its relA/spoT mutant were cultivated to mid- to late log phase (OD600 = 5.0–6.0) in KMB at 28 °C for RNA sampling; each strain was repeated three times. RNA sequencing was performed in triplicate for each strain. RNA isolation and sequencing were carried out by Shanghai Bohao Biotechnology Company (Shanghai, China). Total RNA was extracted using the Qiagen (Dusseldorf, Germany) RNeasy Mini Kit (Cat. 4106), quantified by a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Waltham, MA), and assessed for quality by the Agilent (California, USA) BioAnalyzer 2100. The isolated total RNA was further purified using the Qiagen (Dusseldorf, Germany) RNeasy micro-kit (Cat. 74004) and the Qiagen (Dusseldorf, Germany) RNase-Free DNase Set (Cat. 79254). mRNA was enriched by removing rRNA and further fragmented to synthesize the first and second cDNA strands for constructing the sequencing library. Cluster generation and de novo sequencing were performed using the Illumina (San Diego, CA) Hiseq 2500.
Sequenced reads were trimmed for adaptor sequences and masked for low-complexity or low-quality sequences by Trimmomatic (v0.30) (Bolger et al. 2014). The quality of the sequenced data was assessed with FastQC (v0.10.1) (Wingett and Andrews 2018). Clean reads were mapped to the P. protegens H78 complete genome using Tophat (v2.0.9) (Trapnell et al. 2012). The expression abundance of genes was quantified by Htseq (v0.6.1) using the reads per kilobase per million mapped reads (RPKM) method (Mortazavi et al. 2008). Mean RPKM values were calculated for the P. protegens H78 relA/spoT mutant and the wild-type strain from their respective repeats. The RNA-seq data normalization and differential gene expression analysis between two groups (relA/spoT mutant and wild-type H78) were carried out using the DESeq2 normalization method (Love et al. 2014) on the bioconductor platform. The transcripts with a fold change cutoff of ≥ 2 (P value < 0.05) were considered to be differentially expressed. The differentially expressed genes were further analysed using the COG (Tatusov et al. 2000), GO (Camon et al. 2004) and KEGG pathway (Ogata et al. 1998) databases.
Construction of in-frame lacZ fusion plasmids
The pltL, phlA, prnA, hcnA, and pstS genes are, respectively, the first gene of the Plt biosynthetic operon pltLABCDEFG, the DAPG operon phlACBDE, the Prn operon prnABCD, the HCN operon hcnABC, and the phosphate transport operon pstSCAB. The in-frame fusions of these genes with lacZ were generated by cloning the first several codons and promoter/operator regions of each gene individually into pME6015. In the pME6015 plasmid, the E. coli lacZ gene lacks its own promoter/operator and the first seven codons (Supplemental Table S1). Moreover, these five in-frame lacZ fusion plasmids were individually introduced into P. protegens H78 and its relA/spoT mutant to reflect the total regulatory effect of relA/spoT on the corresponding operons and validate the transcriptomic data.
Assessment of bacterial growth with optical density and colony forming units
The growth of P. protegens H78 and its relA/spoT mutant was assayed at 28 °C in KMB, LB, and M9 media. In the M9 medium, 5 g l−1 glycerol was added as the sole carbon source. The total biomass (optical density (OD600)) and viable cell number (colony forming units (CFU)) of bacterial cultures were measured at different time points.
In this study, the number of viable cells in cultures of P. protegens H78 and its derivative strains was assessed using CFU per millilitre. At the selected time points, 1 ml of cell suspension was sampled and diluted by serial 10-fold (10−1) dilutions. Then, 50 μl of sample from each dilution gradient was spread on LB agar plates. After overnight culture, colony counting was carried out on the plate in which a sufficient number of colonies were isolated well enough to ensure reliable counting.
Quantification of antibiotic production and β-galactosidase expression
Fresh overnight cultures of H78 and its derivatives were inoculated into 100 ml KMB (for Plt production) in a 500-ml Erlenmeyer flask, with a final OD600 of 0.05, and cultivated at 28 °C and 200 rpm. Plt was extracted using ethyl acetate and DAPG with trifluoroacetic acid (TFA) and ethyl acetate (v/v = 1:9). HPLC quantification of Plt and DAPG production was performed according to our method (Huang et al. 2004) and that of Zhang et al. (2014), respectively.
P. protegens H78 and its derivatives carrying the in-frame lacZ fusion vectors were inoculated and cultured under the same conditions used for antibiotic fermentation. The cultures were sampled at different time points, and β-galactosidase activity was quantified by referring to the Miller method (Sambrook and Russell 2001) and our previous description (Huang et al. 2004).
Agar plate assay for siderophore production
The chrome azurol S (CAS) method (Schwyn and Neilands 1987) was utilized to assess siderophore production as previously described (Wei et al. 2013). Approximately 5 μl of diluted overnight cultures (OD600 = 0.1) of P. protegens H78, the relA/spoT mutant and its complemented strain were separately inoculated at the centre of the CAS plate and grown at 28 °C for 56 h. The size of the fluorescent orange zone formed on the CAS plate indicated the siderophore production level.
Assessment of iron, carbohydrate, phosphorus, cysteine, and urea utilization
The iron-utilization capacity of P. protegens H78 and its relA/spoT mutant was assessed in iron-deficient succinate minimal medium (5.41 g glycerol, 9.13 g K2HPO4, 2.99 g KH2PO4, 0.92 g (NH4)2SO4, 0.06 g MgSO4, 5.4 g sodium succinate, pH 8.0, and 1 l deionized H2O) (Zolotarev et al. 2008), which was supplemented without or with (i) 100 μM FeCl3, (ii) 3.3 μM haemin and 150 μg ml−1 EDDHA (ethylenediamine-di(o-hydroxyphenylacetic) acid), or (iii) 3.0 μM haemoglobin and 150 μg ml−1 EDDHA (Zhao and Poole 2000).
To evaluate the influence of relA/spoT on the carbohydrate utilization of P. protegens H78, succinate (3 g l−1), mannitol (5 g l−1), or phenethylamine (1 ml l−1) was chosen as the sole carbon source in the M9 medium (17.17 g Na2HPO4·12H2O, 2.99 g KH2PO4, 0.58 g NaCl, 5.95 g (NH4)2SO4, 0.246 g MgSO4·7H2O, 0.01 mg vitamin B1, 4.3 mg CaCl2, 16.7 mg FeCl3·6H2O, 1 mg MnCl2·4H2O, 1.7 mg ZnCl2, 0.43 mg CuCl2·2H2O, 0.6 mg CoCl2·6H2O, 0.6 mg H4MoNa2O6, and 1 l H2O).
For the phosphate utilization assay, 2 mM KH2PO4, as the sole phosphate source, was added to the medium (5 g MgCl2, 0.25 g MgSO4·7 H2O, 0.2 g KCl, 1 g (NH4)2SO4, 1.0 g glycerol, and 1 l H2O, pH 7.0). To assess the ability of P. protegens H78 to utilize cysteine as the sole sulphur source, 0.5 mM cysteine was added into the M9 medium supplemented with 5 g l−1 glucose. Similarly, the ability of P. protegens H78 to utilize urea as the sole nitrogen source was assayed in M9 medium supplemented with 20 mM urea and 5 g l−1 glucose and containing no NH4Cl.
In the above media, overnight cultures of H78 and its relA/spoT mutant were inoculated at a final concentration of OD600 = 0.03 and cultured at 28 °C and 200 rpm. Bacterial growth was measured over time.
Assay for antibiotic resistance in P. protegens H78
Three antibiotics, including gentamicin, kanamycin, and tetracycline, were selected to determine the influence of relA/spoT on the antibiotic resistance of P. protegens H78. The wild-type strain H78 and its relA/spoT mutant were grown in LB medium containing increasing antibiotic concentrations. After 12 h of shaking culture at 28 °C, the OD600 and CFU counts were monitored.
Statistical analysis
In this study, every experiment was separately carried out more than three times. In each experiment, at least triplicate parallel samples were included. Each value was described as the average ± SD of three repeats. Two groups of data were compared by Student’s t test in Excel (Microsoft, Washington, USA). When the P value was < 0.05, the results were considered statistically significant.
Accession number of nucleic acid sequences and RNA-seq data
In this study, the DNA sequences of relA, spoT, and other related genes were extracted from the P. protegens H78 complete genome (GenBank accession No. CP013184). The raw reads and analysed data from RNA-seq have been deposited in the Sequence Read Archive database (Accession No. SRP091880) and the Gene Expression Omnibus database (Accession No. GSE89004) in NCBI.
Results
Effects of relA/spoT deletion on the transcriptomic profile of P. protegens H78
Whole-genome sequencing analysis of P. protegens H78 showed that two (p)ppGpp biosynthesis- and hydrolysis-related genes, relA and spoT, are single-copy genes, and no other homologous genes exist in the H78 genome (GenBank No. CP013184) (Huang et al. 2017). The relA/spoT deletion mutant of P. protegens H78 was constructed by homologous recombination (Supplemental Fig. S1) and validated by PCR and sequencing. The relA/spoT mutant exhibited an almost complete disappearance of the biosynthesis of Plt, an antibiotic we studied extensively. This outcome encouraged us to use RNA-seq to investigate the influence of the RelA/SpoT-mediated stringent response on genome-scale transcriptional expression in P. protegens H78. Transcriptomic profiles were compared between H78 and its relA/spoT mutant, and the strains were cultured until mid- to late log phase at an OD600 of 5.0–6.0 in KMB at 28 °C. The RNA-seq data was validated using quantitative real-time PCR (qRT-PCR) and lacZ reporter analyses (Supplemental Fig. S2).
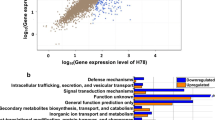
As shown in Supplemental Tables S3 and S4, the transcript abundance of 1463 genes, which accounted for nearly 23% of all annotated genes in the H78 genome, was significantly modified by more than 2-fold in the relA/spoT mutant compared with that in the wild-type strain H78. Among the relA/spoT regulons, 767 genes were downregulated, and 696 genes were upregulated. The functional categories of the relA/spoT regulons were plotted relative to downregulation and upregulation according to PseudoCAP (Fig. 1a). The relA/spoT-inhibited genes were involved in DNA replication and cell division, amino acid metabolism, energy and coenzyme metabolism, translational and posttranslational modification, and cell motility. However, the (p)ppGpp-mediated stringent response predominantly activated the expression of genes related to secondary metabolism, cell envelope biogenesis, transcriptional factors, and regulators (Fig. 1a).
Transcriptomic profiles of P. protegens H78 and its relA/spoT mutant. a Functional classification of the relA/spoT regulon (fold change ≥ 2; P value < 0.05). b Scatter plot comparing the transcriptomes of P. protegens H78 and its relA/spoT mutant. The x-axis indicates gene number in the H78 genome; the y-axis indicates the log2 fold change of the transcript abundance of each gene in the relA/spoT mutant relative to its parental strain H78. Significantly regulated gene clusters with known functions are marked
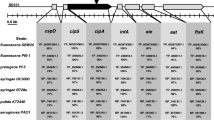
Most of the operons or gene clusters under the control of relA/spoT are indicated in Fig. 1b. The biosynthetic gene clusters of secondary metabolites, including Plt (plt), DAPG (phl), Prn (prn), pyochelin (pch), pyoverdine (pvd), haemophore (has), Fit (fit), and orfamide (ofa), were globally upregulated by RelA/SpoT. The negative regulons of relA/spoT included numerous primary metabolism-related gene clusters, such as DNA replication- and cell division-related gene clusters (dnaK-dnaJ-grpE and mur-fts-mra); energy and carbohydrate metabolic gene clusters (pnt, atp, ped, and mtl); amino acid metabolic gene clusters (met, cys, leu, arn, and glt); nitrogen, phosphate and sulphur transport and utilization gene clusters (ure-urt, phoR/phoB-pst, and cys); membrane transport and secretion gene clusters (tonB-exbD-exbB4–6 and gsp); and a flagellum and chemotaxis gene cluster (flg-fli-che). Therefore, the RelA/SpoT-mediated stringent response mainly inhibits primary metabolism and activates secondary metabolism in P. protegens H78.
The relA/spoT mutant exhibits upregulated expression of the dnaK-dnaJ-grpE and fts-mur-mra clusters
Transcriptomic profiling showed that the expression levels of two chaperone systems, DnaK-DnaJ-GrpE and GroEL/ES, which are involved in protein quality control and DNA replication (Calloni et al. 2012; Konieczny and Zylicz 1999), were significantly upregulated in the relA/spoT mutant compared with those in the parental strain H78 (Fig. 2a). Another gene cluster involved in cell division and peptidoglycan biosynthesis, fts-mur-mra, showed a more than 2-fold increase in mRNA levels in the relA/spoT mutant (Fig. 2b). The negative regulatory effect of relA/spoT on fts-mur-mra expression was further confirmed by qPCR with the method and primers described in Supplemental experimental procedure and Table S2. The results indicated that three selected genes (ftsQ, murF, and mraZ) in this gene cluster were significantly upregulated at the transcriptional level by the relA/spoT mutation (Fig. 2c).
a, b Negative regulatory effect of relA/spoT on the expression of the grpE-dnaK-dnaJ and groL/groS chaperone gene clusters (a) and the cell division-related fts-mur-mra gene cluster (b). The fold change of the transcript abundance in the relA/spoT mutant relative to wild-type H78 is shown at the top of the arrows. c The relative fold change in the mRNA levels (quantified by qPCR) of three genes (ftsQ, murF and mraZ) in the fts-mur-mra cluster in the relA/spoT mutant relative to the wild-type H78
The well-known cell division gene ftsZ showed a 2.3-fold increase at the transcriptional level in the relA/spoT mutant relative to the wild-type H78 (Fig. 2b). The negative targets of relA/spoT include other genes that are involved in DNA replication and cell division, such as the DNA gyrase subunit B-encoding gene gyrB, the single-stranded DNA-binding protein-encoding gene ssb, the HU family gene hupA, and the cell division gene ftsX. Moreover, the RelA/SpoT-mediated stringent response participates in the regulation of DNA repair. The DNA repair gene recO was upregulated by 2.8-fold in the relA/spoT mutant. Most significantly, the H78_01844 gene encoding the helicase subunit of the DNA excision repair complex was dramatically upregulated by 219.1-fold after relA/spoT mutation (Supplemental Table S4).
The relA/spoT deletion partially increases bacterial growth and significantly reduces the cell survival rate of the H78 strain
The involvement of RelA/SpoT in DNA replication and cell division brings into question about the influence of RelA/SpoT on bacterial growth and cell survival in P. protegens H78. The OD600 and viable cell number (CFU ml−1) of H78 and its relA/spoT mutant were monitored in three media at 28 °C: KMB (the medium used for RNA-seq analysis), LB, and M9. As expected, the growth of the relA/spoT mutant was moderately upregulated compared with that of its parental strain H78 in the three media (Fig. 3a, c, and e). However, the number of live cells was significantly reduced in the relA/spoT mutant relative to the wild-type strain H78 (Fig. 3b, d, and f). These results suggested that the (p)ppGpp-mediated stringent response plays an important role in reducing bacterial growth and increasing the cell survival rate in P. protegens H78.
Influence of relA/spoT on bacterial growth and cell survival in P. protegens H78. Bacterial growth (optical density (OD600)) and viable cell number (colony forming units per millitre (CFU ml−1)) of P. protegens H78 and its relA/spoT mutant, which were inoculated at an OD of 0.03 and were measured at 28 °C in three media: LB (a, b), KMB (c, d), and M9 (e, f). In the M9 medium, 5 g l−1 glycerol was added as the sole carbon source
Negative targets of RelA/SpoT include some core transcriptional and translational elements and are involved in amino acid metabolism
In the transcriptomic profiles of H78 and its relA/spoT mutant, relA/spoT negatively regulated some general bacterial transcriptional and translational factors. Two genes, rpoB and rpoC, which respectively encode the core RNAP subunits ß and ß′, were significantly upregulated by 2.2-fold in the relA/spoT mutant compared with H78. Similarly, relA/spoT inhibited the expression of genes encoding general translational factors, such as the 50S ribosomal proteins L17 (H78_05744, rplQ), L25 (H78_05366), and L31 (H78_00458, rpmE); the 30S ribosomal protein S20 (H78_05530, rpsT); 16S rRNA methyltransferases (H78_00022, rsmB, and H78_00436, rsmE); the translational elongation factors Ts (H78_01218, tsf) and TU (H78_05788, tuf); an aminoacyl-tRNA hydrolase (H78_05367, pth); and a methionyl-tRNA formyltransferase (H78_00023, fmt) (Supplemental Table S4). These data further confirmed that the (p)ppGpp-mediated stringent response can regulate DNA replication, transcription, and translation. Among the relA/spoT regulons related to amino acid transport and metabolism, 103 genes were significantly upregulated by at least 2-fold in the relA/spoT mutant compared with H78 (Fig. 1a). These genes that are negatively regulated by RelA/SpoT were mainly involved in the transport and metabolism of amino acids, including arginine, histidine, glutamate, leucine, methionine, and tryptophan.
The RelA/SpoT-mediated stringent response globally activates antibiotic biosynthesis and gene expression
The results from RNA-seq revealed that the most significant downregulation caused by the relA/spoT mutation was observed for the Plt gene cluster. The transcript abundance of all 14 genes in the Plt biosynthesis (pltLABCDEFG) and transport operons (pltIJKNOP) decreased by 11.7- to 78.2-fold in the relA/spoT mutant compared with that in the wild-type H78 strain. The other two regulators, the LysR-type transcriptional activator PltR and the TetR-type transcriptional regulator PltZ, were respectively downregulated by 3.2- and 6.9-fold at the transcript level in the relA/spoT mutant (Fig. 4a). Similarly, three gene clusters that are respectively responsible for the biosynthesis of pyrrolnitrin (prn), orfamide (ofa), and P. fluorescens insecticidal toxin (fit) were significantly downregulated by more than 2-fold in the relA/spoT mutant (Fig. 4c–e). Unexpectedly, no significant influence on the expression of the phlACBDE operon involved in DAPG biosynthesis was observed (Fig. 4b). However, expression of the DAPG hydrolase-encoding gene phlG and the tetR family repressor gene phlF was reduced by 20.2- and 2.2-fold, respectively, at the transcript level in the relA/spoT mutant compared with the H78 strain (Fig. 4b).
Positive regulatory effect of RelA/SpoT on antibiotic gene expression and biosynthesis in P. protegens H78. a–e Fold change in the transcript levels of antibiotic biosynthetic gene clusters, including plt (a), phl (b), prn (c), ofa (d), and fit (e), in the relA/spoT mutant relative to the wild-type H78. f–h Bacterial growth (open symbols) and β-galactosidase expression (solid symbols) of P. protegens H78 and the relA/spoT mutant, which carry the pltL'-'lacZ (f), phlA'-'lacZ (g), or prnA'-'lacZ (h) in-frame fusion in pME6015, were assayed in KMB at 28 °C. i, j Plt (i) and DAPG production (j) in the H78 strain, the relA/spoT mutant and their derivative strains harbouring the empty plasmid pME6032 or the relA expression plasmid p6032-relA were quantified in KMB at 28 °C. *P ˂ 0.05, significant difference
Three in-frame lacZ fusions, pltL′-′lacZ, phlA′-′lacZ, and prnA′-′lacZ, were constructed to reflect the total regulatory activities of the pltLABCDEFG, phlACBDE, and prnABCD operons, respectively, at both the transcriptional and posttranscriptional levels. ß-Galactosidase expression from these three lacZ fusions in pME6015 was comparatively measured between H78 and its relA/spoT mutant grown in KMB at 28 °C. Expression of the pltL'-'lacZ and prnA'-'lacZ fusions was almost completely inhibited by the relA/spoT deletion (Fig. 4f, h). These results suggested that the (p)ppGpp-mediated stringent response globally activates the expression of the pltLABCDEFG and prnABCD operons. In addition, expression of the phlA'-'lacZ fusion was greatly downregulated in the relA/spoT mutant compared with that in H78 (Fig. 4g). This result seemed to contradict the above RNA-seq data, wherein the transcript abundance of the phlACBDE operon was not significantly influenced by the relA/spoT deletion. This outcome might be caused by the indirect regulation of the phlACBDE operon by relA/spoT at the posttranscriptional level.
Plt and DAPG production in H78 and its derivative strains were measured in KMB at 28 °C, and the results are described in Fig. 4i, j. The relA/spoT mutation caused complete inhibition of Plt production in the H78 strain. Moreover, the Plt production of the relA/spoT mutant could be recovered and even exceed the wild-type level due to the heterologous expression of the relA gene in pME6032. Here, the introduction of the empty plasmid pME6032 as a control caused a small increase in Plt production in the wild-type H78 strain (Fig. 4i). Given the relA/spoT mutation, DAPG production was greatly reduced in H78. Moreover, the inhibition of DAPG production in the relA/spoT mutant could be reversed by introducing the relA gene in pME6032 (Fig. 4j). These results suggested that the (p)ppGpp-mediated stringent response plays a key role in globally activating antibiotic biosynthesis in P. protegens H78.
Positive regulatory effects of the RelA/SpoT-mediated stringent response system on siderophore biosynthetic gene expression, siderophore production, and iron utilization
P. protegens H78 can produce two siderophores (pyochelin and pyoverdine) and one haemophore. The transcriptomic profiles of H78 and its relA/spoT mutant revealed that the most significant reduction of transcript levels occurred for the pyochelin biosynthetic gene cluster (pchDHIEFKCBA) in the relA/spoT mutant. The transcript abundance of all genes within the pch cluster decreased more than 8-fold in the relA/spoT mutant compared with that in the wild-type H78 (Fig. 5a). The biosynthetic gene clusters of pyoverdine (pvd) and haemophore (has) were moderately downregulated by the relA/spoT mutation at the transcript level (Fig. 5b, c). Interestingly, ten pairs of FecI/FecR (sigma factor/regulator) related to iron uptake were positively regulated by relA/spoT (Supplemental Table S5).
Positive regulatory effect of relA/spoT on iron carrier biosynthesis and iron utilization in P. protegens H78. a–c Fold change in the transcript levels of iron carrier biosynthetic gene clusters, including pch (a), pvd (b), and has (c), in the relA/spoT mutant relative to the wild-type H78. d Siderophore (pyochelin and pyoverdine) production in H78, relA/spoT mutant and its derivative strain the relA expression plasmid was assayed in a CAS plate. e The fluorescent halo diameter (cm) from in H78, relA/spoT mutant and its derivative strain the relA expression plasmid. Every value represents mean ± SD. *P ˂ 0.05, significant difference. The experiment was repeated independently three times with similar results. f–i Cell growth of P. protegens H78 and its relA/spoT mutant at 28 °C in iron-deficient succinate minimal medium supplemented without (f) or with (1) 100 μM FeCl3 (g), (2) 3.3 μM haemin and 150 μg ml−1 EDDHA (h), and (3) 3.0 μM haemoglobin and 150 μg ml−1 EDDHA (i)
The positive regulatory effect of relA/spoT on the abovementioned iron carrier gene clusters and iron utilization activators implies that iron utilization may be positively controlled by the (p)ppGpp-mediated stringent response system. Siderophore production by P. protegens H78, the relA/spoT mutant, and complemented strains was measured on a solid plate using the chrome azurol S method (Fig. 5d, e). Deficiency of relA/spoT reduced the size of the fluorescent circle, indicating reduced level of siderophore production. Siderophore production in the relA/spoT mutant can be restored to the wild-type level by the exogenous expression of the (p)ppGpp synthetase gene relA in pME6032 (Fig. 5d, e). To further assess the influence of relA/spoT on the iron utilization ability of P. protegens H78, the wild-type H78 strain and its relA/spoT mutant were cultivated in iron-deficient succinate minimal medium with FeCl3, haemin, or haemoglobin as the sole source of iron (Fig. 5f–i). Growth of the H78 strain was significantly inhibited by the deficiency of relA/spoT in all three tested media that were individually supplemented with three different iron sources. Moreover, bacterial growth of the relA/spoT mutant rapidly declined after 21 h. However, the wild-type strain H78 maintained a steady growth rate (Fig. 5g–i). These results indicated that the (p)ppGpp-mediated stringent response positively regulates iron carrier production and iron utilization in P. protegens H78.
The RelA/SpoT regulon is involved in carbon and energy metabolism
Four gene clusters related to carbohydrate metabolism, ped (2-phenylethanol catabolism), mtl (mannitol transport and utilization), suc (succinate metabolism), and iol (inositol metabolism), were greatly upregulated at the transcript level because of relA/spoT deletion (Supplemental Fig. S3a–d). Similarly, three energy metabolic gene clusters, atpBEFHAGDC, pntAB, and cyoABCDE, which are individually responsible for the biosynthesis of ATP, NAD(P)H, and cytochrome O ubiquinol oxidase, were also remarkably inhibited by relA/spoT in H78. The relA/spoT mutation significantly increased the mRNA abundance of all genes in the atp, pnt, and cyo operons (Supplemental Fig. S3e–g). These results implied that the (p)ppGpp-mediated stringent response predominantly plays a negative regulatory role in carbohydrate and energy metabolism in P. protegens H78.
The RelA/SpoT-mediated stringent response regulates the ability of P. protegens H78 to utilize phenylethanol, mannitol, or succinate as the sole carbon source
Based on the negative regulatory effect of relA/spoT on the mtl, suc, and ped gene clusters, we predicted that relA/spoT may similarly inhibit the ability of P. protegens H78 to utilize mannitol, succinate, or phenylethanol as a carbon source. Correspondingly, we compared the bacterial growth measured for the H78 strain and its relA/spoT mutant in M9 medium supplemented with 5 g l−1 mannitol, 3 g l−1 succinate, or 1 ml l−1 phenylethanol as the sole carbon source. As shown in Fig. 6a, the growth of the relA/spoT mutant was strongly inhibited until 50 h in M9 medium with mannitol as the sole carbon source. However, after 60 h, the relA/spoT mutant became significantly superior to the wild-type H78 strain in bacterial growth (Fig. 6a). When succinate was used as the sole carbon source, an obvious upregulation of growth was observed in the relA/spoT mutant (Fig. 6b). In addition, in the M9 medium with phenylethanol as the sole carbon source, bacterial growth of the relA/spoT mutant was almost completely inhibited until 96 h. However, the growth of the relA/spoT mutant unexpectedly increased after 100 h of culture (Fig. 6c). In contrast, the growth of the wild-type H78 strain was steady after peaking at 24 h and began decreasing after 96 h (Fig. 6c). These results suggested that the utilization of different carbon sources is differentially controlled by the (p)ppGpp-mediated stringent response in P. protegens H78.
The relA/spoT mutant exhibits upregulated expression of multiple ion transport and protein secretion operons
(i) Phosphate (pst), cysteine (cys), and urea (urt) transport operons
The transcriptomic profiles of P. protegens H78 and the relA/spoT mutant demonstrated that the mRNA levels of the phosphate-specific transport operon pstSCAB-phoU and the PhoR/PhoB TCS genes were enhanced by more than 25-fold in the relA/spoT mutant compared with those in the H78 strain (Supplemental Fig. S4a). The cysTWA ABC transporter operon and other related genes (cysDN and cysI), which are involved in the transport and metabolism of cysteine as a sulphur source, were significantly upregulated owing to the relA/spoT mutation (Supplemental Fig. S4b). Similarly, a strong inhibitory effect of relA/spoT on the urtABCDE operon involved in urea transport and utilization was observed (Supplemental Fig. S4c). In addition, some urease metabolic genes were also negatively regulated by relA/spoT in H78 (Supplemental Fig. S4c).
(ii) Three TonB-dependent transport systems: ExbB/ExbD/TonB4–6
Like most bacteria, P. protegens H78 possesses multiple TonB-dependent transporters, ExbB/ExbD/TonBs, which are involved in the binding and transport of siderophores, vitamin B12, nickel complexes, and carbohydrates (Noinaj et al. 2010). Transcriptomic data showed that the mRNA levels of the fourth to sixth TonB transporter gene clusters exbB/exbD/tonB4–6 and their individual flanking genes were much higher in the relA/spoT mutant than in the parental strain H78 (Supplemental Fig. S4d).
(iii) Bacterial secretion system Gsp (general secretion pathway)-T2SS
In gram-negative bacteria, a large number of secretion systems, including types I to VI secretion systems (T1SS to T6SS), deliver proteins out of the cells. The secreted proteins include toxins, virulence or biocontrol factors, cytochromes, and exoenzymes (Abby et al. 2016). In P. protegens H78, the expression of Gsp-T2SS gene cluster was significantly enhanced by the relA/spoT deletion (Supplemental Fig. S4e). In addition, five genes (impB2, impC2, hcp, tssE, and clpV) within the gene cluster (H78_06283 to H78_06304), which is homologous to P. aeruginosa H1-T6SS (Wei et al. 2013), were moderately inhibited by RelA/SpoT (Supplemental Table S4).
Regulatory effect of relA/spoT mutation on phosphate, cysteine, and urea utilization
The repressive activity of relA/spoT on the pst, cys, and urt operons led us to further assess the influence of relA/spoT on the ability of P. protegens H78 to take up and utilize phosphate, cysteine, and urea. H78 and its relA/spoT mutant strains were grown at 28 °C in minimal medium with KH2PO4, cysteine, and urea as the sole sources of phosphate, sulphur, and nitrogen, respectively. Bacterial growth was monitored at different time points, and the results were displayed in Fig. 7. In the minimal medium with 2 mM KH2PO4, the bacterial growth of the relA/spoT mutant was superior to that of the wild-type H78 after 12 h of culture (Fig. 7a), demonstrating that the (p)ppGpp-mediated stringent response moderately inhibits phosphate utilization in P. protegens H78.
Regulatory effect of relA/spoT on phosphate, cysteine, and urea utilization. Cell growth of P. protegens H78 and the relA/spoT mutant was monitored at 28 °C in minimal medium with the addition of KH2PO4 (a), cysteine (b), and urea (c) as the sole sources of phosphor, sulphur and nitrogen, respectively
In the M9 medium with 0.5 mM cysteine as the sole source of sulphur, the relA/spoT mutant showed obviously inferior bacterial growth compared with the H78 strain until 60 h (Fig. 7b). This result seemed to be inconsistent with the repressive effect of relA/spoT on the cysteine transport operon cysTWA. This may be because cysteine utilization have been influenced by other operons in addition to cysTWA. In addition, in the minimal medium supplemented with 20 mM urea as the sole source of nitrogen, the bacterial growth of the relA/spoT mutant was significantly inhibited compared with that of the wild-type H78 strain before 30 h of culture. Conversely, the relA/spoT mutant showed obvious superiority to its parental strain H78 in terms of growth after 36 h of culture (Fig. 7c).
Reduction of antibiotic resistance by the relA/spoT mutation
Three common antibiotics, gentamicin, kanamycin, and tetracycline, were chosen to compare the antibiotic resistance levels of P. protegens H78 and its relA/spoT mutant. Bacterial growth was monitored in LB with or without increasing concentrations of antibiotics, and the results are summarized in Fig. 8. Without antibiotics in LB, the bacterial growth of the relA/spoT mutant was significantly superior to that of the wild-type strain H78. However, when grown in LB medium with increasing concentrations (2–12 μg ml−1) of gentamicin, kanamycin, or tetracycline, the bacterial growth of the relA/spoT mutant was significantly inhibited compared with that of H78 (Fig. 8a–c). The results from the CFU assay indicated that the number of live cells was reduced more significantly (relative to OD600) by the relA/spoT mutation in the medium with the adopted concentration of antibiotics (Fig. 8d). These results confirmed that antibiotic resistance is positively regulated by the (p)ppGpp-mediated stringent response in P. protegens H78.
Effect of relA/spoT on antibiotic resistance in P. protegens H78. a–c The bacterial growth (OD600) of P. protegens H78 and the relA/spoT mutant was assessed after 12 h of culture at 28 °C in LB medium with increasing concentrations (0 to 12 μg ml−1) of gentamicin (a), kanamycin (b), or tetracycline (c). d Viable counts (CFU ml−1) of H78 and its relA/spoT mutant after 12 h of culture in LB medium supplemented with gentamicin (2 μg ml−1), kanamycin (2 μg ml−1), or tetracycline (4 μg ml−1). *P ˂ 0.05, significant difference
The RelA/SpoT regulon is involved in flagellum, pilus, and peptidoglycan biosynthesis and chemotaxis
As shown in Supplemental Fig. S5, three gene clusters required for flagellar biogenesis and assembly, H78_01720 to H78_01729, H78_01754 to H78_01762, and H78_04786 to H78_04789, were upregulated by approximately 2-fold at the transcript level in the relA/spoT mutant compared with the H78 strain (Supplemental Fig. S5a). Similarly, the relA/spoT mutation caused a 2-fold increase in the expression of the che chemotaxis operon (Supplemental Fig. S5b). These results suggested that relA/spoT may negatively control flagellar biosynthesis and chemotaxis. In contrast, two exopolysaccharide biosynthetic operons, psl and pel, were significantly downregulated by approximately 2-fold at the transcript level by the relA/spoT mutation (Supplemental Fig. S5c). Similarly, the type I chaperone-usher pilus biogenesis operon, csu, displayed a nearly 2-fold decrease at the transcript level in the relA/spoT mutant relative to the wild-type H78 (Supplemental Fig. S5d). These results implied that the (p)ppGpp-mediated stringent response is involved in flagellum and pilus biogenesis, exopolysaccharide production, biofilm formation, and motility in P. protegens H78.
The RelA/SpoT regulon includes multiple transcriptional factors/regulators and three sRNAs (RsmY, RsmZ, and CrcZ)
Transcriptomic data showed that in the relA/spoT regulon of P. protegens H78, 144 genes encode regulatory systems or factors, including numerous transcriptional factors and regulators, two-component regulatory systems or elements, and three sRNAs (RsmY, RsmZ, and CrcZ). Among the 144 regulators, 93 regulatory genes were significantly downregulated, and 51 genes were upregulated in the relA/spoT mutant relative to the wild-type H78 (Supplemental Tables 6 and 7). Among the positive targets of relA/spoT, ten pairs of FecI/FecR family sigma factors/regulators, such as FecR/FecI (H78_01021/H78_01020) and HasI/HasS, are required for the regulation of iron utilization (Supplemental Table S5). Moreover, four pathway-specific transcriptional regulators (pltR, pltZ, phlG, and pchR) related to the biosynthesis of antibiotics and iron carriers are positively regulated by relA/spoT. The global regulation of relA/spoT appears to be mediated by multiple pathway-specific regulators.
Two important sRNAs, RsmY and RsmZ, which, together with another sRNA, RsmX, form a vital node of the Gac/Rsm cascade, were subject to strong positive regulation by relA/spoT. In the present transcriptomic sequencing study, most of the sRNAs were not sequenced because they are too short. However, RsmY and RsmZ transcripts were detected in the transcriptomic profiles of wild-type strain H78. These two sRNAs completely disappeared at the transcript level in the relA/spoT mutant (Supplemental Table S6). Therefore, the RelA/SpoT-mediated stringent response may activate the expression of the RsmXYZ sRNAs in the Gac/Rsm cascade and further activate the production of secondary metabolites, including antibiotics. Moreover, the transcript levels of gacS and gacA were unchanged between the relA/spoT mutant and its parental strain H78.
Among the negative targets of relA/spoT, the largest class of regulators is TCSs, including glnG/glnL, pedS1/R1, pedS2/R2, rpeA/B, colS/R, and phoR/B (Supplemental Table S7). In addition, the global transcriptional regulator Fis gene exhibited 2.5-fold upregulation at the transcript level in the relA/spoT mutant relative to the wild-type H78 (Supplemental Table S7). The CrcZ sRNA gene in the CbrA/B-CrcYZ-Crc signal transduction cascade was upregulated by 2.5-fold because of the relA/spoT mutation (Supplemental Table S7).
Discussion
The (p)ppGpp-mediated stringent response is widely distributed in bacteria and utilized to sense and respond to starvation and stresses for survival in adverse environments (Hauryliuk et al. 2015). In this study, we investigated the genome-wide regulation of RelA/SpoT in P. protegens H78 grown to the mid- to late log phase with an OD600 of 5.0–6.0 in KMB at 28 °C. Systematic phenotype assays were conducted to assess the influence of relA/spoT deletion on the physiology and metabolism of P. protegens H78. A global regulatory model of the (p)ppGpp-mediated stringent response in P. protegens H78 is summarized in Fig. 9. In P. protegens H78, the stringent response plays an important role in inhibiting bacterial growth and maintaining cell survival. The (p)ppGpp-mediated stringent response globally inhibits bacterial primary metabolism, including DNA replication, transcription, and translation; amino acid metabolism; carbohydrate and energy metabolism; ion transport and secretion systems; and flagellar biogenesis, assembly, and chemotaxis. However, secondary metabolism, including the biosynthesis of antibiotics, iron carriers, and exopolysaccharides, is globally activated by the (p)ppGpp-mediated stringent response. Moreover, the global regulation of the stringent response system may be mediated by numerous transcriptional factors/regulators, TCSs, and sRNAs.
In P. protegens H78, the (p)ppGpp-mediated stringent response system is involved in regulating the entire process of genetic information transfer: DNA replication and cell division (grpE-dnaK-dnaJ, fts-mur-mra, gyrB, ssb, hupA, ftsX, recO, and H78_01844), transcription (rpo, rpoC, and multiple ECF sigma factors/regulators), and translation (rplQ, rpmE, rpsT, rsmB, rsmE, tsf, tuf, pth, and fmt). The negative regulatory effect of the (p)ppGpp-mediated stringent response on DNA replication and cell division corresponds to the upregulation of bacterial growth in the relA/spoT mutant of H78. The (p)ppGpp-mediated stringent response, as a major factor in growth control (Potrykus et al. 2011), is involved in growth and cell cycle arrest (Bokinsky et al. 2013; Ferullo and Lovett 2008), morphological differentiation (Hesketh et al. 2007), and cell persistence (Hauryliuk et al. 2015). In addition, the gene clusters for amino acid, carbohydrate and energy, and inorganic ion transport and metabolism constitute the largest functional groups of the RelA/SpoT regulon. These gene clusters related to nutritional utilization are negatively regulated by relA/spoT, confirming that the (p)ppGpp-mediated stringent response is an important strategy by which bacteria metabolically adapt to nutritional starvation. In this study, P. protegens H78 and its relA/spoT mutant were grown in the rich nutritional medium KMB for RNA-seq analysis. The nutritional level started to decline gradually during mid- to late log phase, at which time the cultures were sampled for RNA-seq. The decrease in nutritional level triggers the bacterial stringent response to globally downregulate bacterial growth and metabolism to save energy and survive the upcoming nutritional depletion.
An interesting phenomenon worth mentioning is that the relA/spoT dual mutant of H78 resumed growth after 50 h of lag phase in M9 medium with mannitol as carbon source (Fig. 6a). Moreover, the lag phase of the relA/spoT mutant still existed when the growth-resuming culture of the relA/spoT mutant after lag phase was re-inoculated into the fresh M9 medium with mannitol as carbon source (Supplemental Fig. S6). It was reported that the relA/spoT dual mutant of E. coli easily acquires suppressors in rpoB and rpoC genes which suppress amino acid auxotrophy phenotype of the relA/spoT mutant in minimal medium (Conrad et al. 2010; Murphy and Cashel 2003). The rpoC mutation can increase growth rate of E. coli in M9 minimal media with glycerol, glucose, or lactate carbon source (Conrad et al. 2010). The growth resumption of the relA/spoT mutant after lag phase may be due to appearance of suppressors. However, DNA sequencing and analysis results showed that no any mutation was found in three genes, including rpoB, rpoC, and rpoZ, in the relA/spoT dual mutant of P. protegens H78 (Supplemental materials).
In contrast with primary metabolism, secondary metabolism, including antibiotic and iron carrier biosynthesis, is globally activated by the (p)ppGpp-mediated stringent response in P. protegens H78. Antibiotics and iron carriers (siderophores and haemophores) mainly contribute to the ecological competency of bacteria. Most antibiotic operons, including plt, prn, ofa, and fit, are positively regulated by relA/spoT. However, the DAPG biosynthetic operon phlACBDE is an exception and its transcript level is not influenced by relA/spoT. In contrast, DAPG production is entirely inhibited by the relA/spoT mutation (Fig. 4). In another report, DAPG production was also found to be positively regulated by relA/spoT in P. protegens CHA0 (Takeuchi et al. 2012). In this study, the downregulation of the transcript level in both the transcriptional repressor gene phlF and the DAPG hydrolase gene phlG seems contradictory with the inhibition of DAPG biosynthesis in the relA/spoT mutant. We speculate that (p)ppGpp may exert indirect posttranscriptional regulation of phlACBDE expression through other underlying posttranscriptional regulatory pathways, such as the Gac/Rsm cascade. This has been implied by the result that the phlA'-'lacZ in-frame fusion expression, which indicated total regulation at both the transcriptional and posttranscriptional levels, was substantially reduced by relA/spoT deficiency (Fig. 4g). Therefore, it can be reasonably deduced that the (p)ppGpp-mediated stringent response globally activates antibiotic biosynthesis directly or indirectly through other regulatory factors or systems.
Here, it is interesting why P. protegens needs to be activated by the (p)ppGpp-mediated stringent response to produce antibiotics at the expense of energy consumption. Pseudomonas spp. inhabit diverse environments, including soil, water, the plant rhizosphere, and animals. Their relatively small core genomes and their flexible and adaptable accessory genomes may provide Pseudomonas spp. with remarkable metabolic and physiologic versatility and an astonishing ecological adaptability (Silby et al. 2011). Each species of Pseudomonas can produce a specific spectrum of secondary metabolites, including antibiotics, contributing to its specific lifestyle and ecological competitiveness in its natural habitat. P. protegens, as a representative of PGPR, can produce a set of antibiotics, including Plt, DAPG, and Prn (Gross and Loper 2009). Under nutritional starvation and other environmental stresses, P. protegens H78 is induced by the (p)ppGpp-mediated stringent response to downregulate primary metabolism for conserving energy and surviving in nutritional starvation and activate secondary metabolism, including antibiotic biosynthesis, for acquiring a competitive advantage over other competitors (such as plant pathogens) and predators in natural habitats.
Specifically, the (p)ppGpp-dependent transcriptomic profile in P. protegens H78 correlates the (p)ppGpp-mediated stringent response with the Gac/Rsm cascade. These two systems similarly activate secondary metabolism, including antibiotic biosynthesis. The Gac/Rsm cascade consists of the GacS/GacA TCS, the RsmY family sRNAs, and the RsmA family translational repressors in Pseudomonas spp. (Haas and Keel 2003). The expression levels of two sRNAs, RsmY and RsmZ, were strongly reduced in the relA/spoT mutant of H78 (Supplemental Table S6), suggesting that the (p)ppGpp-mediated stringent response may activate the expression of RsmYZ sRNAs. Another sRNA, RsmX, was not detected by RNA-seq in either P. protegens H78 or its relA/spoT mutant. In the Gac/Rsm cascade, the GacS/GacA TCS was not significantly influenced by relA/spoT at the transcript level in H78. In a previous report, RsmYZ expression in P. protegens CHA0 was drastically reduced, whereas the expression levels of gacS and gacA were relatively moderately downregulated, in the relA/spoT mutant compared with the wild-type CHA0. Moreover, the gacA mutation upregulated the expression of the (p)ppGpp synthetase gene relA in CHA0 (Takeuchi et al. 2012). However, the potential molecular mechanism by which (p)ppGpp regulates the Gac/Rsm cascade and vice versa remains unknown. This study may provide some information for further clarifying the overlapping activation pattern of secondary metabolism induced by the (p)ppGpp-mediated stringent response and the Gac/Rsm cascade in Pseudomonas and other related bacteria.
In conclusion, the (p)ppGpp-mediated stringent response system plays an important role in downregulating bacterial growth and improving cell survival in P. protegens H78. This system systematically downregulates primary metabolism, including DNA replication, transcription and translation, amino acid metabolism, and carbohydrate and energy metabolism, to save energy and survive starvation. In contrast, it globally activates the biosynthesis of secondary metabolites, including antibiotics, iron carriers, and exopolysaccharides, contributing to bacterial ecological competence. Further research should focus on clarifying the molecular regulatory mechanisms of (p)ppGpp on secondary metabolism, including antibiotic biosynthesis, that are directly related to biocontrol ability in the rhizobacterium P. protegens H78. We should also ascertain whether (p)ppGpp directly targets the RsmXYZ sRNAs or other factors and whether (p)ppGpp functions as a potential environmental cue to stimulate phosphorylation of the sensor kinase GacS and thus activate the Gac/Rsm cascade in Pseudomonas spp. The identification and characterization of direct target sites and regulatory mechanisms of (p)ppGpp in the Gac/Rsm cascade, as well as the clarification of the regulatory mechanism of the Gac/Rsm cascade on the expression of (p)ppGpp biosynthetic or regulatory genes, will help clarify the mutual regulatory relation between these two systems.
References
Abby SS, Cury J, Guglielmini J, Neron B, Touchon M, Rocha EP (2016) Identification of protein secretion systems in bacterial genomes. Sci Rep 6:23080. https://doi.org/10.1038/srep23080
Atkinson GC, Tenson T, Hauryliuk V (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6(8):e23479. https://doi.org/10.1371/journal.pone.0023479
Bokinsky G, Baidoo EE, Akella S, Burd H, Weaver D, Alonso-Gutierrez J, Garcia-Martin H, Lee TS, Keasling JD (2013) HipA-triggered growth arrest and beta-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J Bacteriol 195(14):3173–3182. https://doi.org/10.1128/JB.02210-12
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Rep 1(3):251–264. https://doi.org/10.1016/j.celrep.2011.12.007
Camon E, Barrell D, Lee V, Dimmer E, Apweiler R (2004) The Gene Ontology Annotation (GOA) database—an integrated resource of GO annotations to the UniProt Knowledgebase. Silico Biol 4(1):5–6
Chancey ST, Wood DW, Pierson LS 3rd (1999) Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl Environ Microbiol 65(6):2294–2299
Chatnaparat T, Li Z, Korban SS, Zhao Y (2015) The bacterial alarmone (p)ppGpp is required for virulence and controls cell size and survival of Pseudomonas syringae on plants. Environ Microbiol 17(11):4253–4270. https://doi.org/10.1111/1462-2920.12744
Chen Y, Wang X, Huang X, Zhang X, Xu Y (2008) Las-like quorum-sensing system negatively regulates both pyoluteorin and phenazine-1-carboxylic acid production in Pseudomonas sp. M18. Sci China C Life Sci 51(2):174–181. https://doi.org/10.1007/s11427-008-0026-8
Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BO (2010) RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A 107(47):20500–20505. https://doi.org/10.1073/pnas.0911253107
Dalebroux ZD, Swanson MS (2012) ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10(3):203–212. https://doi.org/10.1038/nrmicro2720
Ferullo DJ, Lovett ST (2008) The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet 4(12):e1000300. https://doi.org/10.1371/journal.pgen.1000300
Gross H, Loper JE (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26(11):1408–1446. https://doi.org/10.1039/b817075b
Gross H, Stockwell VO, Henkels MD, Nowak-Thompson B, Loper JE, Gerwick WH (2007) The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem Biol 14(1):53–63. https://doi.org/10.1016/j.chembiol.2006.11.007
Haas D, Keel C (2003) Regulation of antibiotic production in root-colonizing Peudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41:117–153. https://doi.org/10.1146/annurev.phyto.41.052002.095656
Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K (2015) Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13(5):298–309. https://doi.org/10.1038/nrmicro3448
Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M (2007) The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol 8(8):R161. https://doi.org/10.1186/gb-2007-8-8-r161
Huang X, Zhu D, Ge Y, Hu H, Zhang X, Xu Y (2004) Identification and characterization of pltZ, a gene involved in the repression of pyoluteorin biosynthesis in Pseudomonas sp. M18. FEMS Microbiol Lett 232(2):197–202. https://doi.org/10.1016/S0378-1097(04)00074-6
Huang X, Zhang X, Xu Y (2008) PltR expression modulated by the global regulators GacA, RsmA, LasI and RhlI in Pseudomonas sp. M18. Res Microbiol 159(2):128–136. https://doi.org/10.1016/j.resmic.2007.10.006
Huang X, Wang Z, Liu Y, Zhang X (2017) Complete genome sequence of Pseudomonas protegens H78, a plant growth-promoting rhizobacterium. Genome Announc 5(16):e00233–e00217. https://doi.org/10.1128/genomeA.00233-17
Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76(1):46–65. https://doi.org/10.1128/MMBR.05007-11
Jousset A, Schuldes J, Keel C, Maurhofer M, Daniel R, Scheu S, Thuermer A (2014) Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0. Genome Announc 2(2):e00322–e00314. https://doi.org/10.1128/genomeA.00322-14
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44(2):301–307
Konieczny I, Zylicz M (1999) Role of bacterial chaperones in DNA replication. Genet Eng (N Y) 21:95–111
Liu Y, Shi H, Wang Z, Huang X, Zhang X (2018) Pleiotropic control of antibiotic biosynthesis, flagellar operon expression, biofilm formation, and carbon source utilization by RpoN in Pseudomonas protegens H78. Appl Microbiol Biotechnol 102(22):9719–9730. https://doi.org/10.1007/s00253-018-9282-0
Loper JE, Henkels MD, Rangel LI, Olcott MH, Walker FL, Bond KL, Kidarsa TA, Hesse CN, Sneh B, Stockwell VO, Taylor BJ (2016) Rhizoxin, orfamide a, and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ Microbiol 18(10):3509–3521. https://doi.org/10.1111/1462-2920.13369
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Lu J, Huang X, Li K, Li S, Zhang M, Wang Y, Jiang H, Xu Y (2009) LysR family transcriptional regulator PqsR as repressor of pyoluteorin biosynthesis and activator of phenazine-1-carboxylic acid biosynthesis in Pseudomonas sp. M18. J Biotechnol 143(1):1–9. https://doi.org/10.1016/j.jbiotec.2009.06.008
Manuel J, Berry C, Selin C, Fernando WG, de Kievit TR (2011) Repression of the antifungal activity of Pseudomonas sp. strain DF41 by the stringent response. Appl Environ Microbiol 77(16):5635–5642. https://doi.org/10.1128/AEM.02875-10
Manuel J, Selin C, Fernando WG, de Kievit T (2012) Stringent response mutants of Pseudomonas chlororaphis PA23 exhibit enhanced antifungal activity against Sclerotinia sclerotiorum in vitro. Microbiology 158:207–216. https://doi.org/10.1099/mic.0.053082-0
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628. https://doi.org/10.1038/nmeth.1226
Murphy H, Cashel M (2003) Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol 371:596–601. https://doi.org/10.1016/S0076-6879(03)71044-1
Noinaj N, Guillier M, Barnard TJ, Buchanan SK (2010) TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. https://doi.org/10.1146/annurev.micro.112408.134247
Ogata H, Goto S, Fujibuchi W, Kanehisa M (1998) Computation with the KEGG pathway database. Biosystems 47(1–2):119–128. https://doi.org/10.1016/s0303-2647(98)00017-3
Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS 3rd, Thomashow LS, Loper JE (2005) Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23(7):873–878. https://doi.org/10.1038/nbt1110
Pechy-Tarr M, Bruck DJ, Maurhofer M, Fischer E, Vogne C, Henkels MD, Donahue KM, Grunder J, Loper JE, Keel C (2008) Molecular analysis of a novel gene cluster encoding an insect toxin in plant-associated strains of Pseudomonas fluorescens. Environ Microbiol 10(9):2368–2386. https://doi.org/10.1111/j.1462-2920.2008.01662.x
Potrykus K, Cashel M (2008) (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. https://doi.org/10.1146/annurev.micro.62.081307.162903
Potrykus K, Murphy H, Philippe N, Cashel M (2011) ppGpp is the major source of growth rate control in E. coli. Environ Microbiol 13(3):563–575. https://doi.org/10.1111/j.1462-2920.2010.02357.x
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160(1):47–56
Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35(4):652–680. https://doi.org/10.1111/j.1574-6976.2011.00269.x
Sinha AK, Winther KS, Roghanian M, Gerdes K (2019) Fatty acid starvation activates RelA by depleting lysine precursor pyruvate. Mol Microbiol 112(4):1339–1349. https://doi.org/10.1111/mmi.14366
Steinchen W, Bange G (2016) The magic dance of the alarmones (p)ppGpp. Mol Microbiol 101(4):531–544. https://doi.org/10.1111/mmi.13412
Takeuchi K, Yamada K, Haas D (2012) ppGpp controlled by the Gac/Rsm regulatory pathway sustains biocontrol activity in Pseudomonas fluorescens CHA0. Mol Plant-Microbe Interact 25(11):1440–1449. https://doi.org/10.1094/MPMI-02-12-0034-R
Takeuchi K, Noda N, Someya N (2014) Complete genome sequence of the biocontrol strain Pseudomonas protegens Cab57 discovered in Japan reveals strain-specific diversity of this species. PLoS One 9(4):e93683. https://doi.org/10.1371/journal.pone.0093683
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28(1):33–36. https://doi.org/10.1093/nar/28.1.33
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc 7(3):562–578. https://doi.org/10.1038/nprot.2012.016
Wang Z, Huang X, Liu Y, Yang G, Zhang X (2017) GacS/GacA activates pyoluteorin biosynthesis through Gac/Rsm-RsmE cascade and RsmA/RsmE-driven feedback loop in Pseudomonas protegens H78. Mol Microbiol 105(6):968–985. https://doi.org/10.1111/mmi.13749
Wei X, Huang X, Tang L, Wu D, Xu Y (2013) Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J Bacteriol 195(15):3387–3400. https://doi.org/10.1128/JB.00214-13
Wingett SW, Andrews S (2018) FastQ screen: a tool for multi-genome mapping and quality control. F1000Res 7:1338. https://doi.org/10.12688/f1000research.15931.2
Winstanley C, Langille MG, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F, Thomson NR, Winsor GL, Quail MA, Lennard N, Bignell A, Clarke L, Seeger K, Saunders D, Harris D, Parkhill J, Hancock RE, Brinkman FS, Levesque RC (2009) Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res 19(1):12–23. https://doi.org/10.1101/gr.086082.108
Wu DQ, Ye J, Ou HY, Wei X, Huang X, He YW, Xu Y (2011) Genomic analysis and temperature-dependent transcriptome profiles of the rhizosphere originating strain Pseudomonas aeruginosa M18. BMC Genomics 12:438. https://doi.org/10.1186/1471-2164-12-438
Yan A, Huang X, Liu H, Dong D, Zhang D, Zhang X, Xu Y (2007) An rhl-like quorum-sensing system negatively regulates pyoluteorin production in Pseudomonas sp. M18. Microbiology 153(1):16–28. https://doi.org/10.1099/mic.0.29211-0
Zhang W, Zhao Z, Zhang B, Wu XG, Ren ZG, Zhang LQ (2014) Posttranscriptional regulation of 2,4-diacetylphloroglucinol production by GidA and TrmE in Pseudomonas fluorescens 2P24. Appl Environ Microbiol 80(13):3972–3981. https://doi.org/10.1128/AEM.00455-14
Zhao Q, Poole K (2000) A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol Lett 184(1):127–132
Zolotarev AS, Unnikrishnan M, Shmukler BE, Clark JS, Vandorpe DH, Grigorieff N, Rubin EJ, Alper SL (2008) Increased sulfate uptake by E. coli overexpressing the SLC26-related SulP protein Rv1739c from Mycobacterium tuberculosis. Comp Biochem Physiol A Mol Integr Physiol 149(3):255–266. https://doi.org/10.1016/j.cbpa.2007.12.005
Funding
This study was sponsored by Natural Science Foundation of Shanghai (19ZR1427600) and National Natural Science Foundation of China (31470196, 31270083). This work was also supported by the National Key Research and Development Program of China (2019YFA0904302).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 4048 kb)
Rights and permissions
About this article
Cite this article
Wu, L., Wang, Z., Guan, Y. et al. The (p)ppGpp-mediated stringent response regulatory system globally inhibits primary metabolism and activates secondary metabolism in Pseudomonas protegens H78. Appl Microbiol Biotechnol 104, 3061–3079 (2020). https://doi.org/10.1007/s00253-020-10421-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10421-5