Abstract
Due to their high secretion capacity, Gram-positive bacteria from the genus Bacillus are important expression hosts for the high-yield production of enzymes in industrial biotechnology; however, to date, strains from only few Bacillus species are used for enzyme production at industrial scale. Herein, we introduce Paenibacillus polymyxa DSM 292, a member of a different genus, as a novel host for secretory protein production. The model gene cel8A from Clostridium thermocellum was chosen as an easily detectable reporter gene with industrial relevance to demonstrate heterologous expression and secretion in P. polymyxa. The yield of the secreted cellulase Cel8A protein was increased by optimizing the expression medium and testing several promoter sequences in the expression plasmid pBACOV. Quantitative mass spectrometry was used to analyze the secretome in order to identify promising new promoter sequences from the P. polymyxa genome itself. The most abundantly secreted host proteins were identified, and the promoters regulating the expression of their corresponding genes were selected. Eleven promoter sequences were cloned and tested, including well-characterized promoters from Bacillus subtilis and Bacillus megaterium. The best result was achieved with the promoter for the hypothetical protein PPOLYM_03468 from P. polymyxa. In combination with the optimized expression medium, this promoter enabled the production of 5475 U/l of Cel8A, which represents a 6.2-fold increase compared to the reference promoter PaprE. The set of promoters described in this work covers a broad range of promoter strengths useful for heterologous expression in the new host P. polymyxa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Industrial biotechnology is a rapidly growing field, since it is associated with the fast and cost-effective production of goods such as chemicals, pharmaceuticals, and biofuels in a resource and environmentally sustainable manner (Ahmann and Dorgan 2007). For decades, members of the genus Bacillus have been used extensively for the industrial production of enzymes such as proteases and amylases (Harwood 1992). Bacillus species owe their success, in particular, to their ability to secrete proteins into the extracellular medium with high selectivity and productivity. This facilitates easy and economical downstream processing by simply recovering the target proteins from the culture supernatant. Under ideal conditions, the secretory system can export proteins at grams per liter concentrations (Schallmey et al. 2004). Well-established industrial microorganisms include Bacillus subtilis, Bacillus licheniformis, and a few other Bacillus species and are mainly used to overexpress Bacillus genes for the production of enzymes typically added to washing or cleaning agents (Schallmey et al. 2004; Küppers et al. 2014; Heinze et al. 2018). Despite the availability of production strains for bulk enzymes, the identification and development of new expression hosts from other genera and species are important to enable efficient production of new enzymes (Küppers et al. 2014) in industrially relevant yields. This applies especially to heterologous expression, which is more challenging and usually significantly less efficient (Bien et al. 2014).
Paenibacillus polymyxa is a soil bacterium with a multitude of useful properties. Some examples include plant growth promotion, for example, through nitrogen fixation or production of phytohormones, production of antimicrobial compounds, such as polymyxins and industrially relevant substances, like 2,3-butanediol or exopolysaccharides (Grady et al. 2016; Rütering et al. 2017). In comparison to other strains of the same species, whose genomes have already been published, P. polymyxa DSM292 shows some distinguishing traits. Most notably, strain DSM292 displays a lower exoprotease activity than strain DSM 365 or the type strain P. polymyxa DSM36T. This can lead to higher stability of heterologously expressed, secreted target proteins. Therefore, we selected P. polymyxa DSM 292 as a promising candidate to establish a new expression strain for heterologous, secretory protein production.
The well-characterized thermophilic endoglucanase Cel8A from Clostridium thermocellum (Schwarz et al. 1986; Leis et al. 2017) was chosen as an exemplary target protein with biotechnological relevance. Due to its ability to hydrolyze carboxymethyl cellulose (CMC) at 60 °C, active Cel8A can be easily detected and quantified in the background of P. polymyxa DSM 292 supernatant, which itself does not degrade CMC. Cel8A is a component of the cellulosome, an extracellular multienzyme complex allowing efficient degradation of plant cell wall polysaccharides by its synergistically acting enzyme activities (Shoham et al. 1999; Schwarz 2001; Zverlov and Schwarz 2004). Hence, cellulosomes have elicited great interest in potential biotechnological applications, such as the production of biofuels from cellulosic biomass (Bayer et al. 2007). Yet, several cellulosomal components have to be heterologously expressed to enable the reconstitution of cellulosomes in vitro, and not all components can be produced in large amounts in the available expression hosts (Leis et al. 2017). Escherichia coli, B. subtilis, and Bacillus stearothermophilus have been used to produce Cel8A recombinantly (Schwarz et al. 1986; Soutschek-Bauer and Staudenbauer 1987; Joliff et al. 1989; Bien et al. 2014; Leis et al. 2017). The reported yields are highly variable between different studies. For example, Schwarz et al. (1986) were able to produce 304.4 U of Cel8A per gram of E. coli cells, while Joliff et al. (1989) reported a yield of 800 U/l of secreted Cel8A using B. subtilis. Very high yields of secreted Cel8A were obtained using B. subtilis 168 (13,900 U/l) and B. stearothermophilus CU21 (11,300 U/l) by Soutschek-Bauer and Staudenbauer (1987).
Heterologous protein production depends on the availability of efficient promoter systems that enable a controlled, high level of target gene expression. Three types of promoters can be used: constitutive, inducer-specific, and auto-inducible promoters (Schumann 2007; Nijland et al. 2007; Lee et al. 2010). The choice of promoter system is dependent on both the expression host as well as the characteristics of the desired target protein. By using constitutive promoters, such as the B. subtilis promoter P43 (Wang and Doi 1984), high levels of target protein have been produced. Inducer-specific promoters are also widely used, such as PxylA, which depends on induction by exogenous addition of xylose (Rygus and Hillen 1991; Malten et al. 2005). While these systems offer high degrees of control, the necessity for an external inducer makes them less applicable to large-scale fermentations. Alternatively, transcriptome analyses have identified auto-inducible promoters whose activities are growth phase- or stress-specific, and therefore, they do not require addition of inducers (Nicolas et al. 2012). One promoter frequently used for large-scale recombinant protein production is the B. subtilis aprE promoter (PaprE). In its native host, PaprE is active in the late exponential and stationary growth phase (Valle and Ferrari 1989) and served as a reference promoter in this study.
By analyzing the secretory production of endoglucanase Cel8A under the control of 11 different promoters, this study presents new insights into the performance of these regulatory sequences in P. polymyxa DSM 292, and therefore, provides a basis for exploiting this new expression strain for heterologous protein production. Furthermore, by optimizing the culture medium and applying a new promoter, the yield of the secreted heterologous target protein was significantly improved.
Materials and methods
Bacterial strains and growth conditions
Escherichia coli strains were grown in 20 ml LB medium (Bertani 1951) shaken at 180 rpm or on LB agar plates at 37 °C overnight. E. coli TOP10 (Life technologies, Carlsbad, USA) was used for cloning and as the donor strain for transmating (Heinze et al. 2018) and was grown with 100 μg/ml carbenicillin (Carl Roth, Karlsruhe, Germany). E. coli HB101 carrying the plasmid pRK2013 was used as a helper strain for transmating and was cultured with 50 μg/ml kanamycin (AppliChem, Darmstadt, Germany). E. coli HB101 pRK2013 (DSM No. 5599) and P. polymyxa DSM 292 were obtained from DSMZ (Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany). P. polymyxa DSM 292 transmates carrying the expression plasmid were streaked onto LB selection agar containing 40 μg/ml polymyxin B (Carl Roth) and 10 μg/ml kanamycin or grown in liquid medium with 10 μg/ml kanamycin. For precultures, 20 ml LB liquid medium was inoculated with a single colony of P. polymyxa DSM 292 and grown overnight at 37 °C and 180 rpm. For expression experiments, 20 ml main cultures of P. polymyxa DSM 292 were inoculated to a starting OD600 of 0.2 using overnight precultures and were incubated for 24 h at 37 °C and 180 rpm, unless indicated otherwise. Main cultures were grown in LBS medium (LB medium supplemented with 0.5 M sorbitol) or medium 2 (5 g/l soy peptone, 30 g/l glucose, 7 g/l (NH4)2SO4, 1 g/l MgSO4 × 7 H2O, 15 mg/l CaCl2 × 2 H2O, 25 mM Na/K-phosphate buffer pH 7.0 [corresponding to 15.25 mM K2HPO4 and 9.75 mM NaH2PO4]). A final amount of 0.06% (v/v) of a trace element solution, prepared as described by Häßler et al. (2012), was added to medium 2; however, it lacked biotin and calcium chloride.
Construction of expression vectors
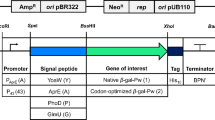
The target gene cel8A from C. thermocellum was synthesized by Eurofins Genomics (Ebersberg, Germany) with codon usage optimization for B. megaterium. The gene was subcloned into pBACOV using NdeI and XbaI restriction enzymes (New England Biolabs, Ipswich, USA), yielding pBACOV-cel8A. To identify a suitable signal peptide (SP) for the secretory production of Cel8A, the B. subtilis secretory protein expression system kit (Takara-Bio Inc., Kusatsu, Japan) was used according to the manufacturer’s instructions to create a SP library. The library was transferred to P. polymyxa DSM 292 for screening and selection of a suitable SP. The resulting plasmid pBACOV-SPLipB-cel8A was used as a chassis for replacing the reference promoter PaprE with alternative promoters by linearizing pBACOV-SPLipB-cel8A with SpeI and MluI-HF (New England Biolabs). In cases where both PaprE and SPLipB were replaced with alternative promoter and SP-sequences, SpeI and EagI-HF (New England Biolabs) were used. In all cases, PaprE was fully replaced, including the original ribosome binding site (RBS) of the exchanged promoter sequence. The promoter sequences were amplified from genomic B. subtilis RIK1285 (Clontech Laboratories, Mountain View, USA) or P. polymyxa DSM 292 genomic DNA. The promoters PxylA and PxylA+ were amplified from plasmid pHIS1525 (Mobitec GmbH, Göttingen, Germany; Table 2). The RBS of PxylA as optimized by Malten et al. (2005) was introduced by PCR amplification from pHIS1525 by miss-matching positions in the reverse primer Pxyl_RBS+_Gibson_rv, thereby generating PxylA+. The oligonucleotide primers used in this study are listed in Supplementary Table S1. The promoter sequences were introduced into vector pBACOV via Gibson assembly (Gibson et al. 2009), and plasmid sequences were confirmed by sequencing.
Plasmid transfer to P. polymyxa DSM 292
Plasmids were transferred to P. polymyxa DSM 292 by transmating as previously described (Heinze et al. 2018).
Azo-CMC assay for measurement of Cel8A activity in supernatants
OD600 of the main cultures was determined after incubation for 24 h. Two milliliters of culture was centrifuged for 5 min at 21,100 g, and the supernatant was carefully collected without disrupting the cell pellet. Cel8A activity in the supernatant was measured using Azo-CM-Cellulose (Azo-CMC, Megazyme, Bray, Ireland). For each sample, 150 μl of 2% substrate solution (dissolved in 200 mM sodium acetate, pH 5.0 (HCl) at 60 °C, 100 mM NaCl, and 20 mM CaCl2) was mixed on ice with 150 μl of culture supernatant. The samples were incubated in a water bath at 60 °C for 5 min. The reaction was terminated with 750 μl of precipitation buffer (294 mM sodium acetate × 3 H2O, pH 5.0 (HCl), 22 mM zinc acetate, and 80% (v/v) ethanol) added, and the samples were cooled at room temperature for 10 min. Samples were centrifuged at 1000 g for 10 min to remove the precipitate, and the absorption of the supernatants was measured at 590 nm. Blanks were prepared by mixing 150 μl of substrate solution with 750 μl precipitation buffer, followed by the addition of 150 μl culture supernatant from the empty vector strain (P. polymyxa DSM 292 pBACOV). The amount of Cel8A was evaluated by comparing the absorption values to a reference curve constructed with known concentrations of Cel8A produced in E. coli BL21 Star™ (Life technologies), as described previously (Leis et al. 2017). The specific activity of the purified Cel8A reference was 15.75 U/mg using the DNSA assay (Wood and Bhat 1988) with 0.5% (w/v) CMC (Sigma-Aldrich, St. Louis, USA) as substrate.
Medium optimization by Plackett–Burman design
Initially, a semidefined medium (termed medium 0) was developed as the basis for further optimization. Medium 0 was optimized using a Plackett–Burman experimental design with 11 factors, resulting in 12 different media for testing. Factors 1–8 correspond to the following medium components: soy peptone, glucose, ammonium sulfate, phosphate buffer, magnesium, calcium, trace elements, and biotin; whereas, factors 9–11 were incubation conditions: temperature, pH, and aeration (using baffled or normal Erlenmeyer flasks). For each factor, a high (1) and low (− 1) level were defined, in addition to the initial condition (0). The specified 1, − 1, and 0 levels for each factor are listed in Table 1. To assess the performance of each medium, P. polymyxa DSM 292 pBACOV-SPLipB-cel8A and the negative control strain carrying pBACOV without the target gene were incubated in triplicate for 24 h under each condition, and the Cel8A activity in the supernatant was determined (Azo-CMC).
Secretome analysis by quantitative mass spectrometry (LC-MS/MS)
P. polymyxa DSM 292 carrying pBACOV-SPLipB-cel8A was incubated in 20 ml medium 2 (37 °C, 180 rpm). After 24 h, 2 ml of the culture was harvested by centrifugation (21,100 g for 5 min at 4 °C). The supernatant was collected without disrupting the cell pellet and stored at − 20 °C until further use for LC-MS/MS analysis.
Three biological replicates of 30 μl supernatant were mixed with LDS sample buffer (Thermo Fisher Scientific, Waltham, USA) for in-gel digestion. The samples were then reduced with 25 mM dithiothreitol, heated for 10 min at 95 °C and alkylated with 55 mM chloroacetamide. Proteins were run on a 4–12% NuPAGE gel (Thermo Fisher Scientific) for about 1 cm to concentrate the sample prior to in-gel tryptic digestion, which was performed according to the standard procedures (Shevchenko et al. 2006). The peptides obtained were dried to completeness and resuspended in 12 μl of buffer A (0.1% formic acid (FA)), and 5 μl of sample was injected per MS measurement.
The samples were analyzed via LC-MS/MS on a nanoLC-Ultra 1D+ (Eksigent, Dublin, CA) coupled online to a Q-Exactive HF mass spectrometer (ThermoFisher Scientific). Peptides were initially loaded onto a 2-cm trap column (75 μm inner diameter, ReproSil-Pur 120 ODS-3 5 μm, Dr. Maisch) at a flow rate of 5 μl/min. Subsequently, they were separated on a 40 cm analytical column (75 μm inner diameter, ReproSil-Gold 120 C18 3 μm, Dr. Maisch) at a flow rate of 300 nl/min and with a gradient of 4–32% buffer B (0.1% FA and 5% DMSO in acetonitrile) in 120 min (buffer A now also contained 5% DMSO).
The eluate from the analytical column was sprayed via a stainless steel emitter (ThermoFisher Scientific) at a source voltage of 2.2 kV into the mass spectrometer. The transfer capillary was heated to 275 °C. The Q-Exactive HF was operated in data-dependent acquisition (DDA) mode, automatically switching between MS1 and MS2 spectrum acquisition. MS1 spectra were acquired over a mass-to-charge (m/z) range of 360–1300 m/z at a resolution of 60,000 (at m/z 200) using a maximum injection time of 50 ms and an AGC target value of 3e6. Up to 20 peptide precursors were isolated (isolation window, 1.7 m/z; maximum injection time, 25 ms; and AGC value, 1e5), fragmented by high-energy collision-induced dissociation (HCD) using 25% normalized collision energy (NCE) and analyzed at a resolution of 15,000 with a scan range from 200 to 2000 m/z. Precursor ions that were singly charged, unassigned, or with charge states > 6+ were excluded. The dynamic exclusion duration of fragmented precursor ions was 20 s.
Peptide and protein identification and quantification were performed using MaxQuant software (version 1.5.7.4; Cox and Mann 2008) by searching the MS data against a P. polymyxa DSM 292 protein database (5339 protein sequences) derived from the P. polymyxa DSM 292 genome sequence (Sequence accession numbers for Contigs 1 to 23: OKC01000001 to OKC01000023; Assembly accession number: GCA_900406265) using the search engine Andromeda (Cox et al. 2011). The Cel8A protein sequence was manually added to the reference database (including the SP sequence) to make it identifiable within the P. polymyxa DSM 292 background proteome. The MaxQuant search was performed using two variable modifications; oxidation of methionine and N-terminal protein acetylation. Carbamidomethylation on cysteines was specified as a fixed modification. Trypsin (Trypsin/P) was specified as the proteolytic enzyme with up to two allowed missed cleavage sites. Label-free quantification (Cox et al. 2014) and match-between-runs (matching time window of 0.7 min and alignment time window of 20 min) were enabled, and the results were filtered for a minimal length of seven amino acids and 1% peptide and protein false discovery rate (FDR). To assess the concentration of the identified proteins relative to each other (i.e., to down-rank the proteins according to their abundance in the sample), we estimated the absolute protein intensities using the intensity-based absolute quantification algorithm (iBAQ; Schwanhäusser et al. 2011), which is implemented within MaxQuant.
Data deposition
Mass spectrometry data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD009882.
Nucleotide sequences of the plasmids used in this study have been deposited in GenBank (accession numbers are given in Table 2).
Results
This study examined the influence of various promoter sequences and medium composition on the secretory production of the clostridial endoglucanase Cel8A by P. polymyxa DSM 292. Promoters from the literature were tested, and the expression medium was optimized. Furthermore, the secretome of P. polymyxa DSM 292 was analyzed by quantitative mass spectrometry in order to identify strongly secreted host proteins, whose promoters were hypothesized to be well-suited for the expression of heterologous target genes. The strategy used in this study to optimize secretory Cel8A production is illustrated in Fig. 1.
Workflow for the optimization of secretory cel8A expression in P. polymyxa DSM 292. To increase the total amount of Cel8A secreted by P. polymyxa DSM 292, a three-tiered strategy was adopted. 1) Well-characterized and published promoters from B. subtilis and B. megaterium were tested, followed by 2) systematic optimization of the expression medium using a Plackett–Burman design. In step 3), host proteins strongly secreted by the production strain under conditions selected from 2) were identified by quantitative LC-MS/MS, and the promoters of the corresponding genes were cloned and tested. The performance of each promoter was evaluated based on Cel8A activity in the supernatant as assessed using the Azo-CMC assay. The depicted protein structure of Cel8A (PDB-ID: 1CEM) was published by Alzari et al. (1996)
Construction of expression vectors
The expression plasmids used in this study are based on the broad host range plasmid pBACOV, featuring the B. subtilis promoter PaprE for heterologous expression in a wide range of bacilli, including P. polymyxa (Heinze et al. 2018). The target gene cel8A, starting from codon 36 of the ORF and lacking its native signal-peptide-coding region, was inserted into pBACOV. To identify a suitable SP for secretory production of Cel8A, an SP library was created and screened (see “Material and methods”). This way, SPLipB was selected, and the resulting plasmid pBACOV-SPLipB-cel8A served as a reference in all expression experiments.
Fourteen plasmids with different promoter and/or SP sequences were derived from plasmid pBACOV-SPLipB-cel8A (Table 2). The plasmids were transferred to P. polymyxa DSM 292 by transmating, and the influence of each promoter on the secretory production of Cel8A was examined. Since P. polymyxa has not yet been used for heterologous secretory protein production, little is known about its endogenous promoter systems. Hence, as a first step, well-characterized promoters originating from other Bacillus species were selected based on their known strength and controllability. These promoters were PxylA, PxylA+, P43, PtrnQ, Pylb, and P43P. Following this, the expression medium was optimized using a Plackett–Burman matrix. Based on a quantitative analysis of the secretome in the optimized medium, additional promoters from the P. polymyxa genome were selected (P01680, P02218, P03466, and P04737) and tested in combination with SPLipB and the respective native SP.
Analysis of the production of Cel8A in LBS medium
In the first round of optimization, the new expression plasmids pPxylA-cel8A, pPxylA+ − cel8A, pP43-cel8A, pPtrnQ-cel8A, pPylb-cel8A, and pP43P-cel8A were introduced into P. polymyxa DSM 292. The resulting strains were cultured in 20 ml of LBS medium for comparison of Cel8A production. The expression levels, which depend on the promoter strengths, were examined in terms of the resulting Cel8A activity in the culture supernatant using the Azo-CMC assay and were compared to the level obtained with the auto-inducible reference promoter PaprE.
First, the xylose-inducible promoters PxylA and PxylA+ were examined. The sequence of PxylA is identical to the promoter of the B. megaterium xyl operon and was used in combination with the gene for the XylR repressor; XylR inhibits transcription in the absence of xylose. PxylA+ is derived from PxylA and features an optimized ribosome binding site (rbs+) for B. megaterium (Malten et al. 2005).
To compare the promoters PxylA and PxylA+, clones carrying the respective plasmids were grown in LBS medium until the OD600 reached 0.3. At this point, expression was induced by addition of xylose to a final concentration of 0.5% (w/v). After incubation for 24 h, the Cel8A activity in the supernatant was measured using the Azo-CMC assay (Table 3). PxylA did not promote cel8A expression under either condition. In contrast, the CMCase activity in the supernatant of cells carrying the pPxylA+ − cel8A plasmid reached about 74% of the reference (PaprE) upon induction by xylose. No CMCase activity was observed without addition of xylose, indicating a tight regulation of the xylose-inducible promoter.
In contrast to the xylose-inducible promoters, P43, PtrnQ, and P43P are constitutive. The constructs with P43 and PtrnQ originating from B. subtilis, as well as P43P from P. polymyxa DSM 292, did not promote cel8A expression in P. polymyxa DSM 292. After incubation in LBS medium, the target protein could neither be detected in the supernatants or cell pellets, by both Western blot analysis (by means of the C-terminal His6-tag and an anti-His/AP antibody-conjugate), nor by Azo-CMC activity assay. False negative results could be excluded since the reference strain (P. polymyxa DSM 292 pBACOV-SPLipB-cel8A) and the negative control (P. polymyxa DSM 292 pBACOV) produced the expected results.
Replacement of PaprE with the auto-inducible Pylb promoter resulted in increased amounts of extracellular Cel8A upon culturing in LBS medium. The Cel8A activity produced by P. polymyxa DSM 292 pPylb-cel8A reached a value of 2082 ± 249 U/l, a 2.3-fold increase over the reference plasmid pBACOV-SPLipB-cel8A (PaprE).
Medium optimization
A second step to increase the total amount of active Cel8A produced was a systematic optimization of the expression medium. By using a Plackett–Burman matrix, 12 expression media covering 11 factors (Table 1) were designed and tested. The Plackett–Burman matrix and the resulting Cel8A activities from P. polymyxa DSM 292 pBACOV-SPLipB-cel8A are presented in Fig. 2. Medium composition had a substantial effect on the Cel8A activity in the supernatant. While productivity was low with most tested media, medium 2 produced an almost fivefold increase in Cel8A production compared to medium 0 (Fig. 2) and a 1.8-fold increase compared to LBS. The composition of medium 2 is listed in “Materials and methods.”
Plackett–Burman matrix for medium optimization and resulting CMCase activities in the supernatant of P. polymyxa DSM 292 pBACOV-SPLipB-cel8A cultures. Twelve media were created to test different combinations of the 11 factors. The specified levels of the 11 factors for optimization are listed in Table 1. The numbers indicate the specified levels of a given factor: 0 = initial level; − 1 = low level; and 1 = high level. To test the productivity in each medium, P. polymyxa DSM 292 pBACOV-SPLipB-cel8A was incubated in each medium for 24 h at 180 rpm, and the Cel8A activity in the supernatant was determined using the Azo-CMC assay. The given values are the means ± standard deviation of three biological replicates
When incubated in medium 2 for 24 h, P. polymyxa DSM 292 pBACOV-SPLipB-cel8A reached a higher final OD600 at 37 °C than at 40 °C, the temperature at which this medium was initially tested. Accordingly, the measured Cel8A activity in the supernatant was significantly higher at 37 °C (data not shown). Therefore, medium 2 at 37 °C was chosen as by far the best expression medium for further experiments.
Since PxylA+ and Pylb performed well in LBS medium, the expression of cel8A under control of these two promoters was also analyzed in medium 2 at 37 °C. Upon induction with xylose, the Cel8A activity in the supernatant of P. polymyxa DSM 292 pPxylA+ − cel8A reached 103 ± 3.26 U/l, whereas the activity without addition of xylose was 34.7 ± 4.08 U/l. In comparison with LBS medium, the amount of Cel8A produced under control of PxylA+ was more than six times lower in medium 2.
For Pylb, the Azo-CMC assay revealed a Cel8A activity in the supernatant of 1904 ± 55.4 U/l in medium 2. This value is in the same range as that observed with Pylb in LBS medium (2082 ± 249 U/l), which indicates that medium 2 did not increase the productivity in this case. Furthermore, the reference promoter PaprE generated a Cel8A activity of 2833 ± 215 U/l (medium 2, 37 °C), indicating a better performance of PaprE over Pylb under these conditions.
Identification of abundant proteins in the supernatant of P. polymyxa DSM 292 for promoter selection
Since the combination of optimized medium 2 and the best promoters from the first round of optimization (i. e., Pylb and PxylA+) did not result in an increased production of Cel8A, a third approach was used to identify promoters that would lead to high levels of Cel8A in medium 2. We hypothesized that promoters regulating the expression of highly abundant host proteins in the supernatant of P. polymyxa DSM 292 may also be well-suited for the secretory production of heterologous enzymes. In order to identify such host proteins, the supernatant of P. polymyxa DSM 292 pBACOV-SPLipB-cel8A grown in medium 2 was analyzed by quantitative mass spectrometry (LC-MS/MS), followed by absolute label-free protein abundance estimation using the intensity-based absolute quantification algorithm iBAQ. The analysis was performed with three biological replicates, and the data were deposited at the Proteome Xchange Consortium (http://proteomecentral.proteomexchange.org, dataset identifier PXD009882). A detailed overview of the quantitative protein results is available in Supplementary Table S2. In total, 1078 proteins were quantified with different abundances (Fig. 3). Secretory SPs were predicted for 60 of the identified proteins using SignalP 4.1 (Petersen et al. 2011), indicating that these proteins are actively secreted by P. polymyxa DSM 292. These 60 proteins (open circles in Fig. 3) were all quantifiable with high concentrations in the supernatant, i.e., they all ranked within the 30% most highly concentrated proteins of the detected P. polymyxa DSM 292 proteome (Fig. 3). The genome of P. polymyxa DSM 292 contains a total of 418 genes with a predicted SP (SignalP 4.1; Petersen et al. 2011; Supplementary Table S2). This means that about 14% of the repertoire of secretory genes was expressed and the respective proteins were detected under the experimental conditions used. The detection of the 1018 proteins lacking a predicted SP was most likely due to the ongoing cell lysis during culturing. These proteins are presumably not actively secreted by P. polymyxa.
Rank plot of proteins detected in the supernatant of P. polymyxa DSM 292 pBACOV-SPLipB-cel8A determined by LC-MS/MS using intensity-based absolute quantification (iBAQ). One thousand seventy-eight proteins were quantified. Open circles indicate the 60 proteins for which a SP was predicted by SignalP 4.1 (Petersen et al. 2011). These secreted proteins rank among the 380 most abundant proteins in the sample. The heterologous target protein Cel8A (triangle) was the fourth most abundant protein in the supernatant. Solid circles indicate intracellular proteins. The four most abundant secreted host proteins (PPOLYM_02218, PPOLYM_01680, PPOLYM_03468, and PPOLYM_04737) are labeled with dashed lines. The promoters regulating the expression of their respective genes were chosen as candidates for heterologous cel8A expression
The six most abundant proteins in the P. polymyxa DSM 292 pBACOV-SPLipB-cel8A supernatant were the host proteins PPOLYM_02218, PPOLYM_00910, PPOLYM_01680, PPOLYM_03468, and PPOLYM_04737 (Table 4) and the heterologous target protein Cel8A. The protein sequences were analyzed by BlastP 2.7.1.+ (Altschul et al. 1997), which revealed that PPOLYM_00910 is a copper amine oxidase-like protein, PPOLYM_01680 has a conserved Peptidase_C92 domain (pfam05708), and PPOLYM_04737 is a pectate lyase homolog belonging to the family PL1 subfamily 8 (determined using dbCAN; Yin et al. 2012). PPOLYM_02218 and PPOLYM_03468 are annotated as hypothetical proteins. An SP was predicted for all proteins except PPOLYM_00910. Therefore, the promoters of ORFs PPOLYM_02218, PPOLYM_01680, PPOLYM_03468, and PPOLYM_04737 were selected as suitable candidates for the regulation of heterologous cel8A expression. The identification of the most abundant secretory proteins of P. polymyxa DSM 292 also provides access to a collection of efficient SP sequences that might be useful for optimization of heterologous protein secretion.
The P. polymyxa DSM 292 promoters P01680, P02218, P03468, and P04737
Fragments of 250–500 bp upstream of the respective coding sequences of PPOLYM_02218, PPOLYM_01680, PPOLYM_03468, and PPOLYM_04737 were amplified from P. polymyxa DSM 292 genomic DNA by PCR and were inserted into pBACOV-SPLipB-cel8A to replace PaprE. Additional versions of the resulting plasmids were created by inserting not only the individual promoter sequences but also the respective predicted SP coding sequences to replace PaprE and SPLipB (Table 2). The cloned regions were assumed to contain all sequences needed for promoter activity. Table 5 provides an overview of hypothetical regulatory elements present in the cloned promoter sequences. Ribosome binding sites were identified by searching for sequences homologous to the P. polymyxa Shine–Dalgarno sequence (5′ UGGAGA 3′) at a distance of 6–11 bp from the start codon. The − 35 elements and − 10 elements were predicted using Bprom (Solovyev and Salamov 2011).
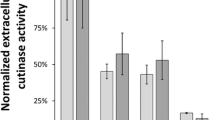
The expression of heterologous cel8A under control of the promoters P01680, P02218, P03468, and P04737 in combination with the original SP or SPLipB was examined in medium 2 at 37 °C (Fig. 4). Interestingly, a strong dependence on the SP was observed for each promoter. While the combination of P01680 with its native SP, SP01680, resulted in a 12.5-fold higher Cel8A activity than P01680SPLipB, SPLipB yielded better results in combination with P02218, P04737 (a 2.7-fold increase in both cases compared to SP02218 and SP04737) and P03468 (an 8.1-fold increase). The highest Cel8A activities were obtained with P01680SP01680 and P03468SPLipB. For P01680SP01680, a Cel8A activity of 2660 ± 245 U/l was measured, which is similar to the amount of Cel8A produced under control of PaprE. The best result was obtained with pP03468-LipB-cel8A (P03468SPLipB) with a specific activity of 5475 ± 411 U/l, representing an increase of 93% over the reference (PaprE, 2883 ± 215 U/l).
Influence of selected promoter sequences and SPs on heterologous cel8A expression in P. polymyxa DSM 292. The promoter sequences were derived from the most abundant secreted host proteins in the P. polymyxa DSM 292 secretome (Fig. 3 and Table 4). Transmates of P. polymyxa DSM 292 carrying pP01680-LipB-cel8A, pP01680-nat-cel8A (= P01680), pP02218-LipB-cel8A, pP02218-nat-cel8A (= P02218), pP03468-LipB-cel8A, pP03468-nat-cel8A (= P03468) or pP04737-LipB-cel8A, and pP04737-nat-cel8A (= P04737) were incubated in medium 2 at 37 °C and 180 rpm, for 24 h. n.c.: negative control, supernatant of P. polymyxa DSM 292 pBACOV. The bars represent the means of three biological replicates (except P03468-SPLipB, 2 replicates), each measured in triplicate, ± standard deviation
Discussion
In this study, we tested new promoter sequences that regulate the expression of abundant proteins in the secretome of P. polymyxa DSM 292 to optimize expression and secretion of the heterologous gene cel8A from C. thermocellum. To the best of our knowledge, this approach is the first report of a quantitative LC-MS/MS analysis for the selection of bacterial promoters for heterologous gene expression. Usually, promoters are identified by dedicated transcriptome analyses, for example, using microarrays (Nicolas et al. 2012; Yu et al. 2015) or RNA-seq (Creecy and Conway 2015). These methods are designed for the examination of gene expression and provide detailed information about the regulation and promoter strength; however, they have two drawbacks. First, unless suitable transcriptome data are published, the acquisition of transcriptome data is expensive and, second, they only provide information about the intracellular process of transcription and promoter strength. However, secretory protein production is a complex process that not only depends on the transcription level but also on the efficient translation and secretion. The rationale was to seek promoter regions not solely based on promoter strength, but to find regulatory elements that are suitable for efficient secretory protein production, involving all steps from transcription to secretion. This was achieved by identifying the most abundant host proteins in the supernatant of P. polymyxa DSM 292 pBACOV-SPLipB-cel8A by quantitative LC-MS/MS. Indeed, five of the six most abundant proteins in the supernatant comprised a predicted secretion SP (SignalP 4.1; Petersen et al. 2011), one of them being the heterologous target protein Cel8A.
Expression was examined with the promoters of the four most abundant secreted proteins (PPOLYM_01680, PPOLYM_02218, PPOLYM_03468, and PPOLYM_04737) in combination with their respective native SPs or with SPLipB, selected from a library of B. subtilis SPs. The results revealed that depending on the promoter sequence, either the native SP or SPLipB provided better performance with regard to Cel8A secretion. It is a well-known fact that the reliable prediction of well-suited SPs for secretion of a given target protein is notoriously difficult to realize (Brockmeier et al. 2006; Degering et al. 2010; Song et al. 2015). We now add the observation that the efficiency of a given SP also depends on the promoter sequence; while SPLipB is suitable for secretion of Cel8A in combination with the promoters PaprE, P02218, P04737, and P03468, it performed poorly with promoter P01680. Conversely, P01680SP01680-mediated cel8A expression led to high amounts of secreted Cel8A. Thus, the efficiency of a given SP does not only depend on the respective target protein but also on the genetic context, that is, the promoter driving expression.
Our hypothesis that the promoter of a highly secreted host protein could be a good candidate for heterologous gene expression, and secretion was strengthened when we tested P03468. In combination with SPLipB, this promoter generated an extracellular Cel8A activity of 5475 ± 411 U/l in the supernatant of P. polymyxa DSM 292 grown in medium 2. In summary, after combining the results from the medium optimization and from the mass spectrometry approach, the secretory production of Cel8A by P. polymyxa DSM 292 could be increased 6.2-fold from 884 ± 29.0 U/l (PaprESPLipB in LBS medium) to 5475 ± 411 U/l (P03468SPLipB in medium 2), surpassing the level of PPOLYM_03468, the protein natively expressed by P03468. The yield of Cel8A in the supernatant of P. polymyxa DSM 292 pP03468-LipB-cel8A in medium 2 can be estimated at about 348 mg/l, by inference from the specific CMCase activity of the purified Cel8A reference produced in E. coli. However, the specific activity of Cel8A produced in E. coli can differ from the one of Cel8A secreted by P. polymyxa DSM 292 which was not determined herein. Accordingly, the actual yield (in mg/l) may vary.
The expression medium was optimized using a Plackett–Burman matrix with 11 factors. Statistical methods, including Plackett–Burman matrices, are commonly used for optimization of culture media for the production of proteins and small molecules (Singh et al. 2017). Yield improvements of factors of 1.45–11.5 have been reported from using Plackett–Burman matrices alone (Narasimhan and Shivakumar 2012; Singh et al. 2017). Plackett–Burman screenings are usually the starting points for further optimization, for example, using response surface methodologies (RSM), such as Box–Behnken design or central composite design, which in most cases leads to further yield improvement by factors of 3.3–13.4 (Fang et al. 2010; Farhat-Khemakhem et al. 2012; Singh et al. 2017). Since this study focused on the identification of promoters for improvement of Cel8A production, we did not include an RSM approach. Nevertheless, the medium optimization here resulted in a production increase from 884 ± 29.0 U/l in LBS with the reference strain to 2883 ± 215 U/l in medium 2 at 37 °C. Thus, the medium optimization alone led to more than a threefold improvement in the productivity.
The well-characterized promoters P43 and PtrnQ from B. subtilis proved to be unsuitable for Cel8A production in P. polymyxa DSM 292. The target protein Cel8A remained undetected, in both the supernatants and the cell pellets. This implies that not the secretion process but cel8A expression itself was unsuccessful. In B. subtilis, P43 controls expression of the cdd gene (Song and Neuhard 1989), which encodes for a cytidine deaminase. A TBLASTN search identified a homologous gene in P. polymyxa DSM 292. The respective promoter was termed P43P and tested; however, it failed to promote cel8A expression in our host strain. The P43P-fragment which was transferred into the expression vector might be a truncated version of the promotor and might lack critical regulatory elements or transcription factor binding sites.
The xylose-inducible promoter PxylA+ from B. megaterium featuring an optimized ribosome binding site (rbs+) supported the secretory cel8A expression in P. polymyxa DSM 292 in LBS medium. In contrast, the native sequence (PxylA) did not promote Cel8A production, suggesting that the ribosome binding site plays a crucial role in the process of cel8A gene expression in P. polymyxa DSM 292. Cel8A activity in the supernatants obtained from cultures using PxylA+ reached about 74% of the reference promoter sample (PaprE). As expected, addition of xylose was essential for target gene expression in P. polymyxa DSM 292, with no detection of leaky Cel8A production in the xylose-deficient cultures. This demonstrates that the xylose-inducible expression system derived from B. megaterium functions in the host strain P. polymyxa DSM 292 when the optimized ribosome binding site (Malten et al. 2005) is employed.
In medium 2, cel8A expression under control of PxylA+ was reduced compared to the reference promoter PaprE regardless of the addition of xylose, which indicates a loss of promoter inducibility. The reduced target gene expression may be attributed to catabolite repression mediated by the glucose present in medium 2. Glucose was reported to be an anti-inducer of xylA expression in B. megaterium as it competes with xylose for the interaction with the xylose repressor XylR (Dahl et al. 1995). Additionally, the prevented uptake of alternative carbon sources is a common mechanism of catabolite repression, as reviewed by Görke and Stülke (2008). Transcriptomic analysis of carbohydrate utilization by Paenibacillus sp. JDR-2 revealed a glucose-dependent downregulation of genes involved in xylose uptake (Sawhney et al. 2016). Similar effects might exist in P. polymyxa DSM 292. Thereby, medium 2 containing glucose is unsuitable for target gene expression under control of a xylose-inducible promoter. If an inducible system is needed, for example, for the production of compounds that are toxic to P. polymyxa, PxylA+ can be used in combination with a glucose-free medium, such as LBS, or in medium 2 with an alternative carbon source.
Another promoter functional in P. polymyxa DSM 292 is Pylb. This auto-inducible promoter was initially discovered in a genome-scale microarray-based screening of B. subtilis promoters by Yu et al. (2015). The promoter activity of Pylb was discovered to be increased by 136% compared to that of PaprE upon growth of the P. polymyxa DSM 292 host in LBS medium. Pylb was therefore the most productive promoter in LBS medium. In medium 2, Cel8A production levels reached by Pylb were similar to those observed in LBS (approximately 2000 U/l), but were less than the PaprE-mediated Cel8A production in medium 2. This indicates that the composition of medium 2 does not have a beneficial effect on the secretory cel8A expression under control of Pylb in P. polymyxa DSM 292.
We were also able to demonstrate the secretory production of three additional C. thermocellum enzymes in P. polymyxa DSM 292, namely Cel9D, Cel9R, and Xgh74A (data not shown). This indicates the suitability of P. polymyxa DSM 292 as a new production host for a variety of enzymes, such as glycoside hydrolases.
In conclusion, we have demonstrated that P. polymyxa DSM 292 can be used as a novel host organism for secretory production of a heterologous enzyme. To optimize the production level, (i) new promoters from the genome of P. polymyxa DSM 292 were identified based on a quantitative secretome analysis, (ii) several characterized promoters from B. subtilis and B. megaterium were tested, and (iii) the expression medium was optimized using a Plackett–Burman matrix. The most significant contributions to productivity enhancement were made by medium optimization and use of the promoter P03468 in combination with SPLipB. Furthermore, a xylose-regulated promoter from B. megaterium was shown to be active and inducible in P. polymyxa DSM 292 after modification of its Shine–Dalgarno sequence. Our results can be the basis for further development of P. polymyxa DSM 292 as a production host for biotechnological enzymes.
References
Ahmann D, Dorgan JR (2007) Bioengineering for pollution prevention through development of biobased energy and materials state of the science report. Ind Biotechnol 3:218–259. https://doi.org/10.1089/ind.2007.3.218
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Alzari PM, Souchon H, Dominguez R (1996) The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4:265–275
Bayer EA, Lamed R, Himmel ME (2007) The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotechnol 18:237–245. https://doi.org/10.1016/j.copbio.2007.04.004
Bertani G (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300
Bien TLT, Tsuji S, Tanaka K, Takenaka S, Yoshida K (2014) Secretion of heterologous thermostable cellulases in Bacillus subtilis. J Gen Appl Microbiol 60:175–182. https://doi.org/10.2323/jgam.60.175
Brockmeier U, Caspers M, Freudl R, Jockwer A, Noll T, Eggert T (2006) Systematic screening of all signal peptides from Bacillus subtilis: a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria. J Mol Biol 362:393–402. https://doi.org/10.1016/j.jmb.2006.07.034
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. https://doi.org/10.1038/nbt.1511
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. https://doi.org/10.1021/pr101065j
Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. https://doi.org/10.1074/mcp.M113.031591
Creecy JP, Conway T (2015) Quantitative bacterial transcriptomics with RNA-seq. Curr Opin Microbiol 23:133–140. https://doi.org/10.1016/J.MIB.2014.11.011
Dahl MK, Schmiedel D, Hillen W (1995) Glucose and glucose-6-phosphate interaction with Xyl repressor proteins from Bacillus spp. may contribute to regulation of xylose utilization. J Bacteriol 177:5467–5472
Degering C, Eggert T, Puls M, Bongaerts J, Evers S, Maurer K-H, Jaeger K-E (2010) Optimization of protease secretion in Bacillus subtilis and Bacillus licheniformis by screening of homologous and heterologous signal peptides. Appl Environ Microbiol 76:6370–6376. https://doi.org/10.1128/AEM.01146-10
Fang TJ, Liao B-C, Lee S-C (2010) Enhanced production of xylanase by Aspergillus carneus M34 in solid-state fermentation with agricultural waste using statistical approach. New Biotechnol 27:25–32. https://doi.org/10.1016/J.NBT.2009.09.008
Farhat-Khemakhem A, Farhat MB, Boukhris I, Bejar W, Bouchaala K, Kammoun R, Maguin E, Bejar S, Chouayekh H (2012) Heterologous expression and optimization using experimental designs allowed highly efficient production of the PHY US417 phytase in Bacillus subtilis 168. AMB Express 2:10. https://doi.org/10.1186/2191-0855-2-10
Ferrari E, Henner DJ, Perego M, Hoch JA (1988) Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol 170:289–295
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. https://doi.org/10.1038/nmeth.1318
Görke B, Stülke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. https://doi.org/10.1038/nrmicro1932
Grady EN, MacDonald J, Liu L, Richman A, Yuan Z-C (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Factories 15:203. https://doi.org/10.1186/s12934-016-0603-7
Harwood CR (1992) Bacillus subtilis and its relatives: molecular biological and industrial workhorses. Trends Biotechnol 10:247–256. https://doi.org/10.1016/0167-7799(92)90233-L
Häßler T, Schieder D, Pfaller R, Faulstich M, Sieber V (2012) Enhanced fed-batch fermentation of 2,3-butanediol by Paenibacillus polymyxa DSM 365. Bioresour Technol 124:237–244. https://doi.org/10.1016/j.biortech.2012.08.047
Heinze S, Kornberger P, Graetz C, Schwarz WH, Zverlov VV, Liebl W (2018) Transmating: conjugative transfer of a new broad host range expression vector to various Bacillus species using a single protocol. BMC Microbiol 18:56. https://doi.org/10.1186/s12866-018-1198-4
Jan J, Valle F, Bolivar F, Merino E (2000) Characterization of the 5′ subtilisin (aprE) regulatory region from Bacillus subtilis. FEMS Microbiol Lett 183:9–14. https://doi.org/10.1111/j.1574-6968.2000.tb08926.x
Joliff G, Edelman A, Klier A, Rapoport G (1989) Inducible secretion of a cellulase from Clostridium thermocellum in Bacillus subtilis. Appl Environ Microbiol 55:2739–2744
Küppers T, Steffen V, Hellmuth H, O’Connell T, Bongaerts J, Maurer K-H, Wiechert W (2014) Developing a new production host from a blueprint: Bacillus pumilus as an industrial enzyme producer. Microb Cell Factories 13:46. https://doi.org/10.1186/1475-2859-13-46
Lee S-J, Pan J-G, Park S-H, Choi S-K (2010) Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J Biotechnol 149:16–20. https://doi.org/10.1016/j.jbiotec.2010.06.021
Leis B, Held C, Bergkemper F, Dennemarck K, Steinbauer R, Reiter A, Mechelke M, Moerch M, Graubner S, Liebl W, Schwarz WH, Zverlov VV (2017) Comparative characterization of all cellulosomal cellulases from Clostridium thermocellum reveals high diversity in endoglucanase product formation essential for complex activity. Biotechnol Biofuels 10:240. https://doi.org/10.1186/s13068-017-0928-4
Malten M, Hollmann R, Deckwer W-D, Jahn D (2005) Production and secretion of recombinant Leuconostoc mesenteroides dextransucrase DsrS in Bacillus megaterium. Biotechnol Bioeng 89:206–218. https://doi.org/10.1002/bit.20341
Narasimhan A, Shivakumar S (2012) Optimization of chitinase produced by a biocontrol strain of Bacillus subtilis using Plackett–Burman design. Eur J Exp Biol 2:861–865
Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Hartig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RAT, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stulke J, Wiegert T, Devine KM, Wilkinson AJ, Maarten van Dijl J, Hecker M, Volker U, Bessieres P, Noirot P (2012) Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. https://doi.org/10.1126/science.1206848
Nijland R, Lindner C, van Hartskamp M, Hamoen LW, Kuipers OP (2007) Heterologous production and secretion of Clostridium perfringens β-toxoid in closely related Gram-positive hosts. J Biotechnol 127:361–372. https://doi.org/10.1016/j.jbiotec.2006.07.014
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Rütering M, Cress BF, Schilling M, Rühmann B, Koffas MAG, Sieber V, Schmid J (2017) Tailor-made exopolysaccharides—CRISPR-Cas9 mediated genome editing in Paenibacillus polymyxa. Synth Biol 2:ysx007. https://doi.org/10.1093/synbio/ysx007
Rygus T, Hillen W (1991) Inducible high-level expression of heterologous genes in Bacillus megaterium using the regulatory elements of the xylose-utilization operon. Appl Microbiol Biotechnol 35:594–599. https://doi.org/10.1007/BF00169622
Sawhney N, Crooks C, Chow V, Preston JF, St John FJ (2016) Genomic and transcriptomic analysis of carbohydrate utilization by Paenibacillus sp. JDR-2: systems for bioprocessing plant polysaccharides. BMC Genomics 17:131. https://doi.org/10.1186/s12864-016-2436-5
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17
Schumann W (2007) Production of recombinant proteins in Bacillus subtilis. Adv Appl Microbiol 62:137–189. https://doi.org/10.1016/S0065-2164(07)62006-1
Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473:337–342. https://doi.org/10.1038/nature10098
Schwarz WH (2001) The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol 56:634–649
Schwarz WH, Gräbnitz F, Staudenbauer WL (1986) Properties of a Clostridium thermocellum endoglucanase produced in Escherichia coli. Appl Environ Microbiol 51:1293–1299
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. https://doi.org/10.1038/nprot.2006.468
Shoham Y, Lamed R, Bayer EA (1999) The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol 7:275–281
Singh V, Haque S, Niwas R, Srivastava A, Pasupuleti M, Tripathi CKM (2017) Strategies for fermentation medium optimization: an in-depth review. Front Microbiol 7:2087. https://doi.org/10.3389/fmicb.2016.02087
Solovyev V, Salamov A (2011) Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW (ed) Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, New York, pp 61–78
Song B-H, Neuhard J (1989) Chromosomal location, cloning and nucleotide sequence of the Bacillus subtilis cdd gene encoding cytidine/deoxycytidine deaminase. MGG Mol Gen Genet 216:462–468. https://doi.org/10.1007/BF00334391
Song Y, Nikoloff JM, Zhang D (2015) Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis. J Microbiol Biotechnol 25:963–977. https://doi.org/10.4014/jmb.1501.01028
Song Y, Nikoloff JM, Fu G, Chen J, Li Q, Xie N, Zheng P, Sun J, Zhang D (2016) Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications. PLoS One 11:e0158447. https://doi.org/10.1371/journal.pone.0158447
Soutschek-Bauer E, Staudenbauer WL (1987) Synthesis and secretion of a heat-stable carboxymethylcellulase from Clostridium thermocellum in Bacillus subtilis and Bacillus stearothermophilus. Mol Gen Genet 208:537–541
Stammen S, Müller BK, Korneli C, Biedendieck R, Gamer M, Franco-Lara E, Jahn D (2010) High-yield intra- and extracellular protein production using Bacillus megaterium. Appl Environ Microbiol 76:4037–4046. https://doi.org/10.1128/AEM.00431-10
Valle F, Ferrari E (1989) Subtilisin: a redundantly temporally regulated gene? In: Smith I, Slepecky RA, Setlow P (eds) Regulation of procaryotic development. American Society for Microbiology, Washington, DC, pp 131–146
Wang P-Z, Doi RH (1984) Overlapping promoters transcribed by Bacillus subtilis sigma55 and sigma37 RNA polymerase holoenzymes during growth and stationary phases. J Biol Chem 259:8619–8625
Wood TM, Bhat KM (1988) Methods for measuring cellulase activities. Methods Enzymol 160:87–112. https://doi.org/10.1016/0076-6879(88)60109-1
Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y (2012) dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. https://doi.org/10.1093/nar/gks479
Yu X, Xu J, Liu X, Chu X, Wang P, Tian J, Wu N, Fan Y (2015) Identification of a highly efficient stationary phase promoter in Bacillus subtilis. Sci Rep 5:18405. https://doi.org/10.1038/srep18405
Zverlov VV, Schwarz WH (2004) The Clostridium thermocellum cellulosome -the paradigm of a multienzyme complex. In: Ohmiya K, Sakka K, Karita S, Kimura T, Sakka M, YO (eds) Biotechnology of lignocellulose degradation and biomass utilization. Uni Pub. Co. Ltd., Tokyo, pp 137–147
Acknowledgments
The authors thank Patricia Krähe and Benedikt Leis for preparing and providing the sample of Cel8A, produced in E. coli, which was used to establish the reference curve for the Azo-CMC assay. We would like to thank Hermine Kienberger for her excellent technical assistance in proteomic sample preparation.
Funding
This work was funded by the German Federal Ministry of Education and Research (FKZ 031A555, Bioeconomy International 2014).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Heinze, S., Zimmermann, K., Ludwig, C. et al. Evaluation of promoter sequences for the secretory production of a Clostridium thermocellum cellulase in Paenibacillus polymyxa. Appl Microbiol Biotechnol 102, 10147–10159 (2018). https://doi.org/10.1007/s00253-018-9369-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9369-7