Abstract
The co-culture system of denitrifying anaerobic methane oxidation (DAMO) and anaerobic ammonium oxidation (Anammox) has a potential application in wastewater treatment plant. This study explored the effects of permutation and combination of nitrate, nitrite, and ammonium on the culture enrichment from freshwater sediments. The co-existence of NO3 −, NO2 −, and NH4 + shortened the enrichment time from 75 to 30 days and achieved a total nitrogen removal rate of 106.5 mg/L/day on day 132. Even though ammonium addition led to Anammox bacteria increase and a higher nitrogen removal rate, DAMO bacteria still dominated in different reactors with the highest proportion of 64.7% and the maximum abundance was 3.07 ± 0.25 × 108 copies/L (increased by five orders of magnitude) in the nitrite reactor. DAMO bacteria showed greater diversity in the nitrate reactor, and one was similar to M. oxyfera; DAMO bacteria in the nitrite reactor were relatively unified and similar to M. sinica. Interestingly, no DAMO archaea were found in the nitrate reactor. This study will improve the understanding of the impact of nitrogen source on DAMO and Anammox co-culture enrichment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Methane is a powerful greenhouse gas, whose global warming potential is about 25 times more than that of carbon dioxide. The ocean covers 70% of the earth’s surface, and more than 90% of escaped methane from marine sediment is consumed by anaerobic oxidation through the methane process (Knittel and Boetius 2009). Anaerobic oxidation of methane (AOM) represents a large methane sink, which considerably reduces greenhouse gas emissions. For a long time, the only electron acceptor during AOM was thought to be sulfate until Raghoebarsing et al. (2006) discovered that nitrate and nitrite could also be reduced by methane as the electron donor. The process became known as the denitrifying anaerobic methane oxidation (DAMO) process. It is widely accepted that DAMO microorganisms are divided to two kinds based on the electron acceptor: one is DAMO archaea transforming nitrate to nitrite, and the other is DAMO bacteria reducing nitrite to nitrogen gas (Ettwig et al. 2010; Haroon et al. 2013). All DAMO archaea are enriched under nitrate conditions as they disappear when only nitrite instead of nitrite and nitrate is provided to DAMO archaea and DAMO bacteria in mixed culture (Ettwig et al. 2008; Hu et al. 2011). Most DAMO bacteria are enriched under nitrite conditions, but one recent study has reported that DAMO bacteria are also successfully enriched when only nitrate is supplied (Wang et al. 2016), but this result needs further confirmation.
DAMO microorganisms are widespread in nature, such as in freshwater lake sediment, halophilic marine environments, paddy soils, agricultural drainage water, wetland ecosystems, wastewater sludge, etc. (Ettwig et al. 2009; He et al. 2015a; He et al. 2015b; Luesken et al. 2011b; Zhu et al. 2010). Furthermore, it is well known that the anaerobic ammonium oxidation (Anammox) process, which couples ammonium oxidation with nitrite reduction under anaerobic conditions, represents 50% of the nitrogen turnover in the marine environment (Schmid et al. 2007). Anammox bacteria are widespread in various natural ecosystems, such as marine sediments, freshwater sediments, wastewater treatment plants, etc. (Hu et al. 2012; Mulder et al. 1995; Schmid et al. 2007). In the traditional denitrifying process, N2O is usually released to the atmosphere and it is a powerful (300 times of CO2) greenhouse gas that can deplete ozone in the stratosphere. However, nitrous oxide is not produced in DAMO and Anammox processes (Ettwig et al. 2010; Kartal et al. 2011).
Nitrate, nitrite, and ammonium are important nitrogen forms in the environment, and these three different nitrogen forms often co-exist in nature. DAMO and Anammox microorganisms inhabit the same environment, which means there may be interactions between them. The Anammox process could interact with the DAMO process via nitrite, because nitrite is not only a substrate for Anammox bacteria, but is also the reduced product of DAMO archaea and the electron acceptor for DAMO bacteria. Furthermore, nitrate is a by-product of the Anammox process and the electron acceptor for DAMO archaea. The DAMO-combined Anammox system is a potential biological nitrogen removal technology, utilizing methane as the only electron donor to completely remove nitrate, nitrite, and ammonium in anaerobic condition. For example, the effluent from an anaerobic digester containing methane and ammonium is suitable for this system.
Different nitrogen source conditions will affect community composition. It has been reported that Anammox bacteria were enriched in DAMO bacterial cultures after ammonium addition (Luesken et al. 2011a) and that DAMO bacteria were enriched in Anammox bioreactors after methane had been provided (Zhu et al. 2011). DAMO archaea, DAMO bacteria, and Anammox bacteria have been simultaneously enriched using a mixed inoculum that included methanogenic sludge and activated sludge under nitrate, ammonium, and methane conditions (Ding et al. 2014), and proportions of the three microorganisms were changed after a 2-year operation in a membrane biofilm reactor (Shi et al. 2013). When ammonium/nitrate/methane or ammonium/nitrite/methane were provided to mixed cultures of DAMO archaea, DAMO bacteria, and Anammox bacteria, the DAMO bacteria disappeared under both conditions after 259 days of operation, which showed that Anammox bacteria outcompeted DAMO bacteria (Hu et al. 2015). However, Chen et al. (2014) reported that DAMO bacteria had a relatively higher affinity for nitrite than Anammox bacteria. The contradiction suggests that different nitrogen sources may play important roles in the DAMO culture enrichment process and in the microbial community and that the nitrogen source effect is far more complicated than previously thought. However, there has been no systematic study of nitrogen source influence on DAMO culture enrichment, such as its effects on nitrogen removal rate or the microbial community.
In this study, we explored the impact of nitrogen source on the DAMO culture and Anammox bacteria enrichment process. Nitrate, nitrite, and ammonium were grouped and provided in different combinations to different reactors. The reactor performances were monitored, and the microbial communities were analyzed using quantitative polymerase chain reaction (qPCR), 16s ribosomal RNA (rRNA) amplicon sequencing, phylogenetic analysis, and statistical analysis. Investigating the effect of nitrogen source on DAMO microorganisms and Anammox bacteria during the enrichment period should provide a better understanding of nitrogen source impact on their enrichment.
Materials and methods
Inoculum and the enrichment process
The inoculum was taken from a freshwater lake (31°16′38.23″N and 120°43′21.17″E) sediment in Suzhou, China, in July 2014. The sediment was sifted with a 280-μm sieve to remove large impurities, and the permeated liquid was washed three times with a mineral medium whose composition has been described in a previous study (Fu et al. 2015) to remove the dissolved organic carbon. The above steps took place in an anaerobic environmental chamber (BACTRON300 SHEL LAB, Sheldon Manufacturing, USA).
To compare the nitrogen source effect on DAMO microorganism enrichment (Table 1), NO3 −-N, NO2 −-N, and NH4 +-N were added to the N1 reactor; NO3 −-N and NH4 +-N were added to the N2 reactor; NO3 −-N was added to the N3 reactor; NO2 −-N and NH4 +-N were added to the N4 reactor; and NO2 −-N was added to the N5 and N6 reactors. The N6 reactor contained nitrogen gas as the control, and the other reactors had a methane atmosphere. The NO sample was the inoculum before enrichment. The inoculum was transferred to the six reactors, which had a total volume of 500 mL and a working volume of 300 mL. The reactors were continually stirred by magnetic stirrers at about 200 rpm. After the reactors were settled for 12 h, a total of 100 mL supernatant was exchanged with fresh anaerobic mineral medium every month. Finally, the reactors were sparged with CH4/CO2 (95/5, v/v) or N2/CO2 (95/5, v/v) for more than 15 min to ensure anaerobic conditions. The initial concentrations of nitrate, nitrite, and ammonium in the reactors were 50, 10, and 50 mg N/L, respectively. The inoculum concentration in the reactors was 1.12 g volatile suspended solids/L. The methane consumption in the reactors was monitored to evaluate DAMO activity over the 140–165-day duration of the experiment.
Chemical analysis
Nitrate, nitrite, and ammonium levels in the liquid were analyzed using a water quality autoanalyzer (Thermo Fisher Scientific, Aquakem 200, Finland), and methane in the headspace was measured by gas chromatography (Fuli, 9790, Zhejiang, China). The detailed methods have been described previously (Fu et al. 2015).
Biological analysis
DNA extraction, PCR, quantitative PCR, and phylogenetic analysis
Liquid samples (12 mL) were sampled from the six reactors on day 0 (NO) and the 165th day (N1–N6) and centrifuged at 8000×g for 5 min to harvest the suspended solids for DNA extraction using a PowerSoil DNA Isolation Kit (Mo Bio Laboratories, USA).
The PCR method was utilized to evaluate the existence of DAMO microorganisms and Anammox bacteria. Primers DP142F and DP779R were used to detect DAMO archaea (Ding et al. 2015), primers 202F and 1043R were used to detect DAMO bacteria (Ettwig et al. 2009), primers Amx368F and Amx820R were used to detect Anammox bacteria (Wang et al. 2015), and primers 27F and 1492R were used to amplify the bacteria (Lane 1991).
The qPCR was performed on a LightCycler480 Software Setup (Roche, Basel, Switzerland) using SG Fast qPCR Master Mix (Bio Basic Inc., Markham, Canada) to estimate the copy numbers for DAMO bacteria and Anammox bacteria. The extracted DNA was used as the template. The primer pairs, qP1F and qP2R, were based on the 16S rRNA gene for DAMO bacteria, and hzsB_396F and hzsB_742R were based on the Anammox hzsB gene for Anammox bacteria (Wang et al. 2012). The double time was roughly calculated using qPCR results based on the equation of DT = T × lg2 / lg(Nt/N0), where DT is the double time, T is the time and its unit is day, and Nt and N0 are copy numbers in the final and initial times, respectively.
The extracted DNA was amplified with 202F and 1043R, which are specific primers for DAMO bacteria (Ettwig et al. 2009). The PCR products were used to conduct TA cloning sequencing utilizing the pUCm-T vector (Bio Basic Inc), and approximately 30 positive clones from each sample (enriched from nitrate or nitrite) were randomly selected and sequenced by Sangon Biotech Company, Shanghai, China. The 16S rRNA sequences were clustered into operational taxonomic units (OTUs) by Mothur win_64 software (Schloss et al. 2009). Phylogenetic analysis was performed using Mega 6.0 (Tamura et al. 2013), and the representative sequences and some related sequences were obtained from a Basic Local Alignment Search Tool (BLAST) search in the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). The sequences were aligned by the muscle algorithm, and the phylogenetic trees were constructed by the neighbor-joining statistical method using the Kimura 2-parameter as the substitution model. The bootstrap replications were set as 1000, and the Acidobacteria served as an outgroup. The representative sequences were submitted to the GenBank database, and the accession numbers were from KU376509 to KU376513.
High-throughput sequencing and statistical analysis
The PCR amplifications used the 16S rRNA gene primer pair PS5 (341b4F-806R) during high-throughput sequencing (Lu et al. 2015). The 16s rRNA amplicon sequencing was performed on the Illumina HiSeq platforms at Novogene, Beijing, China. Effective tags were clustered into OTUs at 97% identity with UPARSE software (Edgar 2013). The sequencing data were submitted to the NCBI Sequence Read Archive database, and the BioProject accession number was PRJNA317910. RDP Classifier and the GreenGene database, along with a representative sequence of each OTU, were used to annotate the taxonomic data. The relationships between the 35 most abundant microorganism genera and environmental factors were evaluated by Canoco 4.5 (ter Braak and Smilauer 2002). First, detrended correspondence analysis was conducted to decide if the ordination method was a unimodal or linear model. Constrained ordination, such as linear model redundancy analysis (RDA) and unimodal model canonical correspondence analysis (CCA), was used to explore the changes in species on specific ordination axes. When the length of the first gradient was below 3, the linear model was reasonable, but when the value was above 4, the unimodal model was more appropriate, and when the value was between 3 and 4, both models were feasible.
Results
Reactor performance during the DAMO enrichment process
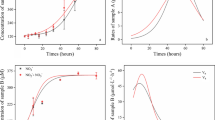
During the enrichment process, the NO3 −-N, NO2 −-N, and NH4 +-N concentrations were measured and their removal rates are shown in Fig. 1. In the N1 reactor, the NO3 −-N, NO2 −-N, and NH4 +-N removal rates gradually increased after a 30-day enrichment and reached 41.2, 37.3, and 28.0 mg/L/day on day 132, respectively. In N4, the nitrogen removal performance significantly increased after a 75-day enrichment, and rNO2 −-N and rNH4 +-N were 59.0 and 43.9 mg/L/day, respectively, on day 75. The rNO2 −-N concentration was 32.8 mg/L/day in N5 on day 132. The nitrogen removal rates were relatively lower in N2 and N3 than in N1, N4, and N5 during the whole enrichment process because rNO3 −-N and rNH4 +-N removal rates were below 2.0 mg/L/day. The rNO2 −-N concentration in N6 quickly increased to 3.6 mg/L/day on day 3, but then decreased to nearly zero (0.2 mg/L/day) on day 132. Therefore, this treatment was removed from the experiment. The total nitrogen removal rates were N1 > N4 > N5 > N2 > N3. N1 and N4 were higher because they were provided with NH4 +-N and the enrichment cultures might have contained Anammox bacteria, which would contribute to nitrogen removal.
The methane consumption test for DAMO activity was conducted between days 140 and 165. As shown in Table 2, the five reactors (N1–N5) consumed methane during this period. The rCH4 consumption ranged between 12.83 ± 0.16 and 4.74 ± 0.18 mmol/day, with a consumption order of N5 > N4 > N1 > N2 > N3. Based on methane, nitrite, and nitrate consumption results, it was found that more methane was oxidized than calculated. For example, in N5, the ratio of nitrite to methane is 2.67 in theory, but the experimental result was 2.02, the same to other reactors. These results suggested that there could potentially be additional methane losses. In N1, the ammonium consumption rate was 82.86 μmol/day, based on Anammox reaction, and 50.43 μmol/day of nitrite should be consumed, indicating that 68.4% of nitrite was converted by Anammox process. Similarly, Anammox bacteria contributed 73.6% of total nitrite removal in N4.
Microbial community composition in the inoculum and enrichment cultures
The PCR results showed that both DAMO archaea and Anammox bacteria were not detectable in the inoculum, but a small amount of DAMO bacteria were detected by the nest PCR (first PCR to amplify bacteria and then PCR to amplify DAMO bacteria results; data not shown). DAMO archaea and Anammox bacteria may also exist in the inoculum, but the contents were too low to be detected. The composition of the microbial communities in the inoculum and the enrichment culture was characterized by the 16S rRNA-based high-throughput sequencing method. The rarefaction curves tended to approach the saturation plateau, and the observed species did not increase significantly after the sequence number reached 26,000 (Fig. 2). This result indicated that the sequence number was reasonable and that the sequences did represent most of the microbial species used for further analysis. For the observed species, the Shannon and Chao1 indexes for sample NO were much higher than the others, which indicated that the enrichment process reduced the diversity of the microbial community compared to the inoculum. The wide coverage revealed that more than 99% of the species in each sample were obtained in all samples (Table S1).
The main microorganisms in NO–N6 were bacteria, the percentages were between 94.8 and 99.9%, and only a few fractions were archaea. All sequences were classified from phylum to genus, and the compositions of the different samples were similar, but the distribution of each phylum and family varied (Fig. 3). In the NO inoculum sample, the most abundant bacteria were Proteobacteria (60.6%). However, after enrichment with the different nitrogen sources, they decreased to 2.5, 23.9, 11.8, 6.1, 14.7, and 47.6% in N1–N6, respectively. NC10 bacteria were successfully enriched after a 165-day cultivation and increased from 2.3% in the inoculum to 51.3, 40.2, 38.9, 46.6, and 64.7% in N1–N5, respectively, which represented 17–28-fold increases compared to the inoculum. However, NC10 bacteria decreased to 0.3% in N6. All NC10 bacteria were classified to the Candidatus Methylomirabilis genus, which indicated that DAMO bacteria were enriched when the different nitrogen sources were added. Proteobacteria, Gemmatimonadetes, and Chloroflexi might have contributed to the small nitrogen removal performance in N6.
NH4 +-N was added to the N1, N2, and N4 reactors, and in addition, NO2 −-N was either provided or produced in these reactors. Thus, Anammox bacterial activity should also be considered in these reactors. In the Planctomycetes phylum, the Brocadiaceae family members that have an Anammox function were 0.10% in NO, but increased to 6.77 and 11.68% in the N1 and N4 reactors, respectively. The percentages of Brocadiaceae in other samples were low and showed little change compared to the inoculum. The Comamonadaceae, a major group of β-Proteobacteria, which are regarded as denitrifying bacteria in activated sludge (Khan et al. 2002), increased in the N2, N3, and N6 reactors and could have slightly contributed to nitrogen removal.
DAMO and Anammox bacterial abundance
DAMO and Anammox bacterial abundances were measured using qPCR. The results revealed that, compared with the inoculum NO, the DAMO bacterial abundance in the N1–N5 reactors increased four or five orders of magnitude after the 165-day enrichment, especially for N1, N4, and N5 (Table 3). DAMO bacteria increased from 8.82 ± 4.03 × 103 copies/L in NO to 1.84 ± 0.22 × 108, 2.37 ± 0.01 × 108, and 3.07 ± 0.25 × 108 in N1, N4, and N5, respectively. They increased because nitrite was provided to N1, N4, and N5. The Anammox bacterial abundance in N2 and N3 did not change much after enrichment, but they increased from 9.38 ± 4.33 × 103 copies/L in NO to 5.01 ± 0.18 × 104 copies/L in N1 and 1.16 ± 0.19 × 105 copies/L in N4. However, they decreased to 5.05 ± 0.74 × 103 copies/L in N5. The Anammox bacterial increase was considerable because nitrite and ammonium co-existed in N1 and N4, and this condition benefited Anammox bacterial growth. The qPCR results showed that the doubling time for DAMO and Anammox bacteria was 11.2 and 46.3 days in N4 (nitrite and ammonium conditions), respectively.
Phylogenetic analysis
The average sequence length of high-throughput sequencing was about 400 bp. Longer sequences are better for phylogenetic analysis, so the PCR products of DAMO bacteria that had 841 bp were purified and then cloned and sequenced to identify the difference in DAMO bacteria between nitrate-enriched (N3) and nitrite-enriched (N5) conditions in the phylogenetic tree. The reconstructed phylogenetic tree, based on the 16S rRNA sequences, is shown in Fig. 4. The DAMO bacteria sequences under the nitrate enrichment condition (N3) were divided into three groups, and the similarity between the sequences in each group was more than 97%. Group 1 was similar (99–100%) to DAMO bacteria enriched by methanogenic sludge and activated sludge mixtures; group 2 was similar (99%) to Candidatus Methylomirabilis oxyfera; and group 3 was similar (99%) to DAMO bacteria enriched by the Rhine sediment (Ettwig et al. 2009). Although the N5 sequences were classified into two groups, the differences between the two groups were slight. Both groups had high similarities (99%) with Candidatus Methylomirabilis sinica sp., which is a new DAMO bacterial species named by He et al. (2015c). The results indicated that DAMO bacteria were quite diverse under the nitrate enrichment condition (N3), and they were similar to the DAMO bacteria existing in the methanogenic sludge and activated sludge from wastewater treatment plants and in freshwater lake sediments. In contrast, DAMO bacteria diversity under the nitrite condition (N5) was relatively simple and similar to the M. sinica found in a paddy soil.
Relationship between the inoculum and different reactor cultures
To compare the similarities and differences between the samples, the shared OTUs were analyzed and the results are shown in a Venn graph in Fig. 5a and summarized in Table S2. NO2 −, NH4 +, and CH4 were added to both N1 and N4. However, NO3 − was also added to N1. They shared 231 OTUs, which indicated that 35.1% of the OTUs in N1 were independent of NO3 −. When comparing reactors N2 and N3, and N1 and N3, there was NO3 − and CH4 enrichment in the three reactors, but NH4 + was added to N2, and NO2 − and NH4 + were added to N1. N2 and N3 shared 451 OTUs and N1 and N3 shared 246 OTUs, which indicated that 52.9% of the OTUs in N2 were independent of NH4 + and 28.9% of the OTUs in N1 were independent of NO2 − and NH4 +. The N4 and N5 and N1 and N5 comparison pairs were all provided with NO2 − and CH4, and the shared OTUs were 179 and 151, respectively, which indicated that 45.2 and 38.1% OTUs were independent of NH4 + in N4, and NO3 − and NH4 + in N1, respectively. The difference between N1 and N2 was that NO2 − was added to N1. They shared 224 OTUs, which indicated that NO2 − led to 72% of the OTU differences. When N1 and N3 were compared, the NO2 − and NH4 + conditions caused more than 70% of the OTUs differences, and when N5 and N6 (N2 instead of CH4) were compared, the shared OTUs revealed that CH4 caused 75.4% of the OTU differences. These results revealed that NO2 − and CH4 had the greatest effects on the microbial community.
The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) is a common clustering method used to analyze the similarity between samples (Sokal and Michener 1958). The structure of the cluster tree is shown in Fig. 5b. The results showed that the seven samples were clustered into five groups at the phylum level. Groups A, D, and E contained only one sample, but groups B and C each contained two samples. The samples divided into two branches. NO was the inoculum (different from the enriched samples) and formed one branch, whereas the others clustered together as the other branch. Sample N6 was distant from N1–N5, because N6 was enriched with nitrogen gas as the control. In the enriched samples, N1 was similar to N4 and N2 was similar to N3. Furthermore, the distance between N5 and N1 and N4 was closer than N2 and N3. The differences between the samples were caused by the enrichment conditions, such as headspace gas and nitrogen source.
Relationship between the microbial community and environmental factors
The 35 most abundant bacteria at the genus level in the seven samples are shown in Fig. S1. Candidatus Brocadia as Anammox bacteria was most abundant in N1, and Candidatus Methylomirabilis as DAMO bacteria was most abundant in N5. The detrended correspondence analysis revealed that the length of the first gradient was 3.486, which lay between 3 and 4. Therefore, both RDA and CCA were reasonable; here we used RDA. The results showed that 93.7% of the cumulative percentage variance in the species data could be explained by environmental factors, which indicated that the model was appropriate. The RDA analysis identified the relationship between environmental factors and the microbial community and the relationship between environmental factors and the samples. Figure 6a shows that the most abundant 18 genera in NO were located in the opposite direction to the environmental factors, which indicated that these bacteria were negatively related to methane and nitrogen source. The seven genera with the greatest numbers in N6 had negative correlations with methane, ammonium, and nitrate and no obvious correlation with nitrite. There was a positive correlation between Candidatus Methylomirabilis and methane, nitrate, ammonium, and nitrite, which meant that methane was the most significant influencing factor. Candidatus Brocadia was positively related to nitrate, ammonium, and nitrite, but the correlation with methane was poor. Acidovor and Hyphomic contents were higher in N2, but they had little correlation with methane. However, other abundant microorganisms, such as Pseudoxa, Diaphoro, Magnetos, and Rhodopla, were positively related to the methane factor. Microorganisms in NO clustered together, and the abundant genera in N6 were centralized, which indicated that the bacterial genera in the NO and N6 microbial communities were highly related to each other.
RDA ordination plots of the relationships between environmental factors. a The main microorganism communities (in Fig. S1). b The inoculum + the six reactor samples
The relationship between the samples is shown in Fig. 6b. N2, N3, N4, and N5 were close to each other, which meant that the differences between them were small. NO, N1, and N6 were distributed away from the other samples, and each of them had larger differences with the others and themselves. There was a small difference in N1 classification between Figs. 5b and 6b because the cluster analysis was based on all microorganisms in each sample, and the environmental factors were not considered in the UPGMA analysis method (Fig. 5b). However, only the most abundant microorganisms in each sample were considered when we explored the relationship between cultures and environmental factors using the RDA analysis method (Fig. 6b). The effect of methane on the cultures, from strong to weak, was N5 > N4 > N1≈N3 > N2 > N6 > NO; the effect of nitrite was N1 > N6 > N5 > N3 > N4 > N2 > NO; and the effects of nitrate and ammonium on the samples were N1 > N5 > N4 > N3 > N2 > N6 > NO.
Discussion
Effects of nitrogen source on DAMO reactor performance
The performance of the six reactors was affected by nitrogen source. Supplying nitrate, nitrite, and ammonium together (N1) shortened the enrichment time and improved nitrogen removal performance. The enrichment time substantially decreased from 75 to 30 day in this study. Generally, DAMO culture enrichment usually takes longer. For example, Raghoebarsing et al. (2006) took 480 days to enrich a DAMO culture when it was supplied with nitrate and nitrite; He et al. (2015b) took 600 days to enrich DAMO bacteria when only nitrite was supplied; and Haroon et al. (2013) took 350 days to enrich DAMO archaea under nitrate and ammonium conditions. The rapid enrichment technique described in this paper will improve DAMO research and application. The nitrite removal rate reached a maximum 34.3 mg/L/day on day 126 for N5. The value was higher than for DAMO bacteria enriched from wastewater sludge, which was 5.1 mg/L/day on 308 days (Luesken et al. 2011b), and DAMO bacteria enriched by paddy soil, which was 12.2 mg/L/day after a 609-day operation in a sequencing batch reactor (He et al. 2015c). The total nitrogen removal rates for DAMO cultures enriched by multiple nitrogen sources were generally higher than for single nitrogen source enrichment in this study. Although the maximum nitrogen removal rate of 106.5 mg/L/day (rNH4 +-N + rNO3 −-N + rNO2 −-N for N1) does not meet standard application requirements, some techniques can improve the DAMO microorganism activity, such as operating the process in a hollow fiber membrane bioreactor (Cai et al. 2015) or a magnetically stirred gas lift reactor (Hu et al. 2014) or by adding a second liquid phase (Fu et al. 2015). Ammonium addition will shorten the enrichment time and improve the nitrogen removal rate. Therefore, the DAMO and Anammox co-culture system might have a potential application in wastewater treatment plants.
The OTU UPGMA and RDA analyses showed that various nitrogen sources had different impacts on the microbial community and that they influenced the extent and the relationship between environmental factors and abundant genera. This type of analysis is a useful way of comparing the differences and similarities between samples and provides information about the effects of environmental factors, including nitrate, nitrite, ammonium, and methane, on the microbial community.
Why DAMO bacteria and not DAMO archaea were enriched under nitrate addition conditions
DAMO archaea were not enriched under the nitrate or nitrate/ammonium conditions in this study. This was unexpected, and some of the possible reasons could be that DAMO archaea are more difficult to enrich than DAMO bacteria, so the relevant DAMO archaea enrichment reports were unusual because most of the DAMO archaea were enriched from original mixed inocula, which can come from freshwater sediments, anaerobic digester sludge, methanogenic sludge, and activated sludge. In this study, the archaea only made up a small proportion (about 0.2–4.9%) of microorganisms in the inoculum and in the enriched cultures under the different nitrogen conditions. DAMO archaea were not detectable in both the inoculum and enriched culture microbial communities, which indicated that it is likely that there were no DAMO archaea in the inoculum rather than the enrichment time not being long enough. A previous study showed that DAMO archaea were also not enriched after 13 months when they were inoculated with freshwater sediment under nitrate and methane conditions (Wang et al. 2016).
It is well known that DAMO bacteria are able to utilize nitrite as an electron acceptor to oxidize methane alone, but whether it can reduce nitrate has not been known until now. When nitrate is provided to DAMO bacteria instead of nitrite, methane oxidizing activity decreased (Ettwig et al. 2008). This result suggests that DAMO bacteria prefer nitrite to nitrate. In denitrifying bacteria, nitrate reductase can reduce nitrate to nitrite. It occurs is in two forms, one of which is in cell membranes, and the other is located at the periplasmic space (Coelho and Romao 2015). Genes narGHJI and napAB are present in the genome of M. oxyfera, but napCDE are absent (Ettwig et al. 2010), which indicates that the nitrate reductase genes inside the membranes are intact. It is possible for DAMO bacteria to utilize nitrate under specific conditions. In this study, DAMO bacteria were successfully enriched in freshwater sediment under nitrate conditions, and this was similar to the results reported by Wang et al. (2016). Although DAMO archaea were not detected in the inoculum, it was hard to exclude the possibility of their existence. After a 165-day enrichment under nitrate conditions, the DAMO bacteria were the main community in N3, which indicated that DAMO bacteria were more competitive than DAMO archaea under N3 conditions.
The reduction process from nitrate to nitrite may also occur in heterotrophic denitrifiers, which utilize a small amount of organic matter from microbial products and microorganism decay. He et al. (2015c) reported that there was a small fraction of heterotrophic denitrifiers that co-existed in DAMO bacterial culture after a long-term operation (609 days). The Comamonadaceae is a group of β-Proteobacteria that can reduce nitrate to nitrite under anoxic conditions by utilizing some carbon sources (Etchebehere et al. 2001). The proportion of Comamonadaceae was small at only 3.0 and 1.6% in the nitrate and nitrite reactors, respectively. The DAMO bacteria dominated under nitrate conditions. The results suggested that DAMO bacteria likely could utilize nitrate but in a low consumption rate. Of course, the reaction of nitrate to nitrite also may be conducted by microorganisms via endogenous respiration, which cannot be excluded.
The phylogenetic analysis revealed that the DAMO bacteria enriched by nitrate and nitrite condition were different. The former were more diverse and one was similar to M. oxyfera, but the latter showed relatively low diversity and one was similar to M. sinica. Although both bacteria are DAMO bacteria, they tended to utilize different electrons. Due to the extra step needed to reduce nitrate to nitrite, DAMO bacteria enriched on nitrate become more diverse. This suggests that the effect of nitrogen source on DAMO microorganisms is more complex than expected.
The interaction between DAMO bacteria and Anammox bacteria
There is a competitive relationship between DAMO bacteria and Anammox bacteria, which compete for the same nitrite electron acceptor. The doubling time for DAMO bacteria is 14 to 25 days (He et al. 2015b), which is longer than the 3–11 days for Anammox bacteria (Lotti et al. 2015; Strous et al. 1998), but in this study, the doubling time for DAMO bacteria decreased to 11.2 days while Anammox bacteria increased to 46.3 days under nitrite and ammonium conditions, which suggested that DAMO and Anammox bacteria were affected under co-culture conditions. It has been reported that DAMO bacteria have a higher nitrite affinity than Anammox bacteria at low nitrite concentrations (Chen et al. 2014). Moreover, previous results have suggested that the main community is made up of DAMO bacteria rather than Anammox bacteria when the NO2 −/NH4 + ratio is below 0.5 (Chen et al. 2014). In our experiments, the NO2 −-N concentration was always below 10 mg/L in order to avoid any inhibition effects and the NH4 +-N concentration was 50 mg/L. Therefore, the NO2 −/NH4 + ratio was less than 0.2, which meant that the DAMO bacteria dominated the microbial population along with a few Anammox bacteria, and these experiment results were consistent with the simulation results reported by Chen et al. (2014). Of course, the ratio between different nitrogen sources can also affect the reactor performance and microbial community, which requires further investigation.
The high-throughput sequencing results showed that the percentage of Anammox bacteria in the Brocadia genus was the highest in N1 (Fig. S1). There are five members of the Anammox bacterial genera (Brocadia, Kuenenia, Scalindua, Anammoxoglobus, and Jettenia) that have been detected in wastewater treatment systems and natural ecosystems (Hu et al. 2012). The Anammox bacteria previously found to co-exist in DAMO cultures are Brocadia, Kuenenia, and Jettenia (Ding et al. 2014; Luesken et al. 2011a). However, in this study, only one genus, Brocadia, co-existed in the DAMO culture. In N1 and N4, Anammox contributed about 70% of nitrite removal (Table 2). This significant contribution of Anammox indicated that there might be other Anammox bacteria possibly co-existing in the DAMO culture, because not all Anammox bacteria, such as Kuenenia, are included in the GreenGene database for high-throughput sequencing.
DAMO and Anammox microorganisms inhabit similar environments, and they are widespread in freshwater, wetland, marine water, and wastewater treatment plants. There are synergetic relationships (nitrate removal is synergistic with DAMO archaea and Anammox bacteria or DAMO bacteria) and competition (nitrite causes competition between DAMO bacteria and Anammox bacteria) between the DAMO and Anammox processes. They make considerable contributions to reducing greenhouse gas emissions and to the nitrogen cycle. The DAMO and Anammox co-culture system also has a potential application in wastewater treatment plant.
In summary, DAMO bacteria were successfully enriched under different nitrogen source conditions and no DAMO archaea were found in nitrate condition. However, DAMO bacteria enriched in nitrate condition were different from that enriched in nitrite condition. Even though ammonium addition led to shorter enrichment time, higher nitrogen removal rate, and the increase of Anammox bacteria, DAMO bacteria still dominated in the reactors. Apparently, nitrogen source effect on the co-culture enrichment system was complex than expected.
References
Cai C, Hu S, Guo J, Shi Y, Xie GJ, Yuan Z (2015) Nitrate reduction by denitrifying anaerobic methane oxidizing microorganisms can reach a practically useful rate. Water Res 87:211–217
Chen X, Guo J, Shi Y, Hu S, Yuan Z, Ni B-J (2014) Modeling of simultaneous anaerobic methane and ammonium oxidation in a membrane biofilm reactor. Environ Sci Technol 48(16):9540–9547
Coelho C, Romao MJ (2015) Structural and mechanistic insights on nitrate reductases. Protein Sci 24(12):1901–1911
Ding ZW, Ding J, Fu L, Zhang F, Zeng RJ (2014) Simultaneous enrichment of denitrifying methanotrophs and anammox bacteria. Appl Microbiol Biotechnol 98(21):10211–10221
Ding J, Ding ZW, Fu L, Lu YZ, Cheng SH, Zeng RJ (2015) New primers for detecting and quantifying denitrifying anaerobic methane oxidation archaea in different ecological niches. Appl Microbiol Biotechnol 99(22):9805–9812
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998
Etchebehere C, Errazquin MI, Dabert P, Moletta R, Muxi L (2001) Comamonas nitrativorans sp nov., a novel denitrifier isolated from a denitrifying reactor treating landfill leachate. Int J Syst Evol Micr 51:977–983
Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, Op den Camp HJM, Jetten MSM, Strous M (2008) Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol 10(11):3164–3173
Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M (2009) Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75(11):3656–3662
Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M (2010) Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464(7288):543–548
Fu L, Ding ZW, Ding J, Zhang F, Zeng RJ (2015) The role of paraffin oil on the interaction between denitrifying anaerobic methane oxidation and Anammox processes. Appl Microbiol Biotechnol 99(19):7925–7936
Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW (2013) Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500(7464):567–570
He Z, Cai C, Shen L, Lou L, Zheng P, Xu X, Hu B (2015a) Effect of inoculum sources on the enrichment of nitrite-dependent anaerobic methane-oxidizing bacteria. Appl Microbiol Biotechnol 99(2):939–946
He Z, Geng S, Cai C, Liu S, Liu Y, Pan Y, Lou L, Zheng P, Xu X, Hu B (2015b) Anaerobic oxidation of methane coupled to nitrite reduction by halophilic marine NC10 bacteria. Appl Environ Microbiol 81(16):5538–5545
He Z, Wang J, Zhang X, Cai C, Geng S, Zheng P, Xu X, Hu B (2015c) Nitrogen removal from wastewater by anaerobic methane-driven denitrification in a lab-scale reactor: heterotrophic denitrifiers associated with denitrifying methanotrophs. Appl Microbiol Biotechnol 99(24):10853–10860
Hu S, Zeng RJ, Keller J, Lant PA, Yuan Z (2011) Effect of nitrate and nitrite on the selection of microorganisms in the denitrifying anaerobic methane oxidation process. Environ Microbiol Rep 3(3):315–319
Hu B, Shen L, Du P, Zheng P, Xu X, Zeng J (2012) The influence of intense chemical pollution on the community composition, diversity and abundance of anammox bacteria in the Jiaojiang Estuary (China). PLoS One 7(3):e33826
Hu B, He Z, Geng S, Cai C, Lou L, Zheng P, Xu X (2014) Cultivation of nitrite-dependent anaerobic methane-oxidizing bacteria: impact of reactor configuration. Appl Microbiol Biotechnol 98(18):7983–7991
Hu S, Zeng RJ, Haroon MF, Keller J, Lant PA, Tyson GW, Yuan Z (2015) A laboratory investigation of interactions between denitrifying anaerobic methane oxidation (DAMO) and anammox processes in anoxic environments. Sci Rep 5.
Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, den Camp HJMO, Harhangi HR, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Keltjens JT, Jetten MSM, Strous M (2011) Molecular mechanism of anaerobic ammonium oxidation. Nature 479(7371):127–130
Khan ST, Horiba Y, Yamamoto M, Hiraishi A (2002) Members of the family Comamonadaceae as primary poly(3-hydroxybutyrate-co-3-hydroxyvalerate)-degrading denitrifiers in activated sludge as revealed by a polyphasic approach. Appl Environ Microbiol 68(7):3206–3214
Knittel K, Boetius A (2009) Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334
Lane DJ (1991) In: Stackebrandt E, Goodfellow M (eds) 16S/23S rRNA sequencing in nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lotti T, Kleerebezern R, Abelleira-Pereira JM, Abbas B, van Loosdrecht MCM (2015) Faster through training: the anammox case. Water Res 81:261–268
Lu YZ, Ding ZW, Ding J, Fu L, Zeng RJ (2015) Design and evaluation of universal 16S rRNA gene primers for high-throughput sequencing to simultaneously detect DAMO microbes and anammox bacteria. Water Res 87:385–394
Luesken FA, Sanchez J, van Alen TA, Sanabria J, Op den Camp HJM, Jetten MSM, Kartal B (2011a) Simultaneous nitrite-dependent anaerobic methane and ammonium oxidation processes. Appl Environ Microbiol 77(19):6802–6807
Luesken FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, Hendrickx TLG, Zeeman G, Temmink H, Strous M, den Camp HJMO, Jetten MSM (2011b) Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol 92(4):845–854
Mulder A, Vandegraaf AA, Robertson LA, Kuenen JG (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol 16(3):177–183
Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Damste JS, Op den Camp HJ, Jetten MS, Strous M (2006) A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440(7086):918–921
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Schmid MC, Risgaard-Petersen N, van de Vossenberg J, Kuypers MMM, Lavik G, Petersen J, Hulth S, Thamdrup B, Canfield D, Dalsgaard T, Rysgaard S, Sejr MK, Strous M, den Camp HJMO, Jetten MSM (2007) Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol 9(6):1476–1484
Shi Y, Hu S, Lou J, Lu P, Keller J, Yuan Z (2013) Nitrogen removal from wastewater by coupling anammox and methane-dependent denitrification in a membrane biofilm reactor. Environ Sci Technol 47(20):11577–11583
Sokal R, Michener C (1958) A statistical method for evaluating systematic relationships. University of Kansas Science Bulletin 38:1409–1438
Strous M, Heijnen JJ, Kuenen JG, Jetten MSM (1998) The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl Microbiol Biotechnol 50(5):589–596
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30(12):2725–2729
ter Braak CJF, Smilauer P (2002) CANOCO reference manual and CanoDraw for windows user’s guide: software for canonical community ordination (version 4.5). Microcomputer Power Ithaca, NY, USA
Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MSM, Yin C, Op den Camp HJM (2012) Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS Microbiol Lett 336(2):79–88
Wang S, Hong Y, Wu J, Xu XR, Bin L, Pan Y, Guan F, Wen J (2015) Comparative analysis of two 16S rRNA gene-based PCR primer sets provides insight into the diversity distribution patterns of anammox bacteria in different environments. Appl Microbiol Biotechnol 99(19):8163–8176
Wang S, Wu Q, Lei T, Liang P, Huang X (2016) Enrichment of denitrifying methanotrophic bacteria from Taihu sediments by a membrane biofilm bioreactor at ambient temperature. Environ Sci Pollut R 23(6):5627–5634
Zhu G, Jetten MS, Kuschk P, Ettwig KF, Yin C (2010) Potential roles of anaerobic ammonium and methane oxidation in the nitrogen cycle of wetland ecosystems. Appl Microbiol Biotechnol 86(4):1043–1055
Zhu B, Sanchez J, van Alen TA, Sanabria J, Jetten MS, Ettwig KF, Kartal B (2011) Combined anaerobic ammonium and methane oxidation for nitrogen and methane removal. Biochem Soc T 39(6):1822–1825
Acknowledgments
The authors would like to acknowledge the financial support from the National Natural Science Foundation of China (51178444), the Program for Changjiang Scholars and Innovative Research Team in University, and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 295 kb)
Rights and permissions
About this article
Cite this article
Fu, L., Ding, J., Lu, YZ. et al. Nitrogen source effects on the denitrifying anaerobic methane oxidation culture and anaerobic ammonium oxidation bacteria enrichment process. Appl Microbiol Biotechnol 101, 3895–3906 (2017). https://doi.org/10.1007/s00253-017-8163-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8163-2