Abstract
In this study, we investigated the use of Illumina high-throughput sequencing of 16S ribosomal RNA (rRNA) amplicons to explore microbial diversity and community structure in raw and secondary treated wastewater (WW) samples from four municipal wastewater treatment plants (WWTPs A–D) across Australia. Sequence reads were analyzed to determine the abundance and diversity of bacterial communities in raw and secondary treated WW samples across the four WWTPs. In addition, sequence reads were also characterized to phenotypic features and to estimate the abundance of potential pathogenic bacterial genera and antibiotic-resistant genes in total bacterial communities. The mean coverage, Shannon diversity index, observed richness (S obs), and abundance-based coverage estimate (ACE) of richness for raw and secondary treated WW samples did not differ significantly (P > 0.05) among the four WWTPs examined. Generally, raw and secondary treated WW samples were dominated by members of the genera Pseudomonas, Arcobacter, and Bacteroides. Evaluation of source contributions to secondary treated WW, done using SourceTracker, revealed that 8.80–61.4% of the bacterial communities in secondary treated WW samples were attributed to raw WW. Twenty-five bacterial genera were classified as containing potential bacterial pathogens. The abundance of potentially pathogenic genera in raw WW samples was higher than that found in secondary treated WW samples. Among the pathogenic genera identified, Pseudomonas and Arcobacter had the greatest percentage of the sequence reads. The abundances of antibiotic resistance genes were generally low (<0.5%), except for genes encoding ABC transporters, which accounted for approximately 3% of inferred genes. These findings provided a comprehensive profile of bacterial communities, including potential bacterial pathogens and antibiotic-resistant genes, in raw and secondary treated WW samples from four WWTPs across Australia and demonstrated that Illumina high-throughput sequencing can be an alternative approach for monitoring WW quality in order to protect environmental and human health.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Water shortage is one of the key challenges due to population growth, urbanization, and climate change (McMichael and Lindgren 2011). To combat the global water crisis, wastewater (WW) and biosolids, the major by-product of the WW treatment process, are being widely used, and such practice is expected to increase in the next few decades (Hamilton et al. 2006). Australia’s urban communities generate large volumes of WW. A wastewater treatment plant (WWTP) collects and then treats domestic and industrial WW to a recommended safe bacterial and chemical level for designated use. Treated WW is often released into the environment and subsequently used for agriculture, ground water recharge, and drinking water production (Harwood et al. 2005; Okoh et al. 2007; Varela and Manaia 2013). Human-derived WW is known to contain pathogenic bacteria, protozoa, viruses, and helminths. Therefore, inadequately treated WW, or that accidentally discharged with a high load of pathogens, pose significant human and environmental health risks.
Direct monitoring of pathogens in WW is an attractive option as their occurrence could be related to health risks. However, due to the complexity of pathogen monitoring, fecal indicator bacteria (FIB), particularly Escherichia coli (E. coli), have been commonly used to monitor the quality of treated WW (Haramoto et al. 2006; Carducci and Verani 2013). This involves monitoring of E. coli in 100-mL treated WW grab samples using a culture-based method. Australian Guidelines for Water Recycling recommends the use of weekly or monthly E. coli monitoring in raw and secondary treated WW samples (AGWR 2006). In addition, in some circumstances, monitoring of Enterococcus spp., coliphages, and Clostridium spores is also recommended depending on the end uses of the treated WW. For example, treated effluent supplied to growers should not contain >1 E. coli, or other FIB, per 100 mL. It has been demonstrated that monitoring FIB does not reflect the occurrence of pathogens, especially protozoa and viruses in treated WW, possibly due to their high susceptibility to chlorination (Miescier and Cabelli 1982; Harwood et al. 2005). In addition, FIB do not correlate well with pathogenic bacteria, protozoa, and enteric viruses in sewage and environmental waters (Baggi et al. 2001; Bonadonna et al. 2002; Harwood et al. 2005).
Culture-based methods historically have been used to enumerate FIB and enteric pathogens. However, culture-based methods for detection of specific pathogens can be challenging and time-consuming, due to the strict growth requirements of certain pathogens (Evangelista and Coburn 2010). In addition, some pathogens may be in a viable but non-culturable state (Bonetta et al. 2010). To circumvent these issues, numerous polymerase chain reaction (PCR) and quantitative PCR (qPCR) assays have been used to detect and quantify specific pathogen(s) in the environmental and WW/biosolid samples (Liang et al. 2015; Gyawali et al. 2016; Haack et al. 2016; Yergeau et al. 2016). However, these assays rely on detecting or quantifying a single pathogen in a sample (Ahmed et al. 2016). Therefore, these approaches do not provide information on the abundance and diversity of a wide array of clinically significant microorganisms present in the total microbial community (Ye and Zhang 2011). This information can be important to establish risks associated with the reuse of treated WW.
In recent years, bacterial community analyses using 454 pyrosequencing or Illumina high-throughput sequencing of 16S ribosomal RNA (rRNA) amplicons have emerged as promising tools to study microbial communities in environmental water and soil with a fairly high taxonomic resolution (Qian et al. 2011; Rosech et al. 2007; Staley et al. 2015). These methods allow not only for the detection and identification of dominant bacterial taxa in a sample but also for the detection of rare species including potential pathogens within bacterial communities (Sogin et al. 2006). Next-generation sequencing has been used to identify potential bacterial pathogens in urban water (Ibekwe et al. 2013), biosolids (Bibby et al. 2010), and raw WW (Ye and Zhang 2011).
The aims of this study were to (i) determine the abundance and diversity of bacterial communities in raw and secondary treated WW samples from four facilities across Australia using an Illumina-based next-generation sequencing approach, (ii) investigate whether the bacterial community structure differs in raw and secondary treated WW samples across the four WWTPs, (iii) determine the fraction of the bacterial population in secondary WW that can be attributed to raw WW in order to determine the percent and composition of the bacterial community removed by treatment, and (iv) estimate the abundance of potential pathogenic bacterial genera and antibiotic-resistant genes in bacterial communities from raw and secondary treated WW samples.
Materials and methods
Sampling raw and secondary treated WW
Four WWTPs, representing four states, Brisbane (WWTP A), Melbourne (WWTP B), Perth (WWTP C), and Hobart (WWTP D), were selected for this study (Table 1). The WWTP A is located in Brisbane and treats raw WW from approximately 250,000 people. The treatment process consists of a primary treatment, a secondary treatment (activated sludge), and disinfection with chlorine and ultraviolet (UV) prior to discharge of the treated WW into the Brisbane River. The WWTP B is located in Melbourne and treats approximately 40% of Melbourne’s total human (approximately 4 million people) WW. The treatment process is similar to that in WWTP A, except that the treated WW is discharged into the Tasman Sea. WWTP C is located in Perth and treats raw WW from approximately 600,000 people. The treatment process is similar to that in WWTP A. However, the treated WW is not subjected to UV disinfection prior to being discharged into the Indian Ocean. The WWTP D is located in Hobart and treats WW from 35,000 people. Prior to being discharged into the Coral River or Derwent River, chlorinated WW is passed through a 10-μm filter.
Raw (approximately 100 mL each) and secondary treated WW samples (approximately 10 L each) were collected from each WWTP in sterile containers. Samples from WWTP A (10 raw and 10 secondary treated WW samples), WWTP B (5 raw and 10 secondary treated WW samples), WWTP C (10 raw and 9 secondary treated WW samples), and WWTP D (7 raw and 5 secondary treated WW samples) were collected over a period of 10–12 weeks in early September to December 2014. Secondary treated WW samples were collected after 55 h (WWTP A), 39 h (WWTP B), 24 h (WWTP C), and 2 h (WWTP D) later to account for the hydraulic retention time (HRT). Samples were transported on ice to the laboratory and stored at 4 °C.
Sample concentration and DNA extraction
Aliquots of raw WW samples (10 mL) were concentrated and purified with an Amicon® Ultra 15-mL centrifugal filter unit with Ultracel-50 membrane (NMWL 50 kDa) (Millipore, Billerica, MA). Briefly, each raw WW sample was centrifuged at 4000g for 10 min to obtain a final volume of 1 mL concentrated sample, which was stored at −80 °C. Secondary treated WW samples (10 L) were concentrated by a hollow-fiber ultrafiltration system (HFUF) using Hemoflow FX 80 dialysis filters (Fresenius Medical Care, Bad Homberg, Germany) as previously described (Hill et al. 2007). The sample was concentrated to approximately 100–120 mL and stored at 4 °C. A new filter cartridge was used for each sample. The concentrated sample was further concentrated using Jumbosep (molecular weight cutoff = 100 kDa) Centrifugal Devices (Pall Corporation, East Hills, NY, USA) to obtain a final volume of 5–6 mL. An aliquot of concentrated raw (200 μL) and secondary treated WW (200 μL) samples was subjected to DNA extraction using a Qiagen DNeasy Blood and Tissue kit, as per the manufacturer’s instructions (Qiagen Inc., Valencia, CA). The extracted DNA samples were stored at −80 °C.
PCR and Illumina MiSeq sequencing
The V5–V6 regions of the 16S rRNA gene were amplified using the barcoded forward (RGG ATT AGA TAC CC) and reverse (CGA CRR CCA TGC ANC ACC T) primer sets described previously (Claesson et al. 2010). Amplicons from each sample were size-selected and pooled in equal amounts. All samples were amplified and paired-end sequenced, at a length of 300 nucleotides (nt) using the dual index method, by the University of Minnesota Genomics Center (Minneapolis, MN), using the HiSeq 2500 platform (Gohl et al. 2016).
Sequence data analysis
Sequence processing was performed using the mothur software (version 1.35.1) (Schloss et al. 2009). Sequences were trimmed to 150 nt and paired-end joined using fastq-join (Aronesty 2013). Quality trimming was performed to remove sequences with average quality scores of <35 over a window of 50 nt, homopolymers of >8 nt, ambiguous bases, or >2 mismatches to primer sequences. High-quality sequences were aligned against the SILVA database (version 119) (Pruesse et al. 2007). Sequences were further quality trimmed by using a 2% precluster error (Huse et al. 2010; Kunin et al. 2010) followed by chimera removal using UCHIME (Edgar et al. 2011). Assignment of operational taxonomic units (OTUs) was performed at 97% identity using the furthest neighbor algorithm. Taxonomic assignments were made against the Ribosomal Database Project (RDP version 14) (Cole et al. 2009). Raw and secondary treated WW samples were grouped by WWTPs and corresponding states [WWTP A (Brisbane), WWTP B (Melbourne), WWTP C (Perth) and WWTP D (Hobart)]. For comparisons, the samples were rarefied by random subsampling to 11,000 reads per sample (Gihring et al. 2012).
The Bayesian classifier software program SourceTracker version 0.9.8 (Knights et al. 2011) was used to determine the proportion of bacterial communities in secondary treated WW samples that could be attributed to raw WW. SourceTracker calculated the probability (α = 0.001) that an OTU present in the bacterial community of secondary treated WW (the sink) was derived from raw WW (the source). The default parameters with rarefaction to 1000 sequence reads were used. OTUs that contributed to the source were also identified by the software.
METAGENassist was used to map bacterial phenotypic information from taxonomy (Arndt et al. 2012). Input files (consensus taxonomy, shared file, and metadata) were created in mothur using all raw and secondary treated WW samples. In all, 1239 taxa were present in the dataset before filtering. After filtering, 171 functionally relevant taxa remained and were normalized over samples by sum (this will adjust for varying sequencing coverage among samples by normalizing to the same total abundance for each sample) and over taxa by Pareto scaling. In the present study, we focused on several phenotype categories including oxygen requirements, temperature range, energy sources, metabolism, habitat, and host.
In order to identify potential pathogenic bacterial genera, the RDP database was used with a bootstrap cutoff of 60%, using the method described previously (Wang et al. 2007). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to infer the prevalence of genes encoding selected antibiotic resistance features in each raw and secondary treated WW sample (Langille et al. 2013). PICRUSt uses 16S rRNA genes to infer metegenome gene fuctional content from phylogenetic information. These predictions are precalcuated for genes in databases including the Kyoto Encyclopedia of Genes and Genomes (KEGG). The output of PICRUSt consists of a table of functional gene counts as KEGG orthologs (KOs). In the present study, we focused on selected KOs among the antibiotic resistance genes (K05595, K07552, K07694, K08170, and K08221) (de Voogd et al. 2015), and the percentages reported in the results reflect the percentage of all genes inferred.
Statistical analysis
Alpha diversity indices, including sample coverage, the number of OTUs observed, the Shannon diversity index (Shannon and Weaver 1949), and abundance-based coverage estimate (ACE) (Chao and Lee 1992), were calculated using mothur. These diversity indices were chosen to provide both parametric and non-parametric estimates of diversity. Ordination of samples was performed via principal coordinate analysis (PCoA). Determination of differences in beta diversity (community composition) between WW types and WWTPs was performed using analysis of similarity (ANOSIM) (Clarke 1993). The significance of group clustering was evaluated in mothur using analysis of molecular variance (AMOVA) (Excoffier et al. 1992). The Kruskal-Wallis test was conducted using LEfSe algorithm to determine which OTUs varied between raw and secondary WW samples from all four WWTPs (Segata and Huttenhower 2011; Acar and Sun 2013).
Sequence data accession number
Raw sequence data, received as fastq files, were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession number SRP078164.
Results
Bacterial α-community diversity
Among all raw WW samples from four WWTPs, a mean bacterial coverage of 97.5 ± 0.41% (mean ± standard deviation) was achieved, with an average of 627 ± 232 OTUs observed in individual samples. Similarly, among all secondary treated WW samples from the four WWTPs, a mean bacterial coverage of 96.8 ± 0.22% (mean ± standard deviation) was achieved, with an average of 770 ± 244 OTUs observed in individual samples. A total of 26,183 and 20,074 OTUs were observed, at 97% similarity, in raw and secondary treated WW samples from the four WWTPs.
The mean coverage, Shannon diversity index, observed richness (S obs), and abundance-based coverage estimate (ACE) of richness for raw and secondary treated WW samples are shown in Table 2. Differences in sample coverage and richness did not differ significantly (P > 0.05) between raw and secondary treated WW samples among the four WWTPs. Similarly, alpha diversity, measured by either the Shannon or ACE indices, did not differ significantly (P > 0.05) between raw and secondary treated WW samples among four WWTPs.
Taxonomic diversity of genera
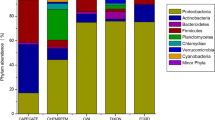
Approximately 98.5, 79.2, 97.3, and 79.7% of raw WW sequences from WWTPs A, B, C, and D were classified into 121, 129, 122, and 133 genera, respectively, while 1.50, 20.8, 2.70, and 20.3% could not be classified to any known genera. Similarly, about 43.1, 68.6, 75.3, and 49.7% of secondary treated WW sequences from WWTPs A, B, C, and D were classified into 128, 133, 129, and 129 genera, respectively, while 56.9, 31.4, 24.7, and 50.3% could not be classified to any known genera. Distributions of the 14 most abundant genera among raw and secondary treated WW samples across four WWTPs are shown in Fig. 1. Raw WW samples from all four WWTPs were dominated by members of the genus Pseudomonas (10.2–38.5%), followed by Arcobacter (8.36–28.6%) and Bacteroides (3.08–8.60%). Similarly, Pseudomonas (0.77–19.1%) was the most abundant genus in the secondary treated WW samples, followed by Arcobacter (0.89–2.77%) and Bacteroides (0.32–0.64%). Among raw WW samples, WWTPs A and C had greater abundances of Pseudomonas and Arcobacter than WWTPs B and D, with WWTP C having greater abundances of both genera than WWTP A (Fig. 1). Secondary treated WW samples from WWTP A were predominated by OTUs that could not be classified to any genera, while WWTP C had greater abundances of Paludibacterium and Conchiformibius than other WWTPs examined. In contrast, WWTPs B and D had similar taxonomic compositions in both raw and secondary treated WWs, with communities predominated by unclassified genera and Pseudomonas.
Raw and secondary treated WW samples collected from WWTPs A and C had significantly different beta diversity from each other (P < 0.05) and other WWTPs (B and D) (P < 0.05). Raw and secondary treated WW samples within and between WWTPs at WWTPs A and C also had significantly different community composition (P < 0.05), and samples clustered separately on the basis of type (raw or secondary) and WWTP location via AMOVA (P < 0.05) (Fig. 2a, b). However, samples from WWTPs B and D tended to cluster together, with less separation of raw and secondary treated WW sample communities, and communities from samples collected at these sites could not be significantly differentiated based on the WW type or WWTP location (ANOSIM P > 0.05; AMOVA P > 0.05) (Fig. 2a, b).
Among all WW samples, differences in beta diversity, as evaluated by ANOSIM, were significantly different between raw and secondary samples (P < 0.05). PCoA ordination of the samples similarly showed significant separation (AMOVA P < 0.05) of raw from secondary treated WW samples along the x-axis (Fig. 2c). Evaluation of the 20 most abundant genera by LEfSe found that 19 were differentially abundant between raw and secondary treated WWs (P < 0.05 for both Kruskal-Wallis and Wilcoxon tests) (Fig. 3) The genera Arcobacter and Pseudomonas, as well as genera within the order Bacteroidales, predominated in raw WW, while secondary treated WW had a great abundance of OTUs that could not be classified to greater-resolution, taxonomic levels (Figs. S1 and S2).
Genera in secondary treated WW samples attributed to raw WW
Evaluation of source contributions to secondary treated WW using SourceTracker revealed that 8.77 ± 1.25% of the bacterial communities in secondary treated WW samples was attributed to raw WW in WWTP A. Similarly, 61.4 ± 7.60, 14.8 ± 1.90, and 52.6 ± 7.00% communities from secondary treated WW samples were attributed to raw WW samples in WWTPs B, C, and D. The predominant OTUs that were maintained following secondary treatment could not be classified into any known genera at all WWTPs (Fig. 4), but at WWTPs B and D, a relatively large proportion of OTUs were also classified to the genus Pseudomonas (18.8 ± 24.0 and 13.3 ± 18.4% of the total sequence reads, respectively). The remaining OTUs were classified among less abundant genera.
Phenotypic mapping
METAGENassist was used to characterize phenotypic features of bacterial genera in pooled raw and treated WW samples. Phenotypic mapping of oxygen requirements revealed that raw and treated WW samples contained both aerobic and anaerobic bacteria. However, the percentage of aerobic and anaerobic bacteria was greater in raw WW, compared to secondary treated WW samples (Fig. 5a). Phenotypic mapping to temperature indicated that raw and treated WW samples contained primarily mesophilic bacteria. The percentage of mesophilic bacteria was greater in secondary treated WW compared to raw WW. In addition, both raw and secondary treated WW samples also contained small percentages of psychrophilic and thermophilic bacteria (Fig. 5b). Energy source mapping indicated that raw WW was enriched with more heterotrophs compared to secondary treated WW. Raw and secondary treated WWs also contained small amounts of autotrophic, chemoheterotrophic, and lithoautotrophic bacteria (Fig. 5c).
With respect to metabolic activity, raw WW had larger fractions of ammonia-oxidizing, chitin-degrading, dehalogenating, and nitrite- and sulfate-reducing bacteria than secondary treated WW. Secondary treated WW contained bacteria with degradation ability for cellulose and xylan. Habitat data suggested that free-living bacteria from freshwater, sediment, and soil, as well as host-associated bacteria, were present at low levels in samples from both raw and secondary treated WW samples. Raw WW samples had more host-associated bacteria than did secondary treated WW (Fig. 5e). Mapping to host revealed that 80% of bacteria in raw WW belonged to human host whereas only 60% in secondary treated WW mapped to this host (Fig. 5f).
Detection of potential pathogenic genera
The RDP classifier was used to identify potential pathogenic bacteria at the genus level. Table 3 shows the percentage of pathogenic genera in raw and secondary treated WW samples from four WWTPs. OTUs from raw and secondary treated WW samples were assigned to 25 potentially pathogenic genera (Table 3). The abundance of potentially pathogenic genera in pooled raw WW samples ranged from 25.6 to 70.0%, whereas genera in pooled secondary treated WW samples ranged from 3.80 to 26.7%. Among the pathogenic genera identified, Pseudomonas and Arcobacter had the greatest percentage of the sequence reads, representing 10.3–38.5 and 8.37–28.6% of reads, respectively, in raw WW samples across four WWTPs. However, Pseudomonas and Arcobacter represented only 0.78–19.1 and 1.00–9.42% of reads, respectively, in secondary treated WW samples.
Detection of antibiotic resistance genes
The abundances of antibiotic resistance genes inferred from the KEGG orthology were generally low (<0.5% of inferred genes), except for genes encoding type ABC transporters, which accounted for approximately 3% of inferred genes (Fig. 6). Among primary WW samples, genes encoding butirosin, neomycin, and streptomycin biosynthesis (P < 0.05) were significantly greater at WWTP D and lower at WWTP C. In contrast, tetracycline biosynthesis genes were significantly greater (P < 0.05) at WWTP C and lower at WWTP D. These patterns were also significant for butirosin and neomycin biosynthesis (P < 0.05) and tetracycline biosynthesis (P < 0.05) in secondary treated WW. In addition, in secondary treated WW, genes encoding type ABC transporters and novobiocin differed among WWTPs (P < 0.05 for both) and tended to be greater at WWTPs A and D. The abundances of every gene class analyzed, except for vancomycin biosynthesis, differed significantly between primary and secondary treated WWs (P < 0.05). Interestingly, genes encoding biosynthesis of butirosin and neomycin, novobiocin, and streptomycin were greater in secondary treated WW than in primary WW.
Discussion
Microbial communities in raw WW are composed of human feces and non-fecal microbes derived from graywater or stormwater (Shanks et al. 2013). The sewerage network carries domestic and, in some cases, industrial WW to the WWTPs. However, factors such as extreme weather events, broken or blocked WW pipes, aging infrastructure, and mechanical failures, such as loss of electrical power and overflow, can discharge harmful human pathogens and other contaminants directly into the environment. Direct and indirect exposure to raw and partially treated WW remains a significant public health concern.
High-throughput sequencing and microarray analysis have emerged as potential tools for monitoring WW streams (Wong et al. 2013). To date, most studies have focused on sludge (Yadav et al. 2014; Yang et al. 2014), bioreactors (Valentin-Vargas et al. 2012; Shrestha et al. 2014), and treated WW (Hu et al. 2016). However, only a small number of studies have provided information about the composition of microbial communities in raw WW, and very little is known about the bacterial composition of raw WW in Australia. In view of this, we determined the abundance and diversity of bacterial communities in raw and corresponding secondary treated WW samples from four WWTPs across Australia. We also estimated the abundance of potential pathogenic bacterial genera and antibiotic-resistant genes along with phenotype mapping.
The Shannon diversity and ACE richness of the raw and secondary treated WW samples from all four WWTPs were similar, although these samples were collected from different geographical areas. Similar Shannon diversity and ACE richness values were also reported in previous studies that investigated bacterial community structure in raw WW and effluent using 454 pyrosequencing of 16S rDNA (McLellan et al. 2010; Ye and Zhang 2011).
The numbers of OTU obtained for raw and secondary treated WW samples tested in this study were less than two previous studies that identified 1667 to 3000 OTUs in raw WW (McLellan et al. 2010; Ye and Zhang 2011) and 1932 OTUs in effluent samples (Ye and Zhang 2011). However, McLellan and colleagues and Ye and Zhang subsampled 18,800 and 17,300 sequences, respectively, compared to 11,000 sequences subsampled in this study. The differences in sequence reads may have contributed to the smaller numbers of OTUs obtained in this study. It is also possible that the composition of raw WW (influent) was different in those studies compared to this study.
Approximately 20% of raw WW sequences from WWTPs B and D could not be classified into any known genera. Similarly, about 31–57% of secondary treated WW sequences from all WWTPs could not be classified into any known genera. These data suggest high bacterial diversity of raw WW and secondary treated WW samples. A large portion of sequence reads could not be assigned to the genus level. These results are in accordance with previous studies that investigated bacterial communities in activated sludge samples (Sanapareddy et al. 2009; Zhang et al. 2012). It is also possible that secondary clarifiers harbor novel bacterial taxa that were not present in the RDP classifier used in this study. In addition, PCR artifacts and/or sequencing errors may also affect classification.
Raw and secondary treated WW samples collected from WWTPs A and C had significantly different beta diversity and community composition (P < 0.05) than did plants B and D. The WWTPs A and C are located in subtropical and Mediterranean regions, respectively, while WWTPs B and D are located in temperate regions. It has been reported that bacterial communities associated with WW infrastructure vary among cities and may exhibit both spatial and temporal dynamics (Shanks et al. 2013). Different plant designs, operational parameters, and composition of influent in each WWTP may have attributed to the differences in bacterial community composition. In addition, different origins of WW (domestic, industrial, farm) may have attributed differences in beta diversity and community composition. For example, WWTP A received 80% domestic and 20% industrial WW compared to WWTP B, which received 100% domestic WW.
The high abundances of Bacteroides, Pseudomonas, and Arcobacter observed in this study have been reported in raw WW samples of two WWTPs in metropolitan Milwaukee (McLellan et al. 2010) and at a municipal WWTP in Moscow (Shchgolkova et al. 2016). Among the genera detected, Pseudomonas and Arcobacter were highly abundant in both raw and secondary treated WW samples in WWTPs B, C, and D. WW treatment systems appear to supply a unique niche where Arcobacter can flourish, and as a result, members of this genus are often found in great abundances (4–11%) in association with raw WW (Collado et al. 2011; Cai et al. 2014; Shchgolkova et al. 2016). In this study, Arcobacter comprised 8.4–29 and 0.9–2.8% of the total bacterial communities in raw and secondary treated WW samples, respectively. Similarly, Pseudomonas was very prevalent in both raw and secondary treated WW samples ranging from 10 to 38 and 0.8 to 19% of reads, respectively. Pseudomonas was previously reported to be present in great abundances in raw WW samples (Wheater et al. 1980; McLellan et al. 2010). The WW environment contains high nutrient concentrations and is rich in biofilms, which may act as a reservoir for WW-specific organisms (Leung et al. 2005; Ort and Gujer 2008). Therefore, it is possible that these groups of bacteria are propagating within these systems.
The activated sludge process is one of the key components of a WWTP. It involves the biological reduction of biochemical oxygen demand and a decrease in suspended solids and pathogens of raw input WW and the production of a more oligotrophic, pathogen-free, environmentally benign outgoing effluent. SourceTracker was used to determine the proportion of bacterial communities in secondary treated WW samples that could be attributed to raw WW. Interestingly, the percentages of bacterial communities in secondary treated WW attributable to primary WW varied considerably between WWTPs, with a greater similarity between primary and secondary WWs at WWTPs B and D, when analyzed both separately and with all WWTPs pooled. This suggests that bacterial communities present in the secondary treated effluents were similar to raw WW at these two WWTPs. Such results suggest the inefficacy of secondary treatment process in these two WWTPs. It is also possible that several of the genera found in secondary treated WW at WWTPs B and D (Pseudomonas, Arcobacter, Flavobacterium, and Polynucleobacter) may be ubiquitous throughout the treatment process, resulting from adaptation, and/or may be associated with sewer infrastructure (Shanks et al. 2013; Rodríguez et al. 2015). On the other hand, bacterial communities present in the secondary treated effluents were different to raw WW at WWTPs A and C. These differences in similarity between raw and secondary treated WWs may be due to differences in influent composition or WWTP treatment efficiency.
Phenotypic mapping revealed that bacterial communities in secondary treated WW had less metabolic diversity and a lower percentage of heterotrophic bacteria than did raw WW. A recent study characterizing carbon metabolism in influent and effluent at three WWTPs similarly saw a shift in carbon metabolism among the microbial communities as a result of treatment (Jałowiecki et al. 2016). There still exists a paucity of information regarding the relationship between functional community shifts and WWTP treatment efficacy, although maximizing functional diversity during the treatment process has been suggested to improve treatment ability for variable influent compositions (Rodríguez et al. 2015). More importantly, the human-host associated percentage of the community, likely to represent the greatest health risk, was reduced by 20% among pooled samples. In contrast, the environmentally associated fraction of the community increased in secondary treated WW, suggesting a reduction in pathogen load following treatment.
In this study, 25 potential pathogenic genera were identified among raw and secondary treated WW samples from various geographical locations. A previous study reported the presence of 16 pathogenic genera in activated sludge, raw WW, and effluent samples using pyrosequencing (Ye and Zhang 2011), and another showed that Illumina sequencing characterized a much more complex pathogenic community in WW and sludge samples than did 454 pyrosequencing (Lu et al. 2015). In agreement with our phenotypic characterization, the abundance of potentially pathogenic genera was reduced in secondary treated WW relative to raw, but not all genera were reduced and some were shown to increase. Similarly, functional inferences of antibiotic resistance genes did not show a consistent trend in reduction or increase as a result of treatment for all categories analyzed. It is unclear whether these results reflect actual biological phenomena as a result of WWTP processing or whether they are artifacts of the sequencing effort.
The percentage reduction of pathogenic genera in secondary treated WW suggests that certain pathogenic genera such as Aeromonas, Arcobacter, and Pseudomonas may aggregate in activated sludge flocs and could be removed from effluent via a sedimentation process. Similar results have been reported by Lu et al. (2015), who also noted removal of pathogenic species through the activated sludge process. However, the reduction was not consistent across all four WWTPs located in different geographical locations. No or minimal reductions were observed for several pathogenic genera such as Acinetobacter and Bordetella; however, the results were not consistent for all WWTPs. The sequence reads of some pathogenic genera were found to be higher in secondary treated WW compared to the raw WW, suggesting that these bacteria may be multiplying under the conditions in the secondary treated WW tank.
Among the pathogenic genera detected, Acinetobacter, Aeromonas, Arcobacter, and Pseudomonas were most abundant in both raw and secondary treated WW samples, indicating similar potential pathogenic bacterial communities. Previous studies also reported that Arcobacter was the most abundant (2.90%) in raw WW (Ye and Zhang 2011; Kumaraswamy et al. 2014). The abundance of pathogenic genera may vary among studies due to different sequencing technology, DNA extraction kit, and inherent amplification biases during PCR (Haft and Tovchigrechko 2012).
There are several limitations of sequencing approaches for the detection of pathogenic bacterial communities in WW and environmental samples. Illumina sequencing of the 16S rRNA gene will also generate bias because of the highly variable copy number of the 16S rRNA gene, ranging from 1 to 15 (Lu et al. 2015). This may lead to some inaccurate estimate as it is difficult to exactly quantify pathogens in terms of cell numbers (Cai and Zhang 2013). In addition, all potential pathogenic genera are not pathogenic. For example, there are 200 species in the Bacillus genus, while only few of them such as Bacillus anthracis and Bacillus cereus are pathogenic. The 16S rRNA approach taken in this study may be less sensitive than qPCR at the current sequencing depth, and because of that, rare pathogens may not be detected especially for samples such as those ones tested in this study. However, there are several advantages of the sequencing approaches such as this technique is high throughput, which can target many pathogens in a single assay. Compared to PCR/qPCR assays, 16S rRNA sequence provides a broader spectrum of occurrence of potential bacterial pathogens (Cai and Zhang 2013). High-resolution taxonomic assignment of sequences using short sequence reads has been shown to be somewhat error-prone, and resultantly, inferring gene abundances from such data may also yield spurious results (Mizrahi-Man et al. 2013). Furthermore, the Illumina data generated here are only relatively quantitative, so shifts in abundance should be interpreted cautiously, but these data suggest promising targets for further, more detailed characterization. To the best of our knowledge, only a few studies identified sequences to the species level, including emerging and reemerging pathogens, which will aid in public health risk assessment.
In summary, alpha diversity, measured by either the Shannon index or ACE algorithm, did not differ significantly between raw and secondary treated WW samples among four WWTPs (A–D), although these samples were collected from different geographical areas. In contrast, beta diversities of WWTP A and C were different than WWTPs B and D, suggesting that bacterial communities associated with WW vary among geographical locations. From our data, it appears that genera such as Pseudomonas and Arcobacter were highly abundant in both raw and secondary treated WW samples. The WW treatment environment appears to supply a unique niche where Pseudomonas and Arcobacter can flourish. In total, 25 potential pathogenic genera were identified among raw and secondary treated WW samples with varying abundance. The abundance of potentially pathogenic genera was reduced in secondary treated WW relative to raw, but not all genera were reduced and some were shown to increase. Similarly, functional inferences of antibiotic resistance genes did not show a consistent trend in reduction or increase as a result of treatment for all categories analyzed. Further research should focus on testing final effluent samples to determine which genera can still persist and deserved more concerns. The sequence reads of some pathogenic genera were found to be greater in secondary treated WW compared to the raw WW, suggesting that these bacteria may be multiplying under the conditions in the secondary treated WW treatment system. Despite several intrinsic limitations of the sequencing technologies, there are several advantages. Compared to current pathogen detection approach, 16S rRNA sequence analysis provides a broader spectrum of occurrence of potential bacterial pathogens, and it has the potential to aid in the risk assessment when the limitations such as sequencing depth and accuracy will be improved.
References
Acar EF, Sun L (2013) A generalized Kruskal-Wallis test incorporating group uncertainty with application to genetic association studies. Biometrics 69:427–435
AGWR (2006) Australian guidelines for water recycling: managing health and environmental risks (phase 1). Natural Resource Management Ministerial Council, EPA-HCA-AHMC, Australia
Ahmed W, Hughes B, Harwood VJ (2016) Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 8:231
Arndt D, Xia J, Zhou Y, Guo AC, Cruz JA, Sinelnikov I, Budwill K, Nesbo CL, Wishart DS (2012) METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res 40(W1):W88–W95
Aronesty E (2013) Comparison of sequencing utility programs. Open Bioinforma J 7:1–8
Baggi F, Demarta A, Peduzzi R (2001) Persistence of viral pathogens and bacteriophage during sewage treatment: lack of correlation with indicator bacteria. Res Microbiol 152:743–751
Bibby K, Viau E, Peccia J (2010) Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Res 44:4252–4260
Bonadonna L, Briaancesco R, Ottaviani M, Veschetti E (2002) Occurrence of Cryptosporidium oocysts in sewage effluents and correlation with microbial, chemical and physical water variables. Environ Monit Assess 75:241–252
Bonetta S, Bonetta S, Ferretti E, Balocco F, Carraro E (2010) Evaluation of Legionella pneumophila contamination in Italian hotel water systems by quantitative real-time PCR and culture methods. J Appl Microbiol 108:1576–1583
Cai L, Zhang T (2013) Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ Sci Technol 47:5433–5441
Cai L, Feng J, Zhang T (2014) Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl Microbiol Biotechnol 98:3317–3326
Carducci A, Verani M (2013) Effects of bacterial, chemical, physical and meteorological variables on virus removal by a wastewater treatment plant. Food Environ Virol 5:69–76
Chao A, Lee S-M (1992) Estimating the number of classes via sample coverage. J Am Stat Assoc 87:210–217
Claesson MJ, Wang QO, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O’Toole PW (2010) Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Collado L, Levican A, Perez J, Figueras MJ (2011) Arcobacter defluvii sp. nov., isolated from sewage samples. Int J Syst Evol Microbiol 61:2155–2161
De Voogd NJ, Cleary DFR, Polónia, Gomes NCM (2015) Bacterial community composition and predicted functional ecology of sponges, sediment and seawater from the thousand islands reef complex, West Java, Indonesia. FEMS Microbiol Ecol 91:fiv019. doi:10.1093/femsec/fiv019
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Evangelista KV, Coburn J (2010) Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol 5:1413–1425
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial DNA restriction data. Genetics 131:479–491
Gihring TM, Green SJ, Schadt CW (2012) Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14:285–290
Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, Knights D, Beckman KB (2016) Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34:942–949
Gyawali P, Ahmed W, Sidhu JPS, Nery SV, Clements AC, Traub R, McCarthy JS, Llewellyn S, Jagals P, Toze S (2016) Quantitative detection of viable helminth ova from raw wastewater, human feces, and environmental soil samples using novel PMA-qPCR methods. Environ Sci Pollut Res. doi:10.1007/211356-016-7039-9
Haack SK, Duris JW, Kolpin DW, Focazio MJ, Meyer MT, Johnson HE, Oster RJ, Foreman WT (2016) Contamination with bacterial zoonotic pathogen genes in U.S. streams influenced by varying types of animal agriculture. Sci Total Environ 563-564:340–350
Haft DH, Tovchigrechko A (2012) High-speed microbial community profiling. Nat Methods 9:793–794
Hamilton AJ, Versace VL, Stagnitti F, Li P, Yin W, Maher P, Hermon K, Premier RR, Ierodiaconou D (2006) Balancing environmental impacts and benefits of wastewater reuse. WSEAS Transactions on Environment and Development. World Sci Eng Acad Soc (WSEAS):71–80
Haramoto E, Katayama H, Oguma K, Yamashita H, Tajima A, Nakajima H, Ohgaki S (2006) Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci Technol 54:301–308
Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB (2005) Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170
Hill VR, Kahler AM, Jothikumar N, Johnson TB, Hahn D, Cromeans TL (2007) Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl Environ Microbiol 73:4218–4225
Hu Q, Zhang XX, Jia S, Huang K, Tang J, Shi P, Ye L, Ren H (2016) Metagenomic insight into ultraviolet disinfection effects on antibiotics resistance in biologically treated wastewater. Water Res 101:309–317
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:889–898
Ibekwe AM, Leddy M, Murinda SE (2013) Potential human pathogenic bacteria in a mixed urban watershed as revealed by pyrosequencing. PLoS One 11:e79490
Jałowiecki Ł, Chojniak JM, Dorgeloh E, Hegedusova B, Ejhed H, Magnér J, Płaza GA (2016) Microbial community profiles in wastewaters from onsite wastewater treatment systems technology. PLoS One 11:e0147725
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST (2011) Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–U107
Kumaraswamy R, Amha YM, Anwar MZ, Henschel A, Rodriguez J, Ahmad F (2014) Molecular analysis for screening human bacterial pathogens in municipal wastewater treatment and reuse. Environ Sci Technol 48:11610–11619
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123
Langille MGI, Zaneveld J, Caporaso JG, McDonald DM, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814
Leung HD, Chaen G, Sharma K (2005) Effect of detached/re-suspended solids from sewer sediment on the sewage phase bacterial activity. Water Sci Technol 52:147–152
Liang L, Goh SG, Vergara GGRV, Gfang HM, Rezaeinejad S, Chang SY, Bayen S, Lee WA, Sobsey MD, Rose JB, Gin KYH (2015) Alternative fecal indicators and their empirical relationships with enteric viruses, Salmonella enterica, and Pseudomonas aeruginosa in surface waters of a tropical urban catchment. Appl Environ Microbiol 81:850–860
Lu X, Zhang X-X, Wang Z, Huang K, Wang Y, Liang W, Tan Y, Liu B, Tang J (2015) Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS One 10:e0125549
McLellan S, Huse S, Mueller Spitz S, Andreishcheva E, Sogin M (2010) Diversity and population structure of sewage derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12:378–392
McMichael AJ, Lindgren E (2011) Climate change: present and future risks to health, and necessary responses. J Intern Med 270:401–413
Miescier JJ, Cabelli VJ (1982) Enterococci and other microbial indicators in municipal wastewater effluents. J Water Pollut Control Fed 54:1599–1606
Mizrahi-Man O, Davenport ER, Gilad Y (2013) Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS One 8:e53608
Okoh AI, Odjadjare EE, Igbinosa WO, Osode AN (2007) Wastewater treatment plants as a source of microbial pathogens in receiving watersheds. Afr J Biotechnol 6:2932–2944
Ort C, Gujer W (2008) Sorption and high dynamics of micropollutants in sewers. Water Sci Technol 57:1791–1797
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Qian P, Wang Y, Lee O, Lau S, Yang J, Lafi F, Al-Suwailem A, Wong T (2011) Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J 5:507–518
Rodríguez E, García-Encina PA, Stams AJM, Maphosa F, Sousa DZ (2015) Meta-omics approaches to understand and improve wastewater treatment systems. Rev Environ Sci Biotechnol 14:385–406
Rosech L, Fulthorpe R, Riva A, Casella G, Hadwin A, Kent A, Daroub S, Camargo F, Farmerie W, Triplett E (2007) Pyrosequencing enumerates and contrasts soil microbial activity. ISME J 1:507–518
Sanapareddy N, Hamp TJ, Gonzalez LC, Hilger HA, Fodor AA, Clinton SM (2009) Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl Environ Microbiol 75:1688–1696
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Segata N, Huttenhower C (2011) Toward an efficient method of identifying core genes for evolutionary and functional microbial phylogenies. PLoS One 6:e24704
Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL (2013) Comparison of the microbial community structure of untreated wastewaters from different geographic locales. Appl Environ Microbiol 79:2906–2913
Shannon CE, Weaver W (1949) The mathematical theory of communication. The University of Illinois Press, Urbana
Shchgolkova NM, Krasnov GS, Belova AA, Dmitriev AA, Kharitonov SL, Klimina KM, Melnikova NV, Kudryavtseva AV (2016) Microbial community structure of activated sludge in treatment plants with different wastewater compositions. Front Microbiol 7:90
Shrestha PM, Malvankar NS, Werner JJ, Franks AE, Elena-Rotaru A, Shrestha M, Liu F, Nevin KP, Angenent LT, Lovely DR (2014) Correlation between microbial community and granule conductivity in anaerobic bioreactors for brewery wastewater treatment. Bioresource Technol 174:306–310
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proceed Natl Acad Sci 103:12115–12120
Staley C, Gould TJ, Wang P, Philips J, Cotner JB, Sadowsky MJ (2015) Species sorting and seasonal dynamics primarily shape bacterial communities in the upper Mississippi River. Sci Total Environ 505:435–445
Valentin-Vargas A, Toro-Labrador G, Massol-Deya AA (2012) Bacterial community dynamics in full-scale activated sludge bioreactors: operational and ecological factors driving community assembly and performance. PLoS One 7:e42524
Varela AR, Manaia CM (2013) Human health implications of clinically relevant bacteria in wastewater habitats. Environ Sci Pollut Res 20:3550–3569
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wheater DWF, Mara DD, Jawad L, Oragui J (1980) Pseudomonas aeruginosa and Escherichia coli in sewage and freshwater. Water Res 14:713–721
Wong MVM, Hashsham SA, Gulari E, Rouillard J-M, Aw TG, Rose JB (2013) Detection and characterization of human pathogenic viruses circulating in community wastewater using multi target microarrays and polymerase chain reaction. J Water Health 11:659–670
Yadav TK, Khardenavis AK, Kapley A (2014) Shifts in microbial community in response to dissolved oxygen levels in activated sludge. Bioresource Technol 165:257–264
Yang Y, Yu K, Xia Y, Lau FTK, Tang DTW, Fung WC, Fang HHP, Zhang T (2014) Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol 98:4709–5718
Ye L, Zhang T (2011) Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing. Environ Sci Technol 45:7173–7179
Yergeau E, Masson L, Elias M, Xiang S, Madey E, Huang H, Brooks B, Beasdette LA (2016) Comparison of methods to identify pathogens and associated virulence functional genes in biosolids from two different wastewater treatment facilities in Canada. PLoS One. doi:10.1371/journal.pone.0153554
Zhang T, Shao M-F, Ye L (2012) 454 pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6:1137–1147
Acknowledgements
The information used in this paper comes from results of a research project funded by the Australian Centre of Excellence (NATVAL 2.2 subproject 3). Sequence processing and analyses were performed in part using resources of the Minnesota Supercomputing Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical statements
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 195 kb)
Rights and permissions
About this article
Cite this article
Ahmed, W., Staley, C., Sidhu, J. et al. Amplicon-based profiling of bacteria in raw and secondary treated wastewater from treatment plants across Australia. Appl Microbiol Biotechnol 101, 1253–1266 (2017). https://doi.org/10.1007/s00253-016-7959-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7959-9