Abstract
ω-Transaminases (ω-TAs) are one of the most popular candidate enzymes in the biosynthesis of chiral amines. Determination of yet unidentified ω-TAs is important to broaden their potential for synthetic application. Taurine–pyruvate TA (TPTA, EC 2.6.1.77) is an ω-TA belonging to class III of TAs. In this study, we cloned a novel thermostable TPTA from Geobacillus thermodenitrificans (TPTAgth) and overexpressed it in Escherichia coli. The enzyme showed the highest activity at pH 9.0 and 65 °C, with remarkable thermostability and tolerance toward organic solvents. Its K M and v max values for taurine were 5.3 mM and 0.28 μmol s−1 mg−1, respectively. Determination of substrate tolerance indicated its broad donor and acceptor ranges for unnatural substrates. Notably, the enzyme showed relatively good activity toward ketoses, suggesting its potential for catalyzing the asymmetric synthesis of chiral amino alcohols. The active site of TPTAgth was identified by performing protein sequence alignment, three-dimensional structure simulation, and coenzyme pyridoxamine phosphate docking. The protein sequence and structure of TPTAgth were similar to those of TAs belonging to the 3N5M subfamily. Its active site was found to be its special large pocket and substrate tunnel. In addition, TPTAgth showed a unique mechanism of sulfonate/α-carboxylate recognition contributed by Arg163 and Gln160. We also determined the protein sequence fingerprint of TPTAs in the 3N5M subfamily, which involved Arg163 and Gln160 and seven additional residues from 413 to 419 and lacked Phe/Tyr22, Phe85, and Arg409.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chiral amines are important intermediates and have been widely used in pharmaceutical and fine chemical industries (Breuer et al. 2004; Gotor-Fernandez and Gotor 2009; Hohn and Bornscheuer 2009). Several biosynthetic methods, with improved optical purity and yield, involving efficient biocatalysts have been developed for performing amination to replace asymmetric chemical synthesis and to reduce environmental impact. These biosynthetic methods mainly involve enzymes such as amine oxidases, ammonia lyases, amine dehydrogenases, and transaminases (TAs) (Breuer et al. 2004; Gotor-Fernandez and Gotor 2009; Hohn and Bornscheuer 2009). TAs catalyze the introduction of amine groups at carbonyl positions. Therefore, these enzymes are preferred for both kinetic resolution and directed stereoselective synthesis of chiral amines (Ward and Wohlgemuth 2010).
TAs are key enzymes in the synthesis and metabolism of amines in most cellular organisms. They use pyridoxal phosphate (PLP) as a cofactor and catalyze its covalent binding to an amino group from a donor to form pyridoxamine phosphate (PMP). The amino group is then transferred to a carbonyl group of an acceptor to complete the transformation (Shin and Kim 2002). According to sequence- and structure-based analyses, TAs are classified into the fold type I and type IV PLP enzymes (Jansonius 1998). TAs in the former type are further divided into five classes (Rausch et al. 2013). Of these, ω-transaminases (ω-TAs), which belong to class III (Rausch et al. 2013), are the most popular candidate enzymes in biocatalysis because of their synthetic capability to transfer amino groups to isolated carbonyl moieties, thus increasing the range of substrates and products for enzymatic transamination (Koszelewski et al. 2010; Malik et al. 2012).
In the past decade, several studies have been performed on the screening, characterization, crystal structure determination, directed evolution, and synthetic application of ω-TAs. Typical ω-TAs such as those isolated from Vibrio fluvialis (Shin et al. 2003), Paracoccus denitrificans (Park et al. 2010), Caulobacter crescentus (Hwang et al. 2008), Chromobacterium violaceum (Kaulmann et al. 2007), Pseudomonas aeruginosa (Ingram et al. 2007), and Burkholderia vietnamiensis (Jiang et al. 2014) have been successfully cloned and used for the asymmetric synthesis of optically pure aliphatic, aromatic, and arylaliphatic amines (Koszelewski et al. 2010; Malik et al. 2012). Besides (S)-specific ω-TAs, (R)-specific ω-TAs have also been identified, especially by using in silico strategy or single point mutations (Hohne et al. 2010; Iwasaki et al. 2003; Iwasaki et al. 2006; Mutti et al. 2011; Svedendahl et al. 2010). This discovery has enabled the asymmetric synthesis of both (S)- and (R)-amines (Iwasaki et al. 2003; Iwasaki et al. 2006; Schatzle et al. 2011).
Although TAs are promising biocatalysts in industry, some properties of ω-TAs limit their broader synthetic application. One of the main disadvantages of ω-TAs is the limited space in their “large-small” active pockets (Park et al. 2012). Ketone acceptors with bulky moieties are not accurately accepted by wild-type ω-TAs (Malik et al. 2012). In addition, TAs catalyze reversible transamination, which results in an unfavorable reaction equilibrium. These problems can be partly overcome by protein engineering to increase the size of the active pocket (Savile et al. 2010) or by removing reaction by-products to decrease product inhibition and to shift the overall reaction equilibrium (Koszelewski et al. 2010). Determination of yet unidentified ω-TAs is a key topic because diversity of native TA substrates indicates the presence of diverse substrate-binding pockets that may show high affinity toward various unnatural acceptors. Identification of such ω-TAs is expected to broaden their potential for synthetic application.
Phylogenetic tree analysis indicates that TAs belonging to class III can be further divided into six clades, of which clade 6 includes most of the identified ω-TAs (Rausch et al. 2013). Taurine–pyruvate TA (TPTA, EC 2.6.1.77), which catalyzes the transamination between taurine and pyruvate in many animals (Fig. 1), is classified under clade 6a (Huxtable 1992). TPTAs are also extensively present in microorganisms that use taurine as carbon, nitrogen, and sulfur sources (Cook and Denger 2002). Although several TPTAs from Bilophila wadsworthia (Laue and Cook 2000), Rhodopseudomonas palustris (Novak et al. 2004), Rhodobacter sphaeroides (Novak et al. 2004), Clostridium pasteurianum (Chien et al. 1997), Klebsiella oxytoca (von Rekowski et al. 2005), Paracoccus denitrificans (Felux et al. 2013), Ruegeria pomeroyi (Gorzynska et al. 2006), and other microorganisms have been identified, their substrate specificities toward unnatural donors and acceptors and their synthetic capabilities are unknown. Therefore, we focused on the enzymatic properties of TPTAs and examined their potential for application in chiral amine synthesis.
Thermostable enzymes normally maintain their catalytic activity for long term and show better tolerance toward organic co-solvents (Bruins et al. 2001). Use of thermostable ω-TAs at higher reaction temperatures can help in the evaporation of ketone by-products arising from consumed amine donors, thus preventing product inhibition and shifting reaction equilibrium toward amine synthesis. Therefore, we cloned a novel thermostable TPTA from Geobacillus thermodenitrificans and identified its enzymatic properties.
Materials and methods
Materials
G. thermodenitrificans (DSM No.: 465) was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Germany). Fast HiFidelity PCR Kit, pZeroBack/blunt vector, and Escherichia coli competent cells were purchased from Tiangen Biotechnology (China). Restriction enzymes and ligase were purchased from TaKaRa (China). Genome extraction kit and PCR primers were obtained from Generay Biotech (China). (S)-1-Methylbenzylamine, (R)-1-methylbenzylamine, acetophenone, and pentanal were obtained from Aladdin (China). Taurine, (R,S)-1-methylbenzylamine, 2-phenylethylamine, glyoxylate, ethanol, butanal, propanal, hexanal, benzylacetone, 1-indane, and acetoin were purchased from J&K (China). Pyruvate, hydroxypyruvate, α-ketoglutarate, cyclohexanone, glycolaldehyde, d-glyceraldehyde, d-erythrose, hydroxyacetone, 1,3-dihydroxyacetone, and l-erythrulose were purchased from Sigma-Aldrich (China). (S)-1-phenylpropylamine was obtained from Alfa Aesar (China), and (S)-1-methyl-3-phenylpropylamine was purchased from Acros (USA). l-Glyceraldehyde was synthesized by Boyle Chemical (China).
Cloning of TPTAgth
G. thermodenitrificans was cultivated in a medium containing 5.0 g tryptone, 3.0 g meat extract, and 10 mg MnSO4 in 1 l water (pH = 7.0) at 55 °C and by shaking at 250 rpm for 2 days. Genomic DNA of G. thermodenitrificans was extracted using the genome extraction kit (Generay Biotech, China). The gene encoding TPTAgth was amplified using the Fast HiFidelity PCR Kit with primers TPTAf (5ʹ-CATATG AAA ACT GAA CAA GCC ATC AAT TAT GGC T-3ʹ [forward]) and TPTAb (5ʹ-CTCGAG TTT GAT TTG TGC TAA TGC CTC CG-3ʹ [reverse]). Restriction sites for NdeI and XhoI were designed at the 5ʹ and 3ʹ termini, respectively, of the gene encoding TPTAgth. PCR products were ligated into the pZeroBack/blunt vector for subcloning, and the vector was inserted into pET21a(+) vector to obtain an expression plasmid pET21a-TPTAgth. The sequence of the gene encoding TPTAgth was confirmed by performing DNA sequencing (Sunny Biotech, China). Next, the pET21a-TPTAgth plasmids were transformed into E. coli BL21(DE)3 competent cells for overexpressing recombinant TPTAgth.
Recombinant E. coli cells were grown at 37 °C in 500-ml LB medium containing 100 μg/ml ampicillin and were treated with 0.1 mM isopropyl β-d-thiogalactoside (IPTG) at 28 °C for 12 h to induce the expression of recombinant TPTAgth. The cells were harvested by centrifugation at 4000g for 20 min and were resuspended in 20 mM potassium phosphate buffer (pH 8.0) supplemented with 50 mM NaCl and 20 μM PLP. The cells were then treated with lysozyme and were ultrasonicated to obtain a crude cell extract. Recombinant TPTAgth was purified by performing Ni-affinity chromatography (GE Healthcare, USA). The buffer solution was replaced by 20 mM potassium phosphate buffer (pH 8.0) supplemented with 20 μM PLP in ultrafiltration tubes (Merck, Germany), and the samples were stored at −40 °C. Purity of the obtained recombinant protein was determined by performing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Kinetic constants of TPTAgth
Kinetic constants of TPTAgth toward taurine were determined at 37 °C in a 1-ml reaction mixture containing 100 mM Tris buffer (pH 9.0), 0.5–80 mM taurine, 10 mM pyruvate, and 20 μM PLP. The reaction was initiated by adding 0.1, 0.05, or 0.025 mg/ml (final concentration) of the purified enzyme to the reaction mixture. The reaction was stopped after 10 min by adding 2 ml methanol. The amount of pyruvate consumed was analyzed by performing HPLC with SinoChrom ODS-BP (Elite, China) and a mobile phase of MeOH/H2O (20:80) containing 0.03 % H3PO4 (flow rate, 0.8 ml/min). UV detection was performed at 210 nm. The reaction rate at each concentration was calculated. The resultant data were applied in a nonlinear fit to Michaelis–Menten equation by using Origin 8.0 software (OriginLab, USA) to obtain enzyme kinetics values.
Determination of enzymatic properties
A general enzyme assay was performed at 37 °C in a 1-ml reaction mixture containing 100 mM Tris buffer (pH 9.0), 10 mM (S)-methylbenzylamine ([S]-MBA), 10 mM pyruvate, and 20 μM PLP. The reaction was initiated by adding 0.125 mg/ml (final concentration) of the purified enzyme to the reaction mixture. The reaction was stopped after 10 min by adding 2 ml methanol. Acetophenone produced was analyzed by performing HPLC with Zorbax SB-C18 column (Agilent, USA) and a mobile phase of MeOH/H2O (50:50) containing 0.1 % trifluoroacetic acid (TFA) (flow rate, 1 ml/min). UV detection was performed at 254 nm.
Optimal pH was determined by using100 mM potassium phosphate buffer (pH 6.0–7.5), Tris-HCl buffer (pH 8.0–9.0), and glycine-NaOH buffer (pH 9.5–10). TPTAgth activity was measured at 60 °C. Optimal temperature was determined by using 100 mM Tris buffer (pH 9.0) at temperatures ranging from 30 to 70 °C. Thermostability of TPTAgth was determined using potassium phosphate buffer (pH 8.0) containing 20 μM PLP at 50, 60, and 70 °C for up to 8 h. The enzyme samples were taken to determine enzyme activity at 50 °C. T m value was measured in a 25-μl reaction mixture containing 100 mM sodium borate (pH 8.0), 150 mM NaCl, 20 μM PLP, 10 μg enzyme, and 5 μl Sypro Orange (StepOnePlus Real-Time PCR; Applied Biosystems, USA). The real-time PCR program was 25–95 °C, 1 °C per min. Fluorescence data were recorded and were processed with real-time PCR software. Tolerance toward organic solvents was determined at 37 °C by adding up to 40 and 20 % concentrations of dimethyl sulfoxide (DMSO) and methanol, respectively, to the reaction mixture.
Determination of substrate tolerance
Donor spectrum of TPTAgth was determined at 37 °C in a 1-ml reaction mixture containing 100 mM Tris buffer (pH 9.0), 10 mM donor, 10 mM pyruvate, and 20 μM PLP. The reaction was initiated by adding 0.06, 0.16, or 0.20 mg/ml (final concentration) of the purified enzyme to the reaction mixture according to the donor examined. The reaction was stopped after 10 min by adding 2 ml methanol. In the assay for taurine, pyruvate consumed was analyzed by performing HPLC with SinoChrom ODS-BP and a mobile phase of MeOH/H2O (20:80) containing 0.03 % H3PO4 (flow rate, 0.8 ml/min). UV detection was performed at 210 nm. In the assay for other donors, aromatic ketone produce was analyzed by performing HPLC with Zorbax SB-C18 column and a mobile phase of MeOH/H2O (50:50) containing 0.1 % TFA (flow rate, 1 ml/min). UV detection was performed at 254 or 210 nm. Enzyme activity was defined as 1 U of the enzyme required for consuming 1 μmol of pyruvate in 1 min under the described reaction conditions.
Acceptor spectrum was determined at 37 °C in a 200-μl reaction mixture containing 100 mM Tris buffer (pH 8.0), 10 mM (S)-MBA, 10 mM acceptor, and 20 μM PLP. The reaction was initiated by adding 0.125 mg/ml (final concentration) of the purified enzyme to the reaction mixture. Increase in UV absorbance was determined at 245 nm by using a plate reader SpectraMax 190 (Molecular Devices, USA). Enzyme activity was defined as 1 U of the enzyme required for producing 1 μmol of acetophenone in 1 min under the described reaction conditions.
Structure simulation of TPTAgth
Computational simulation was created using SWISS-Model web tool (http://swissmodel.expasy.org/), which recommended Protein Data Bank (PDB) entry 3N5M as an optimum template for aligning protein sequence data and PDB database search. PMP was docked into the active pocket of TPTAgth, and energy minimization was optimized using Discovery Studio 3.5 software (with default parameters) to obtain a complex of TPTAgth with PMP. The structure with the lowest energy was chosen for the structural analysis of the enzyme. PyMOL software (Schrodinger, USA) was used as a visual tool to show and align the simulated structure.
Nucleotide sequence accession number
The nucleotide sequence of TPTAgth has been deposited in the GenBank database under accession number KT719298.
Results
Identification of TPTA from G. thermodenitrificans
To isolate the genes expressing thermostable ω-TAs in G. thermodenitrificans, we chose the protein sequences of two well-studied ω-TAs from V. fluvialis (ω-TAvfl) (Shin et al. 2003) and Paracoccus denitrificans (ω-TApde) (Park et al. 2010) as the starting templates. A BLAST search (http://blast.ncbi.nlm.nih.gov) identified six TA candidates in G. thermodenitrificans NG80-2 (GenBank ID: NC_009328.1), of which the top-rated candidate showed 51 and 52 % identity with ω-TAvfl and ω-TApde, respectively. The information of this candidate enzyme in the BLAST search results only indicated that it was a TA and did not provide any further details, which increased our curiosity to identify its specificity. To predict its potential reactivity, we aligned the obtained amino acid sequence in the B6 database (http://bioinformatics.unipr.it/cgi-bin/bioinformatics/B6db/home.pl) (Percudani and Peracchi 2009) by performing another BLAST search. Our result indicated that this sequence showed 63 and 62 % identity with TPTAs of Bacillus anthracis (TPTAban) and Bacillus subtilis (TPTAbsu), respectively, suggesting that this protein was a TPTA. Therefore, this potential TPTA was chosen for further cloning and characterization.
The obtained DNA sequence of TPTAgth (Fig. S1) showed 99.9 % identity with the sequence of G. thermodenitrificans NG80-2; however, the translated amino acid sequence was completely identical. Soluble recombinant TPTAgth was purified by performing Ni-affinity chromatography to obtain a homogenous protein with an apparent size of 45 kDa, as identified by performing SDS-PAGE (Fig. S2). The kinetic constants of TPTAgth were determined by using taurine as the donor and pyruvate as the acceptor. The K M and v max values for taurine were 5.3 mM and 0.28 μmol s−1 mg−1, respectively. The K M value obtained in the present study was similar to that of TPTAbwa (7.1 mM) determined by performing coupled method (Laue and Cook 2000).
Enzymatic properties of TPTAgth
Enantioselectivity of TPTAgth was determined using (S)-MBA and (R)-MBA as the donors and pyruvate as the acceptor in a reaction in which TPTAgth could only accept the (S)-enantiomer. Conversion of (S)-MBA as a donor results in the formation of acetophenone. The increase in the specific UV absorbance of this compound is suitable for both HPLC determination (Shin and Kim 1997) and high-throughput assay in 96-well microplates (Schatzle et al. 2009). Therefore, unless otherwise specified, (S)-MBA and pyruvate were used for determining the enzymatic properties of TPTAgth.
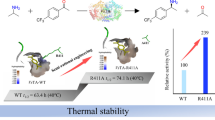
Enzymatic transamination is normally performed under alkaline conditions. The apparent optimum pH of TPTAgth was found to be approximately 9.0 (Fig. 2a), which was similar to that of other TPTAs (pH 8.5–9.5) (Chien et al. 1997; Laue and Cook 2000; Novak et al. 2004). However, the optimum pH range of TPTAgth was very narrow. When pH was <8 or >10, TPTAgth activity was only approximately 20 % or even less. The apparent optimum temperature of TPTAgth was approximately 65 °C (Fig. 2b). However, TPTAgth activity decreased very quickly to <30 % at 70 °C, indicating that it was unstable at temperatures higher than 65 °C. TPTAgth activity at 37 °C was only approximately 17 % of its activity at its optimum temperature.
Enzymatic properties of TPTAgth. The error bars represent the standard deviation (SD) of the mean. a Optimum pH range of TPTAgth. b Optimum temperature range of TPTAgth. c–e Thermostability of TPTAgth (c 50 °C, d 60 °C, and e 70 °C). f Organic co-solvent tolerance of TPTAgth. The reaction system contains 10 mM (S)-MBA, 10 mM pyruvate, 20 μM PLP, and 0.125 mg/ml TPTAgth. The optimum temperature and thermostability used 100 mM Tris buffer (pH = 9.0). For the determination of thermostability, the enzymes in potassium phosphate buffer (pH = 8.0) with 20 μM PLP were incubated at 50, 60, and 70 °C for up to 8 h before the activity determination
Because the optimum temperature for TPTAgth activity ranged between 50 and 70 °C and because the optimum growth temperature of G. thermodenitrificans is approximately 60 °C (Cihan et al. 2011), we determined the thermostability of TPTAgth at 50, 60, and 70 °C (Fig. 2c–e). TPTAgth showed remarkable thermostability similar to other thermophilic enzymes. Incubation of TPTAgth at 50 °C for 8 h increased its activity by up to 2.5-fold (Fig. 2c). The same activity increase was also observed after incubating the enzyme at 60 °C for 30 min (Fig. 2d). This increase in activity was probably caused by temperature-related folding that increased stabilizing interactions, thus improving its reactivity (Bruins et al. 2001). However, TPTAgth lost its activity almost completely after incubation at 70 °C for 15 min (Fig. 2e). Thermostability of TPTAgth was also determined by using a ThermoFluor-based method by using the hydrophobic fluoroprobe SYPRO orange as a dye. Denaturation of a protein at high temperatures in an aqueous solution exposes its hydrophobic interior to the probe, leading to a sharp decrease in the quenching of the dye. This readily detectable fluorescence emission can be determined by performing RT-PCR as a function of temperature (Ericsson et al. 2006; Niesen et al. 2007). This ThermoFluor-based determination showed that the T m value of TPTAgth was approximately 70 °C.
Tolerance of TPTAgth toward organic co-solvents was determined by adding DMSO and methanol (Fig. 2f). TPTAgth activity decreased very quickly after adding >5 % methanol. Interestingly, an increase in DMSO concentration up to 10 % enhanced TPTAgth activity by up to 135 %. The further increase in DMSO concentration decreased the enzyme activity. However, interestingly, TPTAgth retained approximately 90 % of its activity in the presence of 20 % DMSO.
Substrate spectrum of TPTAgth
To evaluate the substrate specificity of TPTAgth, we determined its reactivity toward a series of aromatic amine donors and several keto acids, aldehydes, ketones, aldoses, and ketoses as acceptors. The results are listed in Table 1. TPTAgth showed the best specific activity (10.2 U/mg) toward taurine compared with that toward aromatic amine donors. Its specific activity toward (S)-MBA was 0.26 U/mg; however, no activity was observed toward the (R)-enantiomer. When racemic MBA at two times the concentration (20 mM) was used as the donor, the specific activity of TPTAgth was found to be 0.29 U/mg, which was very similar to its activity toward 10 mM (S)-MBA. This result indicated that (R)-MBA did not inhibit TPTAgth activity. Extension of side chains of aromatic amines decreased the donor affinity of TPTAgth. It showed only 25 % activity (0.065 U/mg) toward (S)-1-methyl-3-phenylpropylamine compared with that toward (S)-MBA. In addition, TPTAgth showed limited activity toward (S)-1-phenylpropylamine (0.001 U/mg). However, TPTAgth showed an activity of 0.080 U/mg toward the aromatic primary amine 2-phenylethylamine; this activity was approximately 8 and 31 % of its activity toward taurine and (S)-MBA, respectively.
Acceptor specificity of TPTAgth was determined by performing a high-throughput UV assay by using 96-well microplates (Schatzle et al. 2009). TPTAgth showed activity toward several keto acids such as pyruvate, hydroxypyruvate, and glyoxylate but showed almost no activity toward 2-oxoglutarate, indicating that TPTAgth was not a taurine:2-oxoglutarate TA (EC 2.6.1.55) (Mikosch et al. 1999). Pyruvate is a natural acceptor, with TPTAgth showing 0.24 U/mg of specific activity toward it. A substituent hydrogen group at the β-position of pyruvate might have formed one or more hydrogen bonds with TPTAgth, thus slightly increasing its activity toward 3-hydroxypyruvate by 0.32 U/mg. The aldehyde group of glyoxylate is more active than its ketone group. Therefore, TPTAgth activity toward glyoxylate (0.71 U/mg) was approximately 3-fold higher than that toward pyruvate. For the same reason, TPTAgth showed higher activity toward aliphatic aldehydes ranging from methanal to hexanal, without any significant difference in the observed activity. Extension of carbon chains had no unfavorable effect on the acceptor affinity of TPTAgth. TPTAgth activity toward aldoses such as glycolaldehyde, d-glyceraldehyde, and l-glyceraldehyde was approximately 0.60–0.75 U/mg, which was similar to its activity toward aliphatic aldehydes. The almost equal activity toward d-glyceraldehyde and l-glyceraldehyde suggested that TPTAgth did not show stereoselection toward the hydroxyl group at α-position. d-Erythrose is mainly present in its furanose form in the aqueous solution. Therefore, the specific activity of TPTAgth toward d-erythrose was only 0.12 U/mg. Unfortunately, TPTAgth showed very low activity toward ketones. Almost no activity was detected toward acetone, butanone, cyclohexanone, benzylacetone, and 1-indanone. This might be because of the low affinity of TPTAgth toward ketones as well as its low activity at low temperature (37 °C). Interestingly, TPTAgth showed activity toward ketoses. Its activity toward hydroxyacetone, 1,3-dihydroxyacetone, acetoin, and l-erythrulose was 0.074, 0.10, 0.038, and 0.13 U/mg, respectively. This may be because of the electrophilic effect of ortho hydroxy moiety, which enhances the reactivity of the ketone group. In addition, the hydroxy moieties of ketoses favor the binding of the polar residues in the active pocket of TPTAgth, which may contribute to the higher activity of TPTAgth with ketoses.
Identification of the active pocket of TPTAgth
To determine the key amino acid residues in the active pocket of TPTAgth, we first aligned the protein sequence of TPTAgth with that of the PDB item 3N5M. We identified three confirmed TPTAs from Bilophila wadsworthia (UniProt ID: Q9APM5) (Laue and Cook 2000), Paracoccus denitrificans (UniProt ID: A1B9Z3) (Felux et al. 2013), Ruegeria pomeroyi (UniProt ID: Q5LVM7) (Gorzynska et al. 2006) in the 3N5M subfamily (Steffen-Munsberg et al. 2015); two potential TPTAs from Bacillus anthracis (UniProt ID: Q81SL2) and Bacillus subtilis (UniProt ID: P33189) in the B6 database; and four well-studied ω-TAs from Pseudomonas putida (UniProt ID: P28269, subclade 6b) (Rausch et al. 2013; Watanabe et al. 1989), Paracoccus denitrificans (UniProt ID: A1B956, subclade 6c) (Park et al. 2010; Rausch et al. 2013), Chromobacterium violaceum (UniProt ID: Q7NWG4, subclade 6c) (Kaulmann et al. 2007; Rausch et al. 2013), and V. fluvialis (UniProt ID: F2XBU9, subclade 6c) (Rausch et al. 2013; Shin et al. 2003) (Fig. S3). A previous study showed that Tyr(Phe)23, Phe88, and Tyr152 (number for ω-TAppu) contributed to hydrophobic interactions in the large pockets of most ω-TAs (Park et al. 2012). Further, Trp60 and Ile262 (number for ω-TAppu) provided a steric barrier in the small pocket (Park et al. 2012). Results of sequence alignment indicated that TPTAgth had identical key residues in the small pocket (Trp57 and Ile255). In TPTAbwa, TPTApde, and TPTArpo, Ile262 (number for ω-TAppu) is replaced by the same nonpolar residue Val but with a shorter carbon chain. However, the key residues in the large pocket of TPTAs are different from those in other ω-TAs, except Tyr152, which is identical in all the TAs listed in this study. Tyr(Phe)23 (number for ω-TAppu) is replaced by Met or Leu in TPTAs. Interestingly, substitution of residues at Phe88 (number for ω-TAppu) in TPTAs does not follow any obvious rules. Thr, Ser, Gly, Ala, and Trp can be found at this position in TPTAs, suggesting that the residue at Phe88 (number for ω-TAppu) did not contribute to the substrate binding of TPTAs. Furthermore, Arg414 (number for ω-TAppu) is the key residue for α-carboxylate recognition of substrates (Park et al. 2012). However, in TPTAs, Arg414 is replaced by Gly without any side chain. Therefore, we presumed that TPTAs represented a different mechanism for α-carboxylate recognition, mainly because of their special donor taurine.
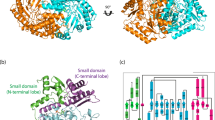
Crystal structure of TPTAs has not been reported in the RCSB PDB thus far. Lack of information on the structure of TPTAs limits the determination of interactions between TPTAs and their substrates. Therefore, we attempted to simulate a three-dimensional structure of TPTAgth by using the SWISS-MODEL web tool (Arnold et al. 2006; Biasini et al. 2014; Guex et al. 2009; Kiefer et al. 2009). Automatic template search based on sequence similarity recommended 3N5M as the template, with 67 % sequence identity. Although the item 3N5M has been marked as adenosylmethionine-8-amino-7-oxononanoate TA (ATA, EC 2.6.1.62), its apparent activity has not been identified by any study thus far. Interestingly, the protein sequence of 3N5M was almost identical to that of TPTAban (99 % identity) in the B6 database. Therefore, we speculated that protein 3N5M was a TPTA instead of ATA. The simulated overall structure of TPTAgth obtained using 3N5M as the template is shown in Fig. 3a. This structure was highly similar to that of 3N5M, with an almost identical active pocket.
Simulated structure and active pocket of TPTAgth. a Overall structure alignment of TPTAgth (orange) and 3N5M (cyan). b Active pocket of TPTAgth. c Substrate tunnel of TPTAgth. d Substrate tunnel of ω-TAvfl. The key residues for the large and small pockets are marked in green and cyan, respectively. The reactive Lys was marked in salmon. The key residues for substrate recognition were marked in slate. The PMP docking into the active pocket is in magenta. Blue and red in the residues represent nitrogen and oxygen, respectively. The additional loop (ATVAGYN) was marked in warm pink. The red arrows show the direction of substrate entrance into the substrate tunnels
Similar to substrate pockets of other ω-TAs, the substrate pocket of TPTAgth contained several flexible loops (Fig. 3b). The coenzyme PMP was located at the bottom of the pocket. Lys281 was the active residue that formed an internal aldimine link with the cofactor PLP. Trp57 and Ile255 formed the small pocket with Glu219 and Gly224, which was very similar to that in ω-TApde, ω-TAvfi, ω-TAcvi, and ω-TAppu. However, Met22 and Thr85 in the large pocket of TPTAgth constricted an uncharged polar pocket with Val315, Asn316, Thr317, and Leu56 compared with a hydrophobic pocket formed by Tyr23 and Phe88 (number for ω-TAppu) in ω-TApde, ω-TAvfi, ω-TAcvi, and ω-TAppu. Compared with Tyr and Phe, smaller side chains of Met22 and Thr85 apparently offered more room to accept a bulky moiety of substrates.
The result of protein sequence alignment showed that Arg414 (number for ω-TAppu) was missing (substituted by Gly) in all TPTAs of the 3N5M subfamily listed in this study. In the TPTAgth structure, we did not observe any alternative Arg residue in the loop containing Arg414 (number for ω-TAppu). However, at the “bottom” of the substrate tunnel, Arg163 showed a side chain in a rational rotation into the active pocket (Fig. 3b). Arg163 may perform the same function as Arg414 (number for ω-TAppu) in ω-TApde, ω-TAvfi, ω-TAcvi, and ω-TAppu. In addition, Gln160 was spatially located between Arg163 and Tyr147, which may offer a hydrogen bond to the sulfonic group of taurine (Fig. 3b). Arg163 and Gln160 are highly conserved in TPTAs belonging to the 3N5M subfamily. Therefore, we presumed that they were key residues for sulfonate recognition and binding.
The structure of TPTAgth also showed an additional sequence (ATVAGYN) between α15 and β13 (Steffen-Munsberg et al. 2015) (Fig. S3), which formed a relatively bulky loop, in the overall structure compared with that in the structures of ω-TApde, ω-TAvfi, ω-TAcvi, and ω-TAppu (Fig. 3b, c). Interestingly, structural alignment suggested that this loop blocked substrate entry in ω-TApde, ω-TAvfi, ω-TAcvi, and ω-TAppu (Fig. 3d). However, substitution of Tyr23, Phe88, and Arg414 (number for ω-TAppu) in TPTAgth by Met, Thr, and Gly, respectively, opened an alternative tunnel that was also present in other TPTAs listed in this study (Fig. 3c). Therefore, we speculated that this tunnel was important for substrate entry in TPTAs.
Discussion
Several thermophilic Geobacillus strains such as Geobacillus stearothermophilus, G. thermodenitrificans, Geobacillus thermoglucosidasius, and Geobacillus thermoleovorans have been isolated from soil (Cihan et al. 2011; Nazina et al. 2001). Genome sequences of 11 Geobacillus spp. are recorded in the NCBI genome database, thus offering a valuable repository for determining genes encoding various thermostable enzymes. We used BLAST search to successfully identify the gene encoding TPTAgth and overexpressed its recombinant form in E. coli. TPTAgth shows the common features of thermophilic enzymes. The high thermostability of TPTAgth can facilitate its simple purification by using a heat shock method, its long-term activity for biosynthesis, especially with weak substrates, and its potential synthetic capability by increasing reaction temperature to evaporate ketone by-products to shift the reaction equilibrium. Low tolerance of ω-TAs toward organic solvents has limited their synthetic application with insoluble acceptors that require organic co-solvents for improving their solubility in aqueous reaction systems (Savile et al. 2010). Therefore, the remarkable tolerance of TPTAgth toward organic solvents can improve the solubility of substrates by increasing co-solvent concentration in the reaction system.
We observed that TPTAgth was a (S)-selective ω-TA. This was also confirmed by performing a novel fluorescent assay for high-throughput discovery and engineering of stereoselective ω-TAs, in which 1-(6-methoxynaphth-2-yl)alkylamines were used as sensitive fluorescent probes (Scheidt et al. 2015). The K M values of TPTAgth were 0.80 and 1.6 mM for (S)- and (R)-1-(6-methoxynaphth-2-yl)ethylamine, respectively, with an E-factor of 38(S) (Scheidt et al. 2015). The substrate spectrum of TPTAgth was relatively wide, which was similar to that of other ω-TAs such as the two well-studied ω-TAs ω-TAvfl (Shin and Kim 2002; Shin et al. 2003) and ω-TAcvi (Kaulmann et al. 2007) (Fig. 4). The donor spectrum of TPTAgth was similar to that of ω-TAvfl. In addition, both the enzymes showed better activity toward (S)-MBA than toward other aromatic amines. However, ω-TAcvi showed much higher activity toward (S)-1-methyl-3-phenylpropylamine. With respect to acceptors, TPTAgth and ω-TAcvi showed increased activity toward pyruvate, hydroxypyruvate, and glyoxylate, while ω-TAvfl preferred pyruvate over glyoxylate. ω-TAcvi showed slight activity toward α-ketoglutarate, while TPTAgth and TAvfl did not. The highest activity of ω-TAvfl was observed toward aldehydes, which was similar to that observed for TPTAgth. However, ω-TAcvi showed slightly lower activity toward aliphatic aldehydes than TPTAgth. Both TPTAgth and ω-TAvfl showed weak activity toward keto acceptors. The relatively high activity toward aldoses and ketoses is a remarkable property of TPTAgth; however, ω-TAcvi showed a similar but slightly lower activity. Therefore, TPTAgth could be a potential biocatalyst for the asymmetric synthesis of chiral amino alcohols.
Fingerprint comparison for TPTAgth, ω-TAvfl, and ω-TAcvi. The relative specific activity data was converted to grayscale values. The specific activity with donor (R,S)-1-methylbenzylamine, (S)-1-methylbenzylamine, and (S)-1-methyl-3-phenylpropylamine was calculated as 100 % reference for TPTAgth, ω-TAvfl, and ω-TAcvi, respectively. The specific activity with acceptor pentanal, butanal, and glyoxylate was calculated as 100 % reference for TPTAgth, ω-TAvfl, and ω-TAcvi, respectively. D1 (S)-1-methylbenzylamine, D2 (R)-1-methylbenzylamine, D3 (R,S)-1-methylbenzylamine, D4 2-phenylethylamine, D5 (S)-1-phenylpropylamine, D6 (S)-1-methyl-3-phenylpropylamine, A1 pyruvate, A2 hydroxypyruvate, A3 glyoxylate, A4 α-ketoglutarate, A5 methanol, A6 ethanol, A7 propanal, A8 butanal, A9 pentanal, A10 hexanal, A11 acetone, A12 butanone, A13 cyclohexanone, A14 benzylacetone, A15 1-indane, A16 glycolaldehyde, A17 d-glyceraldehyde, A18 l-glyceraldehyde, A19 d-erythrose, A20 hydroxyacetone, A21 1,3-dihydroxyacetone, A22 acetoin, A23 l-erythrulose (n.d. no data)
The active site of ω-TAs for recognizing substrates can be divided into a large and a small pocket (Park et al. 2012). However, the diversity of amino acid residues in these two pockets leads to different substrate recognition by ω-TAs. A recent systematic bioinformatics analysis of TAs, especially class III TAs, provided details of the key amino acid residues for the functional prediction of these enzymes from their protein sequences and provided a better understanding of the mechanism underlying substrate recognition by these enzymes (Steffen-Munsberg et al. 2015). Unfortunately, the fingerprint of TPTAs in the 3N5M subfamily could not be obtained because of the lack of structure information, which becomes special interest in the studies on ω-TAs (Steffen-Munsberg et al. 2015). We identified the active pocket of TPTAgth by performing protein sequence alignment, three-dimensional structure simulation, and coenzyme PMP docking. Although the large and small pockets of TPTAgth were similar to those of other ω-TAs, TPTAgth also contained a special substrate tunnel, mainly composed of Met22, Thr85, Val315, and Arg163. Interestingly, this substrate tunnel was located closer to the large pocket than to the small pocket in other kinds of ω-TAs. This apparently enlarged the large pocket, which might help in the entry of substrates with bulky moieties. Our results showed that TPTAgth had a unique mechanism for sulfonate/α-carboxylate recognition and binding, which was contributed by Arg163 and Gln160. The results of sequence and simulated structure alignment showed that other TPTAs in the 3N5M subfamily had the same sequence and structural features as TPTAgth. Therefore, we concluded that the protein sequence fingerprint of TPTAs in the 3N5M subfamily involved Arg163 and Gln160 and seven additional residues from 413 to 419 and lacked Phe/Tyr22, Phe85, and Arg409.
In conclusion, we cloned and characterized a novel thermostable TPTA from G. thermodenitrificans that showed remarkable thermostability, organic solvent tolerance, and relatively broad donor and acceptor spectra, including unnatural substrates. The active site of TPTAgth was found to be its special large pocket and substrate tunnel. In addition, TPTAgth showed a unique mechanism of sulfonate/α-carboxylate recognition. We also determined the protein sequence fingerprint of TPTAs in the 3N5M subfamily. We expect that this sequence and structural fingerprint could be used as a reference for identifying more TPTAs belonging to the 3N5M subfamily from gene and protein databases and for performing rational design and evolution studies on TPTAs to obtain valuable variants for chiral amine synthesis.
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi:10.1093/bioinformatics/bti770
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi:10.1093/nar/gku340
Breuer M, Ditrich K, Habicher T, Hauer B, Kesseler M, Sturmer R, Zelinski T (2004) Industrial methods for the production of optically active intermediates. Angew Chem Int Ed Engl 43:788–824. doi:10.1002/anie.200300599
Bruins ME, Janssen AE, Boom RM (2001) Thermozymes and their applications: a review of recent literature and patents. Appl Biochem Biotechnol 90:155–186. doi:10.1385/ABAB:90:2:155
Chien C, Leadbetter ER, Godchaux W (1997) Taurine-sulfur assimilation and taurine-pyruvate aminotransferase activity in anaerobic bacteria. Appl Environ Microbiol 63:3021–3024, Retrieved from http://aem.asm.org/content/63/8/3021.abstract?sid=4806b8f9-eb22-433d-ba1a-d40b2e2f161c
Cihan AC, Ozcan B, Tekin N, Cokmus C (2011) Geobacillus thermodenitrificans subsp. calidus, subsp. nov., a thermophilic and α-glucosidase producing bacterium isolated from Kizilcahamam, Turkey. J Gen Appl Microbiol 57:83–92. doi:10.2323/jgam.57.83
Cook AM, Denger K (2002) Dissimilation of the C2 sulfonates. Arch Microbiol 179:1–6. doi:10.1007/s00203-002-0497-0
Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem 357:289–298. doi:10.1016/j.ab.2006.07.027
Felux AK, Denger K, Weiss M, Cook AM, Schleheck D (2013) Paracoccus denitrificans PD1222 utilizes hypotaurine via transamination followed by spontaneous desulfination to yield acetaldehyde, and finally, acetate for growth. J Bacteriol 195:2921–2930. doi:10.1128/JB.00307-13
Gorzynska AK, Denger K, Cook AM, Smits TH (2006) Inducible transcription of genes involved in taurine uptake and dissimilation by Silicibacter pomeroyi DSS-3T. Arch Microbiol 185:402–406. doi:10.1007/s00203-006-0106-8
Gotor-Fernandez V, Gotor V (2009) Biocatalytic routes to chiral amines and amino acids. Curr Opin Drug Discov Devel 12:784–797, Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/19894190
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–173. doi:10.1002/elps.200900140
Hohn M, Bornscheuer UT (2009) Biocatalytic routes to optically active amines. Chemcatchem 1:42–51. doi:10.1002/cctc.200900110
Hohne M, Schatzle S, Jochens H, Robins K, Bornscheuer UT (2010) Rational assignment of key motifs for function guides in silico enzyme identification. Nat Chem Biol 6:807–813. doi:10.1038/nchembio.447
Huxtable RJ (1992) Physiological actions of taurine. Physiol Rev 72:101–163, Retrieved from http://physrev.physiology.org/content/72/1/101
Hwang BY, Ko SH, Park HY, Seo JH, Lee BS, Kim BG (2008) Identification of ω-aminotransferase from Caulobacter crescentus and site-directed mutagenesis to broaden substrate specificity. J Microbiol Biotechnol 18:48–54, Retrieved from http://www.jmb.or.kr/journal/viewJournal.html?year=2008&vol=18&num=1&page=48
Ingram CU, Bommer M, Smith MEB, Dalby PA, Ward JM, Hailes HC, Lye GJ (2007) One-pot synthesis of amino-alcohols using a de-novo transketolase and β-alanine: pyruvate transaminase pathway in Escherichia coli. Biotechnol Bioeng 96:559–569. doi:10.1002/bit.21125
Iwasaki A, Yamada Y, Ikenaka Y, Hasegawa J (2003) Microbial synthesis of (R)- and (S)-3,4-dimethoxyamphetamines through stereoselective transamination. Biotechnol Lett 25:1843–1846. doi:10.1023/A:1026229610628
Iwasaki A, Yamada Y, Kizaki N, Ikenaka Y, Hasegawa J (2006) Microbial synthesis of chiral amines by (R)-specific transamination with Arthrobacter sp. KNK168. Appl Microbiol Biotechnol 69:499–505. doi:10.1007/s00253-005-0002-1
Jansonius JN (1998) Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol 8:759–769. doi:10.1016/S0959-440X(98)80096-1
Jiang JJ, Chen X, Feng JH, Wu QQ, Zhu DM (2014) Substrate profile of an ω-transaminase from Burkholderia vietnamiensis and its potential for the production of optically pure amines and unnatural amino acids. J Mol Catal B Enzym 100:32–39. doi:10.1016/j.molcatb.2013.11.013
Kaulmann U, Smithies K, Smith MEB, HaileS HC, Ward JM (2007) Substrate spectrum of ω-transaminase from Chromobacterium violaceum DSM30191 and its potential for biocatalysis. Enzyme Microb Tech 41:628–637. doi:10.1016/j.enzmictec.2007.05.011
Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387–392. doi:10.1093/nar/gkn750
Koszelewski D, Tauber K, Faber K, Kroutil W (2010) ω-Transaminases for the synthesis of non-racemic α-chiral primary amines. Trends Biotechnol 28:324–332. doi:10.1016/j.tibtech.2010.03.003
Laue H, Cook AM (2000) Biochemical and molecular characterization of taurine:pyruvate aminotransferase from the anaerobe Bilophila wadsworthia. Eur J Biochem 267:6841–6848. doi:10.1046/j.1432-1033.2000.01782.x
Malik MS, Park ES, Shin JS (2012) Features and technical applications of ω-transaminases. Appl Microbiol Biotechnol 94:1163–1171. doi:10.1007/s00253-012-4103-3
Mikosch CA, Denger K, Schafer EM, Cook AM (1999) Anaerobic oxidations of cysteate: degradation via L-cysteate:2-oxoglutarate aminotransferase in Paracoccus pantotrophus. Microbiology 145(Pt 5):1153–1160. doi:10.1099/13500872-145-5-1153
Mutti FG, Fuchs CS, Pressnitz D, Sattler JH, Kroutil W (2011) Stereoselectivity of four (R)-Selective transaminases for the asymmetric amination of ketones. Adv Synth Catal 353:3227–3233. doi:10.1002/adsc.201100558
Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, Lysenko AM, Petrunyaka VV, Osipov GA, Belyaev SS, Ivanov MV (2001) Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int J Syst Evol Microbiol 51:433–446. doi:10.1099/00207713-51-2-433
Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi:10.1038/nprot.2007.321
Novak RT, Gritzer RF, Leadbetter ER, Godchaux W (2004) Phototrophic utilization of taurine by the purple nonsulfur bacteria Rhodopseudomonas palustris and Rhodobacter sphaeroides. Microbiology 150:1881–1891. doi:10.1099/mic.0.27023-0
Park E, Kim M, Shin JS (2010) One-pot conversion of L-threonine into L-homoalanine: biocatalytic production of an unnatural amino acid from a natural one. Adv Synth Catal 352:3391–3398. doi:10.1002/adsc.201000601
Park ES, Kim M, Shin JS (2012) Molecular determinants for substrate selectivity of ω-transaminases. Appl Microbiol Biotechnol 93:2425–2435. doi:10.1007/s00253-011-3584-9
Percudani R, Peracchi A (2009) The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinformatics 10:273. doi:10.1186/1471-2105-10-273
Rausch C, Lerchner A, Schiefner A, Skerra A (2013) Crystal structure of the ω-aminotransferase from Paracoccus denitrificans and its phylogenetic relationship with other class III aminotransferases that have biotechnological potential. Proteins 81:774–787. doi:10.1002/prot.24233
Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ (2010) Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329:305–309. doi:10.1126/science.1188934
Schatzle S, Hohne M, Redestad E, Robins K, Bornscheuer UT (2009) Rapid and sensitive kinetic assay for characterization of ω-transaminases. Anal Chem 81:8244–8248. doi:10.1021/ac901640q
Schatzle S, Steffen-Munsberg F, Thontowi A, Hohne M, Robins K, Bornscheuer UT (2011) Enzymatic asymmetric synthesis of enantiomerically pure aliphatic, aromatic and arylaliphatic amines with (R)-selective amine transaminases. Adv Synth Catal 353:2439–2445. doi:10.1002/adsc.201100435
Scheidt T, Land H, Anderson M, Chen YJ, Berglund P, Yi D, Fessner WD (2015) Fluorescence-based kinetic assay for high-throughput discovery and engineering of stereoselective ω-transaminases. Adv Synth Catal 357:1721–1731. doi:10.1002/adsc.201500215
Shin JS, Kim BG (1997) Kinetic resolution of α-methylbenzylamine with ω-transaminase screened from soil microorganisms: application of a biphasic system to overcome product inhibition. Biotechnol Bioeng 55:348–358. doi:10.1002/(SICI)1097-0290(19970720)55:2<348::AID-BIT12>3.0.CO;2-D
Shin JS, Kim BG (2002) Exploring the active site of amine: pyruvate aminotransferase on the basis of the substrate structure-reactivity relationship: how the enzyme controls substrate specificity and stereo selectivity. J Org Chem 67:2848–2853. doi:10.1021/jo016115i
Shin JS, Yun H, Jang JW, Park I, Kim BG (2003) Purification, characterization, and molecular cloning of a novel amine:pyruvate transaminase from Vibrio fluvialis JS17. Appl Microbiol Biotechnol 61:463–471. doi:10.1007/s00253-003-1250-6
Steffen-Munsberg F, Vickers C, Kohls H, Land H, Mallin H, Nobili A, Skalden L, van den Bergh T, Joosten HJ, Berglund P, Höhne M, Bornscheuer UT (2015) Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol Adv 33:566–604. doi:10.1016/j.biotechadv.2014.12.012
Svedendahl M, Branneby C, Lindberg L, Berglund P (2010) Reversed enantiopreference of an ω-transaminase by a single-point mutation. Chemcatchem 2:976–980. doi:10.1002/cctc.201000107
von Rekowski KS, Denger K, Cook AM (2005) Isethionate as a product from taurine during nitrogen-limited growth of Klebsiella oxytoca TauN1. Arch Microbiol 183:325–330. doi:10.1007/s00203-005-0776-7
Ward J, Wohlgemuth R (2010) High-yield biocatalytic amination reactions in organic synthesis. Curr Org Chem 14:1914–1927. doi:10.2174/138527210792927546
Watanabe N, Sakabe K, Sakabe N, Higashi T, Sasaki K, Aibara S, Morita Y, Yonaha K, Toyama S, Fukutani H (1989) Crystal structure analysis of ω-amino acid:pyruvate aminotransferase with a newly developed Weissenberg camera and an imaging plate using synchrotron radiation. J Biochem 105:1–3, Retrieved from https://www.jstage.jst.go.jp/article/biochemistry1922/105/1/105_1_1/_article
Acknowledgments
This work was funded by the National Natural Science Funds of China (Grant No. 21406069).
Conflict of interest
Mr. Yujie Chen declares that he has no conflict of interest.
Dr. Dong Yi declares that he has no conflict of interest.
Ms. Shuiqin Jiang declares that she has no conflict of interest.
Prof. Dr. Dongzhi Wei declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yujie Chen and Dong Yi contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Nucleotide sequence of TPTAgth. The nucleotide sequence of TPTAgth has been deposited in the GenBank database under accession number KT719298. (PDF 99 kb)
Fig. S2
SDS-PAGE analysis of purified recombinant TPTAgth. (PDF 151 kb)
Fig. S3
Protein sequence alignment of TPTAgth. ω-TApde: ω-TA from Paracoccus denitrificans (UniProt ID: A1B956), ω-TAvfl: ω-TA from Vibrio fluvialis (UniProt ID: F2XBU9), ω-TAcvi: ω-TA from Chromobacterium violaceum (UniProt ID: Q7NWG4), ω-TAppu: ω-TA from Pseudomonas putida (UniProt ID: P28269), 3N5M: PDB item 3N5M, TPTAban: TPTA from Bacillus anthracis (UniProt ID: Q81SL2), TPTAbsu: TPTA from Bacillus subtilis (UniProt ID: P33189), TPTAbwa: TPTA from Bilophila wadsworthia (UniProt ID: Q9APM5), TPTApde: TPTA from Paracoccus denitrificans (UniProt ID: A1B9Z3), TPTArpo: TPTA from Ruegeria pomeroyi (UniProt ID: Q5LVM7). The sequence alignment was achieved by NTI Vector software 11.5 (ThermoFischer, USA). The identical residues are marked as *. The key residues are highlighted in bold. Arg163, Gln160 and the additional loop between α15 and β13 are highlighted in grey. (PDF 143 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Yi, D., Jiang, S. et al. Identification of novel thermostable taurine–pyruvate transaminase from Geobacillus thermodenitrificans for chiral amine synthesis. Appl Microbiol Biotechnol 100, 3101–3111 (2016). https://doi.org/10.1007/s00253-015-7129-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7129-5