Abstract
Peroxiredoxins (Prxs), also termed thioredoxin peroxidases (TPXs), are a family of thiol-specific antioxidant enzymes that are critically involved in cell defense and protect cells from oxidative damage. In this study, a putative chloroplastic 2-Cys thioredoxin peroxidase (OsTPX) was identified by proteome analysis from leaf tissue samples of rice (Oryza sativa) seedlings exposed to 0.1 M NaCl for 3 days. To investigate the relationship between the OsTPX gene and the stress response, OsTPX was cloned into the yeast expression vector p426GPD under the control of the glyceraldehyde-3-phosphate dehydrogenase (GPD1) promoter, and the construct was transformed into Saccharomyces cerevisiae cells. OsTPX expression was confirmed by semi-quantitative reverse transcription-polymerase chain reaction and western blot analyses. OsTPX contained two highly conserved cysteine residues (Cys114 and Cys236) and an active site region (FTFVCPT), and it is structurally very similar to human 2-Cys Prx. Heterologous OsTPX expression increased the ability of the transgenic yeast cells to adapt and recover from reactive oxygen species (ROS)-induced oxidative stresses, such as a reduction of cellular hydroperoxide levels in the presence of hydrogen peroxide and menadione, by improving redox homeostasis. OsTPX expression also conferred enhanced tolerance to tert-butylhydroperoxide, heat shock, and high ethanol concentrations. Furthermore, high OsTPX expression improved the fermentation capacity of the yeast during glucose-based batch fermentation at a high temperature (40 °C) and at the general cultivation temperature (30 °C). The alcohol yield in OsTPX-expressing transgenic yeast increased by approximately 29 % (0.14 g g−1) and 21 % (0.12 g g−1) during fermentation at 40 and 30 °C, respectively, compared to the wild-type yeast. Accordingly, OsTPX-expressing transgenic yeast showed prolonged cell survival during the environmental stresses produced during fermentation. These results suggest that heterologous OsTPX expression increases acquired tolerance to ROS-induced oxidative stress by improving cellular redox homeostasis and improves fermentation capacity due to improved cell survival during fermentation, especially at a high temperature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Redox balance is important in cellular physiology and is affected by maintaining a steady-state balance between oxidants and antioxidants. However, it has been proposed that oxidative stress may disrupt redox signaling and control (Rouhier et al. 2008). Aerobic organisms have evolved a complex network of mechanisms to maintain redox homeostasis under the unfavorable environmental alterations imposed by their natural habitats (Ashraf 2009). The main components of antioxidant defense systems are low molecular mass antioxidants such as ascorbate, glutathione (GSH), thioredoxin (Trx), and tocopherol, as well as the enzymes of antioxidant systems (mainly the Halliwell–Asada cycle in plants), including superoxide dismutase, catalases, and peroxidases (Dietz 2007), which can be used to sequentially neutralize reactive oxygen species (ROS) (Rouhier and Jacquot 2005). Trx and GSH are important antioxidant molecules that are maintained in their reduced forms by enzymes involved in the thioredoxin–glutathione cycle. These enzymes, glutathione peroxidase, glutathione reductase, thioredoxin reductase, and thioredoxin peroxidase (TPX), are critical in protecting cells from oxidative stress and minimizing their subsequent degradation (Kingston-Smith and Foyer 2000). Ascorbate peroxidases, which are involved in the ascorbate-dependent water/water cycle in plants, are heme-containing enzymes with very high specificity and very high turnover with hydrogen peroxide (H2O2), but are unable to reduce the highly toxic alkyl hydroperoxides that affect cell metabolism by causing changes in ion homeostasis, increases in protein and lipid peroxidation, or decreases in the mitochondrial membrane potential (Dietz 2007). Conversely, the recently characterized peroxiredoxins (Prxs) reduce a broader range of peroxide substrates and constitute an ascorbate-independent pathway (Rouhier et al. 2008).
Ubiquitous thiol-specific peroxidases, Prxs, have recently been characterized as novel antioxidant systems. Prxs exist in multiple isoforms that catalyze the reduction of peroxynitrites and a broad range of different peroxides, via catalytic cysteine and thiol-containing proteins that act as reductants (Rouhier and Jacquot 2005; Tripathi et al. 2009). These peroxidases can be divided into three groups, typical 2-Cys Prx, atypical 2-Cys Prx, and 1-Cys Prx, which can be distinguished by the number and position of conserved catalytic cysteine residues (Kim et al. 2011b; Rouhier and Jacquot 2005). 2-Cys Prxs can resolve two cysteines, including to the second redox-active cysteine, which localizes to the C-terminal region of the enzyme. In plants, the expression levels of 2-Cys Prxs are modified under different conditions. Specifically, their expression is regulated during salt or ascorbate treatment or by changes in the light/dark cycle and oxygen concentration, whereas it is not modified or only very slightly modified in response to oxidative stress (diamide and H2O2), hormones (abscisic acid and gibberellic acid), heavy metals (Cd, Cu, Ni, Zn, and As), ozone, and deficiencies in nutrients such as nitrogen, phosphorus, and sulfur in photosynthesizing leaves (Dietz 2007). In contrast, the expression of 2-Cys Prxs decreases with leaf age (Bhatt and Tripathi 2011; Rouhier and Jacquot 2005).
Prxs, including 2-Cys Prxs, play different roles in cells exposed to abiotic and biotic stresses. Specifically, their functions are as follows: antioxidant defense, detoxification of ROS and reactive nitrogen species (RNS), cellular redox signaling and differentiation, and defense against pathogen attack (Rouhier and Jacquot 2005; Tripathi et al. 2009). Regeneration of the reduced form of Prx is facilitated by various reducing systems including Trx or glutaredoxin (Grx) systems, cyclophilin (Cyp) in eukaryotic organisms such as plants, and AhpF and AhpD in bacteria (Tripathi et al. 2009). Based on these features, it is not surprising that the enzymatic Prx equipment used for antioxidant repair is considerably enhanced and diversified in plants, compared to those of bacteria, yeast, fungi, or animal systems (Bhatt and Tripathi 2011; Fomenko et al. 2011). Many studies have been performed to examine Prx overexpression in a wide variety of organisms, including plants and mammals (Kim et al. 2011b; Morgan and Veal 2007); however, the main roles of the chloroplastic 2-Cys peroxiredoxin BAS1 from Oryza sativa (OsTPX) in yeast cells under fermentation and abiotic stress conditions are very poorly understood.
During the fermentation process, yeast cells are dynamically challenged by mixed and interrelated stresses, including oxygen concentration, decreasing pH, osmotic pressure, oxidative stress, high temperature, ethanol concentration, and nutrient starvation (Ma and Liu 2010; Stanley et al. 2010; Zhao and Bai 2009). These conditions affect the cell population and fermentation yield, including ethanol production, which is dependent on the ability of yeast to adapt to such changes, particularly during batch fermentation (Gibson et al. 2007; González-Párraga et al. 2008). In this regard, studies of stress tolerance are of fundamental scientific importance and significant economic interest. At present, there is increasing demand for alternative energy sources because the supply of fossil fuels is dwindling and gas prices increase every year. One of the alternative options is renewable energy sources such as biofuel for industrial biotechnology (Ding et al. 2009; Shima and Takagi 2009). The use of biofuel attenuates the problems caused by the use of fossil fuels, especially with respect to greenhouse gas emissions (Zhao and Bai 2009). Among biofuels, the greatest issues have been experienced with bioethanol. Elucidating the underlying mechanisms of the yeast stress response or resistance, or obtaining a stress-tolerant yeast strain, could help increase the ethanol yield during bioethanol production and facilitate the use of this biofuel in a wide range of applications (Ding et al. 2009; Ma and Liu 2010).
In this study, the heterologous expression of the salt-induced OsTPX encoding a 2-Cys Prx from O. sativa in genetically modified Saccharomyces cerevisiae was found to improve fermentation capacity and acquired tolerance by enhancing redox homeostasis to abiotic stresses. Furthermore, these findings provide insight into the fermentation of glucose-based agricultural biomass to ethanol for use as an industrial biotechnology biofuel.
Materials and methods
Plant materials and growth conditions
The Ilmi variety of O. sativa L. japonica was used as the source of the OsTPX gene that was overexpressed in yeast. Rice seeds were germinated in half-strength Murashige–Skoog (MS) solid medium in a dark growth chamber for 2 days at 32 °C. The seeds were then transplanted into paddy soil pots and grown in a greenhouse (16 h light/8 h dark cycle) at 28–32 °C. Three-week-old rice seedlings were subsequently placed in 100 mM NaCl solution for 8 days and then used for the following experiments. The plants that continued to grow were photographed to record their phenotype.
Construction of the recombinant plasmid harboring OsTPX
Total RNA was isolated from the leaves of 4-week-old rice seedlings using an RNeasy Plant Mini kit (Qiagen, Hilden, Germany). The cDNA probe was synthesized according to the manufacturer’s instructions using reverse transcription-polymerase chain reaction (RT-PCR) premix kit (Bioneer, Daejeon, South Korea). The OsTPX (accession no. AK068919.1 or Os02g0537700) coding region was amplified from cDNA by PCR using ExTaq polymerase (Takara Bio Inc., Shiga, Japan). The PCR reaction conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 1 min, and a final extension for 7 min at 72 °C. PCR cloning of the OsTPX gene was conducted using 5′-AATCCCCTCCTCATCCACTCC-3′ and 5′-GATCAGACGAGCACACGATA-3′ as the sense and antisense primers, respectively. The PCR product was purified using a gel extraction kit (Nucleogen, Siheung, South Korea) and then inserted into the TOPO TA cloning vector (Invitrogen, Carlsbad, USA). The cloned plasmid was sequenced using a T7 primer set to confirm that no PCR-induced mutations had been introduced, after which it was digested with the EcoRI restriction enzyme. The digested plasmid DNA fragment was ligated into the yeast expression vector p426GPD (EUROSCARF, Frankfurt, Germany), and the resulting plasmid was named p426GPD::OsTPX. Competent Escherichia coli DH5α cells that had been transformed with the p426GPD260::OsTPX plasmid were used to amplify the plasmid DNA in DifcoTM Luria–Bertani broth (BD, Sparks, USA) containing ampicillin (100 μg ml−1). The plasmid DNA was isolated using a plasmid isolation kit (Nucleogen) according to the manufacturer’s protocol. The cloned plasmid was sequenced using the PF primer (5′-TGTTTTCTTCACCAATCAG-3′) complementary to the promoter region, to confirm proper gene ligation and gene direction. The resulting plasmid was used to transform yeast.
Yeast transformation
BY4741 (accession no. Y00000) and ahp1-deficient (accession no. Y02720) cells of S. cerevisiae (Table 1) were grown overnight in DifcoTM YPD broth (BD), inoculated into a fresh YPD broth, and then incubated for 4 h at 30 °C with shaking (160 rpm) until the cultures achieved an OD600 of approximately 1.5. The yeast was then transformed with the p426GPD::OsTPX plasmid using the PEG/LiCl method (Gietz and Woods 2001) as previously described. The transformants were then plated on yeast synthetic dropout agar medium containing a 0.67 % yeast nitrogen base without amino acids containing ammonium sulfate and 0.192 % yeast synthetic dropout medium supplement without uracil (Sigma, Saint Louis, USA), 2 % glucose, and 2 % agar, and incubated at 30 °C for 3 days. Positive colonies were grown in medium lacking uracil. The strains containing the transformed plasmid (p426GPD or p426GPD::OsTPX) (Table 1) were used for subsequent experiments.
Semi-quantitative RT-PCR
Total RNA from plant tissues and from mid-log phase yeast cells (OD600 = 2.0) was obtained using a total RNA isolation kit (Promega, Madison, USA) according to the manufacturer’s instructions. Semi-quantitative RT-PCR was performed with a one-step RT-PCR PreMix kit (iNtRON, Seongnam, South Korea) according to the manufacturer’s instructions using primer sets specific for the sequence of each gene. Each semi-quantitative RT-PCR reaction contained 50 ng of total RNA and 1 μg of primers in a total reaction volume of 20 μl. The primers OsTPX-F (5′-TCTACCCGTTGGACTTCACC-3′) and OsTPX-R (5′-GAAGGGTCCTCATGGTCTCA-3′) were used for this procedure. Semi-quantitative RT-PCR was performed by subjecting the reaction mixture to the following conditions: 1 cycle at 42 °C for 60 min, followed by 20 –24 cycles at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min, and a final extension step at 72 °C for 5 min. The PCR products were separated on a 1.2 % agarose gel in 0.5× TBE buffer, and stained with ethidium bromide for visualization. The housekeeping genes TUB for tubulin and PDA1 (YER178W) encoding the E1 alpha subunit of the pyruvate dehydrogenase complex (Kim et al. 2012) were used as controls for plants and yeast, respectively. The TUB and PDA1 gene primer sets were as follows: TUB-F—5′-TACCGTGCCCTTACTGTTCC-3′ and TUB-R—5′-CGGTGGAATGTCACAGACAC-3′; PDA1-F—5′-CTTCATTCAAACGCCAACCA-3′ and PDA1-R—5′-GAGGCAAAACCTTGCTTTTTG-3′.
2D electrophoresis analysis of rice plants
Rice leaves (3 g) were ground in liquid nitrogen and suspended in five volumes of cold extraction buffer containing 0.7 M sucrose, 0.1 M KCl, 0.5 M Tris–HCl, pH 7.5, 50 mM EDTA, 2 % β-mercaptoethanol (ME), and 1 mM phenylmethylsulfonyl fluoride (PMSF), as well as an equal of volume of phenol saturated with Tris–HCl at pH 7.5. The mixture was incubated for 30 min at 4 °C and then centrifuged at 5,000 rpm for 30 min. An equal volume of extraction buffer was then added to the upper phenolic phase, after which the sample was incubated for 30 min at 4 °C and centrifuged at 5,000 rpm for 30 min at 4 °C. This step was repeated twice. To precipitate the protein, five volumes of cold 0.1 M ammonium acetate in methanol were added to the collected phenol phase, and the mixtures were stored at −20 °C overnight and then centrifuged at 5,000 rpm for 30 min at 4 °C. To wash the pellet, two volumes of ice-cold methanol were added to the cleared supernatant and mixed gently, and the solution was centrifuged at 5,000 rpm for 10 min at 4 °C. This step was repeated three times. The pellet was then vacuum-dried and resuspended in sample buffer containing 9.5 M urea, 4 % CHAPS, 40 mM Tris, 0.1 M dithiothreitol (DTT), and 0.2 % Bio-Lyte (3-10; Bio-Rad, Hercules, USA) at room temperature with shaking. The sample was subsequently centrifuged at 15,000 rpm for 30 min, after which the cleared supernatant was carefully collected, and the protein concentration was measured using a modified Bradford assay (Ramagli and Rodriguez 1985). Next, 250 μg of proteins was loaded onto preparative gels and analyzed with pH 4–7 immobilized pH gradient (IPG) strips (7 cm, linear; Bio-Rad) under the following focusing conditions: 250 V for 1 h, 250–4,000 V for 3 h, and 28,000 V/h at 4,000 V. After isoelectric focusing (IEF), the gel strips were equilibrated and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on 12 % gels at 10 mA per gel for the first 1 h followed by 25 mA per gel, stained with Coomassie Brilliant Blue R-250 (CBB R-250; Sigma), and then destained. Spots with significant changes were considered to be accumulated proteins. Overexpressed spots of interest were excised from the gel and subjected to in-gel digestion and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) analysis (Kim et al. 2012). Finally, protein identification was conducted using the MASCOT software (http://www.matrixscience.com).
Western blot analysis
In plants, protein extraction was performed using a modified version of the trichloroacetic acid (TCA)/acetone precipitation method. Leaf samples (0.2 g fresh weight) were finely ground in a mortar with liquid N2 to make a powder, which was then suspended with 5 ml ice-cold acetone containing 10 % TCA and 0.07 % DTT per gram fresh weight. The resulting protein-containing suspension was allowed to precipitate overnight at −20 °C and then centrifuged at 15,000 rpm for 30 min. The pellet was then rinsed three times for 1 h each time, with ice-cold acetone containing 0.07 % DTT and 1 mM PMSF at −20 °C. Finally, the protein pellet was air-dried and solubilized in 50 mM Tris–HCl, pH 7.5, 5 % glycerol, 1 % Triton X-100, 0.1 % deoxycholic acid, 50 mM DTT, and protease inhibitor cocktail for 30 min at room temperature. Insoluble material was removed by centrifugation at 13,000 rpm for 30 min. The protein concentrations were then determined using a Pierce BCA protein assay kit (Thermo Scientific, Waltham, USA) (Komatsu et al. 2009; Peng et al. 2011). In yeast, crude protein extracts were prepared using glass beads. Briefly, cells grown to the mid-log phase were exposed to 30 mM H2O2, 0.2 mM 2-methyl-1,4-naphtoquinone (also called menadione, MD; water-insoluble form), a superoxide-generating agent used extensively for studies of cellular oxidative stress (Kim et al. 2012), 20 mM tert-butylhydroperoxide (t-BOOH), 1 % SDS, 15 % ethanol (EtOH), 60 mM sulfuric acid (H2SO4), 5 mM cadmium chloride (CdCl2), 10 mM copper chloride (CuCl2), 0.1 M zinc sulfate (ZnSO4), 0.1 M ferric chloride (FeCl2), salicylic acid (SA), and 5.0 M sodium chloride (NaCl) for 1 h at 30 °C (while shaking). Cells were also heated for 10 min at 55 °C. The cells were then washed three times with cold phosphate-buffered saline (PBS, pH 7.3 to 7.5; made from 10× tablets) (Invitrogen) and resuspended in a lysis buffer containing 20 mM HEPES, pH 7.5, 5 % glycerol, 1 mM DTT, 1 mM PMSF, and EDTA-free protease inhibitor cocktails (Roche, Mannheim, Germany) with an equal volume of glass beads (425–600 μm; Sigma). After vigorously vortexing five times for 1 min each at 2-min intervals on ice, the protein extracts were cleared by centrifugation at 13,000 rpm for 20 min at 4 °C. Finally, protein concentrations were determined by Bradford assay using a Protein Dye Reagent (Bio-Rad). Protein extracts (20 μg) were loaded onto a 12 % SDS-PAGE gel and separated at 50 V. The proteins were then transferred to PVDF membranes (Bio-Rad), which were incubated in blocking buffer containing 5 % non-fat skim milk and 0.02 % sodium azide in TBST (0.05 % Tween-20, 10 mM Tris–HCl, pH 7.6; and 150 mM NaCl) for 1.5 h at room temperature, and then incubated overnight at 4 °C with anti-Prx (Ab Frontier, Seoul, South Korea), anti-tubulin (Tub; Santa Cruz Biotechnology, Santa Cruz, USA), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Ab Frontier), and anti-Tsa1 and anti-Ahp1 (Kim et al. 2011a) antibodies diluted appropriately with blocking buffer. The blots were then washed three times for 30 min with TBST, after which they were incubated with anti-rabbit or anti-rat secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, USA) diluted with blocking buffer lacking 0.02 % sodium azide for 1.5 h at room temperature. After washing with TBST, binding of antibodies was visualized using the Enhanced Chemiluminescence western blotting detection reagent (GE Healthcare, Piscataway, USA). Tub and GAPDH were used as loading controls.
Stress tolerance assay
Yeast cells were cultured at 30 °C with shaking (160 rpm). Fresh overnight cultures of yeast cells in YPD medium (containing 1 % yeast extract, 2 % peptone, and 2 % glucose) were resuspended in fresh YPD medium containing H2O2 at concentrations ranging from 0 to 8 mM and MD at 0 – 0.1 mM. The initial optical density was adjusted to 0.1 at 600 nm. After culturing for 18 h at 30 °C with shaking, aliquots of each sample were taken and their absorbances were measured at 600 nm. When A 600 > 1.0, aliquots were diluted and new measurements were taken. To measure the response to stress under oxidative conditions, yeast cells (1 × 105 cells ml−1) grown overnight were used to inoculate YPD medium. Upon reaching the mid-log phase (A 600 = 4.0), the cells were exposed to 20 mM H2O2 and 0.2 mM MD for 1 h at 30 °C with shaking, after which they were serially diluted 10-fold with distilled water. Five microliters of diluted solution was loaded onto YPD agar medium (YPD plus 1.5 % agar). One-hundred microliters of diluted solution was spread onto YPD agar. The plates were then incubated for 3 days at 30 °C, after which they were photographed or counted. Relative cell viability was considered to be 100 % in wild-type (WT) cells containing an empty vector and grown in the absence of any stressors (20 mM H2O2 and 0.2 mM MD). To monitor the growth rate under abiotic stress, cells (1 × 105 cells ml−1) were grown overnight and inoculated into YPD medium supplemented with 5.0 mM H2O2 or 50 μM MD. The optical density (OD) at 600 nm was then measured at 2 h intervals. Mid-log phase yeast cells were streaked onto YPD agar supplemented with 5.0 mM H2O2, 50 μM MD, and 1.5 mM t-BOOH. To induce heat shock, cells were streaked after incubation for 5 min at 55 °C. To examine the stress response to ethanol, mid-log phase (A 600 = 4.0) and stationary phase (A 600 = 8.0) yeast cells were serially diluted with distilled water, after which 5 μl of the diluted cells was loaded onto YPD agar plates containing 0, 12, 16, or 20 % ethanol. All agar plates were incubated for 3 days at 30 °C and then photographed.
Redox state analysis
The intracellular hydroperoxide level was determined using the FOX reagent (100 μM xylenol orange, 250 μM ammonium ferrous sulfate, 100 mM sorbitol, and 25 mM sulfuric acid) by ferrous ion oxidation in the presence of a ferric ion indicator, xylenol orange (Kim et al. 2011a). To measure cytosolic ROS using an oxidant probe, exponential phase cells were incubated for 20 min at 30 °C with 50 μM dichlorodihydrofluorescein diacetate (DCFHDA; Invitrogen), exposed to 20 mM H2O2 or 0.2 mM MD for 1 h, washed twice with PBS buffer, and then resuspended in the same PBS buffer. Cells loaded with fluorescent probes were subsequently viewed by fluorescence microscopy (excitation, 488 nm; emission, 525 nm).
Laboratory-scale batch fermentation
To evaluate fermentation ability, cells were grown under aerobic fermentation conditions on a shaker (160 rpm) for 84 h at 30 and 40 °C in YG medium containing 20 % (w/v) glucose and 1 % (w/v) yeast extract. The alcohol concentration was then determined based on the percentage (v/v) of alcohol in the distillate after fermentation, measured using an alcohol hydrometer (REF 503; Korins, Seoul, South Korea). Residual total sugar concentrations were measured using a hand-held Refractometer N1 (Atago, Tokyo, Japan). Alcohol and residual glucose concentrations were obtained after removing cell debris by centrifugation (2,000 rpm, 3 min). To measure cell survival during the fermentation process, the cells were harvested at various time points (24, 48, and 72 h) and serially diluted to 10−9 with distilled water, after which 5 μl of the diluted cells were loaded onto YPD agar plates, incubated for 3 days at 30 °C and then photographed.
Amino acid sequence alignment and molecular building
OsTPX was aligned with known TPX sequences using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) software (http://clustalw.ddbj.nig.ac.jp/). The amino acid sequences were as follows: O. sativa TPX (OsTPX; accession no. BAD27915.1), Hordeum vulgare (HvTPX; accession no. Q96468.1), Arabidopsis thaliana TPX (AtTPX; accession no. AAM62760.1), Brassica rapa TPX (BrTPX; accession no. AAF00001.1), S. cerevisiae TSA1 (ScTSA1; accession no. NP_013684.1), S. cerevisiae TSA2 (ScTSA2; accession no. NP_010741.1), Homo sapiens TPX (HsTPX; accession no. P32119.1), Populus tricocharpa TPX (PtTPX; accession no. EEF03570.1), and S. cerevisiae AHP1 (ScAHP1; accession no. NP_013210.1). To predict the OsTPX three-dimensional (3D) structure based on this homology, we used bioinformatics tools, including the Protein Data Bank (PDB; http://www.rcsb.org/pdb) and Chimera software (http://www.cgl.ucsf.edu/chimera/index.html) from the University of California, San Francisco (UCSF) to obtain information regarding the OsTPX protein. Homology modeling was performed with the target sequence and the 3D structure of thioredoxin peroxidase B from red blood cells (accession no. P32119). The FASTA sequences of the query (accession no. BAD27915.1) and PDB template (1QMV_G) were uploaded onto the ESyPred3D Web Server 1.0 (http://www.fundp.ac.be/sciences/biologie/urbm/bioinfo/esypred/) to construct an OsTPX PDB file. The PDB files for the query and homologous target sequence were further utilized for 3D model building. Visualization of 3D protein structures was conducted using UCSF Chimera (http://www.cgl.ucsf.edu/chimera/index.html), which enabled assessment of the positions of different amino acids present in the active sites of the proteins.
Statistical analysis
All experiments were repeated at least three times independently, and the results were expressed as the mean ± standard deviation (SD). The results of a spotting assay, growth kinetics, redox state, and fermentation ability were representative of at least two independent experiments carried out under identical conditions. In addition, all spotting assays have been carried out with distilled water under identical conditions; therefore, any possible stress effects by the distilled water can be neglected in the analysis. Expression intensity measured by semi-quantitative RT-PCR and immunoblotting analysis was determined using ImageJ software (http://rsbweb.nih.gov/ij/).
Results
Identification of a putative thioredoxin peroxidase of rice with increased expression under salt stress
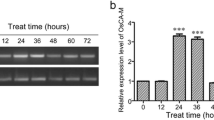
We screened salt stress-responsive proteins in the leaves of rice (O. sativa L. Ilmi), because the understanding of stress responses via a proteome approach will be greatly helpful in improving tolerance to abiotic stresses through genetic engineering. To accomplish this, 3-week-old rice seedlings were treated with 100 mM NaCl for 8 days. The treated seedlings displayed stress symptoms such as shriveled leaves, but survived the salt stress challenge (Fig. 1a). Comparative proteomic analysis was used to investigate the protein profiles under salt stress. To distinguish stress responses from protein upregulation, both normal and salt-treated leaves were harvested. The total protein content of the leaves was then extracted and separated by 2-DE using pH 4–7 IPG strips and IEF (Fig. 1b). One protein spot showing significant changes in abundance (Fig. 1b, indicated by an arrow) was then analyzed using the Mascot software after MALDI-TOF MS analysis (Fig. 1c) and identified as putative chloroplastic 2-Cys thioredoxin peroxidase (OsTPX; accession no. BAD27915.1). The predicted molecular weight and pI of the identified protein OsTPX were 28.079 kDa, and 5.57, respectively. We then performed semi-quantitative RT-PCR and immunoblotting analyses to determine if OsTPX expression was strongly induced at both the transcription and translation levels under salt stress conditions. As shown in Fig. 1d, e, the transcription and translation levels of endogenous OsTPX increased by at least 2.3- and 2.1-fold in the salt-treated sample compared to the control sample, although changes in transcription and translation differed somewhat among samples. These results demonstrate that the expression of the OsTPX protein was mildly upregulated in the presence of high salinity in rice leaves.
Induction of the OsTPX gene in the presence of salt stress. a Three-week-old rice seedlings were incubated in a 100 mM NaCl solution for 8 days. Upregulation of the OsTPX gene was confirmed by 2-DE (b) and MALDI-TOF analysis (c). Expression of this gene was identified by semi-quantitative RT-PCR (d) and western blot (e). The product of the rice TUB gene was used as a housekeeping standard for transcription and translation. Expression intensity was expressed as the ratio of OsTPX to TUB (right). 1 Without salt treatment, 2 with salt treatment
Construction and expression of OsTPX recombinant yeast
Immunoblotting analysis was performed to evaluate the expression patterns of endogenous yeast thioredoxin peroxidases (Tsa1 and Ahp1) in the presence of different stressors. TPX expression was downregulated in the presence of various stressors, including heat shock, H2O2, MD, t-BOOH, EtOH, SDS, H2SO4, CdCl2, CuCl2, ZnSO4, FeCl2, SA, and NaCl. As shown in Fig. 2a, the expression of Tsa1 and Ahp1 was unchanged or downregulated in response to various stressors, including heat shock. However, the Tsa1 protein was induced in the presence of CuCl2 and NaCl, while Ahp1 was upregulated under FeCl2 and SA stress. To examine the effects of thioredoxin peroxidase downregulation, an ahp1 mutant strain unable to the produce thioredoxin peroxidase was examined. The ahp1 mutant cells (A1 cells) were hypersensitive to heat shock, H2O2, MD, t-BOOH, and EtOH, but resistant to H2SO4, FeCl2, SA, and NaCl (Fig. 2b). These findings demonstrate that A1 cells are sensitive to ROS-induced oxidative stress, but not to acids, metals, or salt. In the presence of ROS-induced abiotic stresses, TPX expression of the yeast cells decreased (Fig. 2a), and the A1 cells were hypersensitive to these stresses (Fig. 2b). Based on our results, we examined whether expression of the exogenous OsTPX gene influenced the acquired tolerance towards ROS-induced oxidative stress in yeast cells since endogenous yeast TPX expression was downregulated in the presence of different stimuli. To accomplish this, cDNA (accession no. NM_001053585) containing the open reading frame (ORF) of the OsTPX gene from O. sativa was cloned into the yeast expression vector p426GPD, which allows constitutive expression of a target gene under control of the yeast GPD1 promoter. Upon digestion with the EcoRI restriction enzyme, the inserted DNA fragment was found to be approximately 940 bp (Fig. 2c). The construct was then sequenced to confirm that the ORF orientation was correct, after which it was introduced into S. cerevisiae strain BY4741. To explore whether the OsTPX gene was effectively expressed, semi-quantitative RT-PCR analysis was conducted. One DNA fragment of 354 bp corresponding to a region within the OsTPX ORF was detected in the OsTPX-expressing transgenic BY4741 cells (TG cells), while no signal was detected in the BY4741 cells transformed with the empty vector (WT cells) (Fig. 2d). We then performed immunoblotting analysis to determine if the OsTPX gene was properly translated. As shown in Fig. 2e, immunoblotting analysis conducted using the plant anti-Prx antibody revealed the presence of a single band produced by the transgenic (TG) cells under normal conditions. No signal was detected in WT cells under the same conditions. Taken together, our results indicate that the OsTPX gene under the control of the GPD1 promoter was properly expressed in genetically modified yeast cells.
Heterologous expression of the OsTPX gene in transgenic yeast cells. a Change in the expression of endogenous yeast thioredoxin peroxidases (Tsa1 and Ahp1) in the presence of various abiotic stressors analyzed by western blot. Tub protein was used as a loading control. b Stress sensitivity in ahp1 mutant cells. 1 BY4741 cells, 2 A1 cells. c Schematic diagram showing the overexpression of OsTPX in yeast. GPD1p glyceraldehyde-3-phosphate dehydrogenase promoter, OsTPX chloroplastic thioredoxin peroxidase gene of O. sativa. Arrows indicate the direction of each gene. Expression of the OsTPX gene was confirmed by semi-quantitative RT-PCR (d) and western blot (e). The PDA1 transcript and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein were used as standard controls in transcriptional and translational analyses, respectively. 1 WT cells, 2 TG cells
Molecular modeling of the OsTPX protein
To examine the molecular properties of OsTPX, multiple sequence alignment (MSA) was performed on OsTPX using the known AtTPX, BrTPX, PtTPX, HvTPX, HsTPX, ScTSA1, ScTSA2, and ScAHP1 sequences as described in “Materials and methods” Pairwise alignment of OsTPX and other TPXs was conducted using the NCBI BLAST software, and conserved redox-active disulfide bridges were observed between Cys114 and Cys236 of OsTPX (red box) as well as the Tre114, Trp149, and Trp240 residues (blue box) (Fig. 3a). According to the MSA, these two cysteines were preserved in ScTSA1 and ScTSA2 (Cys48 and Cys171), as well as in ScAHP1 (Cys62 and Cys120) (Figs. 3a and S 1). The molecular weight of the OsTPX enzyme is about 28 kDa. It has been reported that OsTPX shows a loss of activity in the absence or mutagenesis of thiol (redox active site Cys114) (Pérez-Ruiz et al. 2006). OsTPX had 91, 87, 84, 50, 50, 62, 33, and 20 % identity and 95, 94, 88, 65, 65, 76, 51, and 39 % similarity when compared to HvTPX, AtTPX, BrTPX, ScTSA1, ScTSA2, HsTPX, PtTPX, and ScAHP1, respectively (Fig. 3b). These results indicate that the molecular structure of the OsTPX protein contains a disulfide bond and conserved domains. Since the 3D structure of yeast TPX has not been elucidated, we introduced the 3D structure of human thioredoxin peroxidase (PPRX2; accession no. P32119; EC 1.11.1.15), which is 45.4 % homologous to OsTPX. The 3D structure of the TPX protein from O. sativa was constructed by homology modeling based on the PDB file obtained from the ESyPred3D Web Server 1.0 using the UCSF Chimera software. This structure consisted of amino acids 65–260 out of the total 261 amino acids (Fig. 3c). The active sites of the OsTPX model fit the human thioredoxin peroxidase template (Cys51 and Cys172) well, whereas some differences were observed between the model and template at the N- and C-termini and in the chloroplast leader peptide (residues 1–60) (Fig. 3d). The high sequence and structural homology of OsTPX should enable this protein to catalyze the thioredoxin-dependent reduction of a variety of substrates and indicates that it is a 2-Cys Prx.
Alignment and predicted structure of OsTPX. a Alignment between OsTPX and eight Prx sequences. 2-Cys Prxs were selected to represent OsTPX. “*” indicates that the residues in that column are identical in all sequences in the alignment, “:” indicates the presence of conserved substitutions, and “.” indicates the presence of semiconserved substitutions. b Phylogenetic trees of OsTPX and the eight Prxs. c Predicted structure of the OsTPX protein. The conserved cysteine residues are depicted and the N- and C-terminal amino acids are indicated. The model was generated using the ESyPred3D Web Server 1.0 based on the structure of 1QMV_G (identity, 45.4 %) and visualized using the Chimera software. d Domain annotation of OsTPX based on MSA and the predicted structure
Cellular response of the transgenic yeast cells expressing OsTPX to oxidative stresses
The effect of stress on cell survival was evaluated in yeast transformed with p426GPD plasmid containing the OsTPX gene or the vector alone to explore the ability of OsTPX expression to enhance resistance to oxidative stresses, including the major ROS-generating agents H2O2 and MD (Kim et al. 2011a), which are primarily used as an oxidant source in S. cerevisiae. Stress tolerance was measured based on cell viability, growth kinetics, and spotting and streaking assays. To measure cell viability, various doses of H2O2 and MD were added to cultures in YPD medium and the samples were then incubated for an additional 18 h at 30 °C with vigorous shaking (160 rpm). Cell viability was then calculated by measuring the optical density at 600 nm. Concentrations of H2O2 greater than 2.0 mM caused a decrease in cell viability, and exposure to 6.0 and 7.5 mM resulted in less than 10 % survival for WT and TG cells, respectively (Fig. 4a). Additionally, exposure to 20 μM MD reduced viability, exposed to 50 and 75 μM resulting in less than 10 % survival for WT and TG cells, respectively (Fig. 5a). Sensitivity to oxidants showed a dose-dependent decrease following treatment with exogenous drugs. Cell viability revealed that OsTPX-expressing TG cells were inhibited by higher concentrations of H2O2 and MD than were WT cells. To further examine this tolerance, we measured the sensitivity of the cells to oxidants in terms of growth rate in liquid medium containing 5.0 mM H2O2 or 50 μΜ MD. To accomplish this, the initial OD was adjusted to A 600 = 0.2, and the OD was then measured at various time points. During the first 4 and 8 h, the optical density of TG yeast cells increased at almost the same rate as that of WT yeast cells in the presence of H2O2 and MD. This was followed by a lag, after which growth resumed more rapidly. TG yeast cells appeared to enter the late log or early stationary phase at 28 and 32 h in the presence of H2O2 and MD, respectively. WT yeast cells had a longer lag phase (16 h in H2O2 and 22 h in MD) compared to the rapid restoration of growth observed in TG yeast cells (Figs. 4b and 5b). However, no difference was observed in the growth rates of TG and WT yeast cells under normal conditions. A streaking assay on YPD supplemented with 5.0 mM H2O2 and 50 μΜ MD clearly confirmed the stress response to H2O2 and MD. TG cells rapidly recovered from exposure to H2O2 and MD, compared to WT cells (boxed in Figs. 4b and 5b), even though no difference between TG and WT cells was observed under normal conditions. Thus, the cell recovery of TG yeast cells was greater than that of the WT cells. In addition to the dynamic stress response, static responses to H2O2 and MD were investigated in both cells. First, cell viability was evaluated in mid-log phase cells (A 600 = 2.0) exposed to 20 mM H2O2 and 0.2 mM MD for 1 h with shaking by measuring the number of colony-forming units. The cell viability of TG cells decreased by up to 20 and 10 % in the presence of H2O2 and MD, respectively, compared to the 100 % cell viability of oxidant-untreated cells, whereas viability in WT cells was reduced by up to 60 and 50 %, respectively, under the same conditions (upper panel in Figs. 4c and 5c). The stress response was then measured using a spotting assay with serial dilutions. Cell recovery in mid-log TG yeast cells (A 600 = 2.0) exposed to 20 mM H2O2 and 0.2 mM MD for 1 h was higher than that seen in WT yeast cells (lower panel in Figs. 4c and 5c). Finally, we determined whether stress tolerance in TG yeast cells conferred by OsTPX expression led to improved redox homeostasis. To accomplish this, the redox state was analyzed by measuring the cellular hydroperoxide level using the ferrous oxidation in xylenol-orange (FOX) assay, which is based on color production following the oxidation of ferrous to ferric iron by hydroperoxide under acidic conditions, as well as using an assay based on the cytosolic oxidant-sensitive probe, DCFHDA, measuring the oxidative conversion of DCFHDA to the highly fluorescent compound dichlorofluorescin (DCF), in the absence and presence of 20 mM H2O2 and 0.2 mM MD. Cellular hydroperoxide levels in TG cells determined using the FOX reagent were 2.0- and 1.6-fold lower than those of WT cells in the presence of H2O2 and MD, respectively. However, no differences were observed between the cells under normal conditions (Figs. 4d and 5d). The intensity of intracellular hydroperoxides evaluated using the DCFHDA probe was more pronounced in WT cells than TG cells, although an increase in DCF fluorescence was observed in both WT and TG cells upon exposure to 20 mM H2O2 and 0.2 mM MD for 1 h (Figs. 4d and 5d). We also observed a moderate release of the probe from WT cells in the presence of MD (Fig. 5d). The alleviation of redox state was inversely proportional to cell viability and recovery under oxidative stress conditions. Furthermore, we examined whether OsTPX expression could complement thioredoxin peroxidase activity in ahp1 mutant cells without any construct (A1 cells). As shown in Fig. S2 (see Supplementary material), OsTPX expression in the ahp1 mutant cells (MG cells) recovered rapidly in the presence of H2O2 when compared to the ahp1 mutant cells with an empty vector (A2 cells). Therefore, our results show that heterologous OsTPX expression in transgenic yeast cells confers acquired tolerance against oxidative stress by improving the intracellular redox state through a reduction in the hydroperoxide levels produced by H2O2 and MD, compared to WT cells, and that OsTPX expression in MG cells functionally complements thioredoxin peroxidase activity in A1 cells.
Stress response and redox state in the presence of hydrogen peroxide (H2O2). a Cell viability was monitored by measuring the optical density at various concentrations of H2O2 after incubation for 18 h with shaking. b Cell recovery was measured by monitoring growth kinetics and by the streaking assay (boxed) in the presence of 5 mM H2O2. The growth rate was spectrophotometrically monitored at 600 nm for 24 h in the presence of 5.0 mM H2O2. c To examine the stress response in the presence of high H2O2 concentrations, yeast cells were exposed to 20 mM H2O2 for 1 h. Cell viability was then measured based on the number of colony-forming units (CFU) (upper panel) and the spotting assay (lower panel). d Redox state analysis was carried out using the FOX reagent and an oxidant-sensitive probe, DCFHDA, in the absence and presence of 20 mM H2O2 for 1 h. WT, yeast cells containing empty vector; TG, OsTPX-expressing yeast cells; circles, TG cells without H2O2 treatment; squares, WT cells without H2O2 treatment; upward triangles, WT cells treated with H2O2; downward triangles, TG cells treated with H2O2; minus (−), agar plate without H2O2; plus (+), agar plate supplemented with H2O2; white bar, yeast cells without 20 mM H2O2 treatment; black bar, yeast cells treated with 20 mM H2O2 for 1 h
Stress response and redox state in the presence of menadione (MD). a Cell viability was monitored by measuring the optical density at various concentrations of MD after incubation for 18 h with shaking. b Cell recovery was measured by monitoring the growth kinetics and by the streaking assay (boxed) in the presence of 50 μM MD. c To examine the stress response in the presence of high MD concentrations, yeast cells were exposed to 0.2 mM MD for 1 h. Cell viability was then measured based on the number of colony-forming units (CFU) (upper panel) and the spotting assay (lower panel). d Redox state analysis was carried out using the FOX reagent and an oxidant-sensitive probe, DCFHDA, in the absence and presence of 0.2 mM MD for 1 h. WT, yeast cells containing empty vector; TG, OsTPX-expressing yeast cells; circles, TG cells without MD treatment; squares, WT cells without MD treatment; upward triangles, WT cells treated with MD; downward triangles, TG cells treated with MD; minus (–), agar plate without MD; plus (+), agar plate supplemented with MD; white bar, 0.2 mM MD-untreated cells; black bar, 0.2 mM MD-treated cells
Response to abiotic stresses in OsTPX-expressing transgenic yeast cells
We evaluated the response to other stressors because TG cells showed enhanced tolerance against H2O2 and MD. The exogenous stimuli tested were mainly sources that inhibited Tsa1 and Ahp1 expression in yeast (Fig. 2a). Mid-log phase yeast cells (A 600 = 2.0) were streaked onto YPD agar plates with and without 1.5 mM t-BOOH. For heat shock, cells from the same phase were exposed to 55 °C for 5 min and then streaked onto YPD agar plates. Cell recovery was more rapid in TG cells than WT cells in the presence of t-BOOH (Fig. 6b) and high temperature (Fig. 6c). No differences in the growth rates of TG and WT yeast cells were detected under normal conditions (Fig. 6a). We also investigated the response to various concentrations of EtOH. To accomplish this, yeast cells were grown to the log phase (A 600 = 2.0) or stationary phase (A 600 = 8.0) and then exposed to 12, 16, and 20 % EtOH for 1 h with shaking, diluted serially, and spotted onto YPD agar plates. The acquired response of the TG yeast cells exposed to different concentrations of EtOH was higher than that of WT cells in both the log and stationary phases, and these responses occurred in a dose- and phase-dependent manner (Fig. 6d). Our results indicate that high OsTPX expression in TG cells enhances tolerance to abiotic stresses, including t-BOOH, high temperature, and ethanol exposure.
Stress response of heterologous OsTPX-expressing yeast cells to the presence of various stressors. Mid-log phase yeast cells were streaked onto YPD agar plates supplemented without (a) or with t-BOOH (b). For heat shock, yeast cells were incubated for 5 min at 55 °C and then streaked onto YPD agar plates (c). All plates were incubated for 3 days at 30 °C and then photographed. d To investigate the stress response to ethanol, log and stationary phase yeast cells were challenged with various concentrations of ethanol (0 to 20 %) for 1 h, serially diluted to 10−4, and then spotted onto YPD agar plates. WT, yeast cells containing empty vector alone; TG, yeast cells expressing OsTPX
Effect of high OsTPX expression under batch fermentation conditions
Since TG cells expressing OsTPX were more resistant to high ethanol concentrations and ROS-induced oxidative stress than were WT cells, laboratory-scale fermentation using TG or WT yeast cells was performed at 30 and 40 °C under aerobic conditions. A distinguishable difference in alcohol yield and residual glucose content was observed between TG and WT cells at 30 °C, which is the temperature generally used for industrial fermentation. During fermentation for 84 h at 30 °C, the alcohol yield of TG cells was approximately 21 % higher than that of WT cells. The final alcohol concentration was approximately 14 and 11 % following fermentation with TG cells and WT cells, respectively. The residual glucose concentration was inversely proportional to the alcohol concentration during fermentation (Fig. 7a). In addition, a distinct difference was observed in cell survival between WT and TG cells during the fermentation period. Specifically, the survival of TG cells was significantly higher than that of WT cells during fermentation for 72 h in YG medium, although cell viability decreased with time (Fig. 7b). Similar results were observed in both WT and TG cells during fermentation at a high temperature (40 °C). The alcohol concentration and residual glucose content differed between WT and TG cells when the same fermentation medium was used. Specifically, the alcohol yield produced by TG cells was approximately 29 % higher than that produced by WT cells. The final alcohol concentrations were approximately 8.5 and 6.0 % in TG cells and WT cells, respectively, following fermentation for 84 h at 40 °C (Fig. 7c). Therefore, our findings indicate that OsTPX expression enhances fermentation at moderate and high temperatures; this is an important factor for the increased alcohol yield of the TG cells during fermentation.
Fermentation ability and cell survival in OsTPX-expressing yeast cells during the batch fermentation process. a Fermentation ability was analyzed by measuring the alcohol (AC) and residual glucose (RG) concentrations after incubation in YG medium for 84 h at 30 °C. Circles, RG in WT cells; squares, RG in TG cells; upward triangles, AC in WT cells; downward triangles, AC in TG cells. b Cell viability was determined by performing a spotting assay. Cells were harvested at 24 h (upper panel), 48 h (middle panel), and 72 h (lower panel) after initiating fermentation. WT, yeast cells containing empty vector; TG, yeast cells expressing OsTPX. c Fermentation ability was analyzed by measuring AC and RG during fermentation for 84 h at 40 °C. Circles, RG in WT cells; squares, RG in TG cells; upward triangles, AC in WT cells; downward triangles, AC in TG cells
Discussion
Abiotic stress causes a series of morphological, physiological, biochemical, and molecular changes that unfavorably affect cell growth and development and productivity in crop plants (Ashraf 2009). To protect themselves from environmental stresses, cells initiate the activation of cascades or network events, which begin with sensing stress and end with the expression of various stress-responsive genes (Dinakar et al. 2010; Rouhier et al. 2008). In this study, we identified a salt-induced putative chloroplastic 2-Cys thioredoxin peroxidase, OsTPX, in rice leaves, and developed an OsTPX-expressing transgenic yeast to investigate the molecular mechanism of the gene, to offset the scarcity of available information regarding the cellular response to abiotic stresses. OsTPX-overexpressing BY4741 yeast cells (TG cells) showed enhanced resistance to the abiotic stressors H2O2 (Fig. 4), MD (Fig. 5), t-BOOH, ethanol, and heat shock (Fig. 6) compared to BY4741 yeast cells containing the empty vector (WT cells). This was because TG yeast cells could improve the redox state by decreasing the cellular ROS levels produced by abiotic stresses such as H2O2 (Fig. 4d) and MD (Fig. 5d), thereby likely minimizing cellular damage caused by ROS. According to studies from the literature, overexpression of the homologous 2-Cys Prx gene encoding ubiquitous thiol-specific peroxidases increased stress tolerance and redox homeostasis in several organisms. For example, salt-induced Debaryomyces hansenii AHP (DhAHP, Prx isoform) expression conferred higher salt tolerance by reducing the cellular ROS level in D. hansenii, S. cerevisiae, and Pichia methanolica (Chao et al. 2009). Overexpression of A. thaliana 2-Cys Prx (At2-Cys Prx; accession no. Y10478) resulted in enhanced tolerance to methyl viologen (MV)-mediated oxidative stress and high temperature in transgenic potato plants (Kim et al. 2011b) and transgenic tall fescue plants (Kim et al. 2010). In mice, overexpression of Prx 3 improved glucose tolerance by reducing the mitochondrial H2O2 level (Jönsson and Lowther 2007). As mentioned above, 2-Cys Prxs, including OsTPX, play an important defense role in response to ROS-induced oxidative stress in aerobic organisms. Oxidized Prx produced during antioxidative reactions needs to be recycled to the reduced form before the next active catalytic cycle can proceed (Dietz 2007). Recently, a large set of various interacting reducing partners, including Trxs, Grxs, CypA and Cyp20-3, GSH, sulfiredoxin (Srx1), and NADPH-dependent thioredoxin reductase, were identified in yeast, plants, and mammals and found to be capable of reversing oxidized Prx to its original reduced form (Dietz 2007). For example, Srx1 catalyzes the reduction of sulfinic acid derivatives of Tsa1 in S. cerevisiae (Morgan and Veal 2007). Plant Srxs induced by H2O2 and drought play a protective role under stress conditions by repairing hyperoxidized 2-Cys Prxs (Jönsson and Lowther 2007). Based on these facts, OsTPX can effectively process ROS neutralization reactions with reducing partners in transgenic yeast cells. On the other hand, OsTPX-expressing ahp1-deficient cells (MG cells) recovered more rapidly in the presence of H2O2 than did ahp1-deficient cells containing the empty vector (A2 cells) (Fig. S2). These results indicate that acquired tolerance to ROS-induced oxidative stress in OsTPX-expressing BY4741 cells (TG cells) results from improved cellular homeostasis and that OsTPX expression functionally complements thioredoxin peroxidase activity in ahp1-deficient cells (A1 cells). Furthermore, our findings suggest new ways of engineering stress tolerance and outline an experimental system with yeast for testing alternative roles for plant Prxs, because many aspects of the functions and structures of these proteins are highly conserved.
In addition to OsTPX overexpression, the physiological importance of 2-Cys Prx has been reported in different organisms. S. cerevisiae Prx (AHP1)-deficient mutant cells (A1 cells) were sensitive to various oxidants (Fig. 2b), as evidenced by the fact that excessive ROS accumulation in the A1 cells showed increased evidence of oxidative damage such as enzyme inactivation by protein carbonylation and membrane instability by lipid peroxidation (Sideri et al. 2010; Wong et al. 2004). Similar physiological phenotypes were observed in photosynthetic organisms. Transgenic 2-Cys Prx mutant plants of Arabidopsis exhibited increased sensitivity to oxidative stress, as evidenced by increased photoinhibition, degradation of chloroplast proteins like D1, light-harvesting protein complex associated with photosystem II (LHCP II) and ATP synthase, and an altered oxidation state of the ascorbate pool (Baier and Diekz 1999; Baier et al. 2000) Additionally, 2-Cys Prx mutants (∆2-CP) of Synechocystis sp. PCC 6803 exhibited decreased growth rates in conditions of light-induced oxidative stress (Dietz et al. 2002). These results show that the defense machinery requires high levels of 2-Cys Prx to protect the cells from oxidative stress and that stress sensitivity is reduced by the accumulation of 2-Cys Prx. According to recent studies, 2-Cys Prx family members are versatile and multifunctional proteins that act as regulators of signal transduction, molecular chaperones, and regulators of responses to DNA damage in yeast (Fomenko et al. 2011; Morgan and Veal 2007). Taken together, it can be concluded that the 2-Cys OsTPX protein could play a central role in cells as a molecular chaperone and regulator of signal transduction and that it might act as an antioxidant in the oxidative stress response.
The function of a protein could stem directly from its structure; therefore, information regarding the predicted 3D structures of proteins plays a crucial role in guiding our understanding of protein function at the molecular level (Karplus and Hall 2007). MSA analysis revealed that the sequence identities of OsTPX to the 2-Cys Prxs analyzed ranged from 20 to 91 %. Similar to other Prxs, OsTPX contains a conserved catalytic peroxidatic cysteine (Cys114) in the N-terminal region, another cysteine residue (Cys236) that likely forms a disulfide bond with Cys114, and a highly conserved active site region, FTFVCPT (blue box) (Fig. 3a). OsTPX also possesses amino acid residues (Thr111, Trp114, and Trp240; blue box) that are likely involved in peroxide reduction (Fig. 3a, d). Mutations in two amino acids that are strictly conserved among nine 2-Cys Prxs, including OsTPX, Thr48 of poplar PtTPX (Thr111 in OsTPX) and Cys64 of barley HvTPX (Cys114 of OsTPX) (Fig. S1) (see Supplementary material), decreased Prx activity (Rouhier and Jacquot 2005), indicating that these residues are essential for catalysis. Moreover, modifications of Thr66, Trp99, and Trp189 of barley HvTPX (Thr116, Trp149, and Trp240 of OsTPX) implicated these residues in the conformational change (dimer to oligomer) that alters protein function (König et al. 2003). In addition, molecular building analysis showed that two conserved cysteine residues (Cys114 and Cys236) of OsTPX could have an important role in neutralizing toxic ROS because the corresponding two cysteine residues (Cys51 and Cys172) of 2-Cys Prx (PDB code 1QMV_G) from H. sapiens (Schröder et al. 2000), which was used as a template, were significantly involved in detoxifying ROS through Trx or Grx regeneration to protect the cells from oxidative stress (König et al. 2003). In an in vitro assay, all plant 2-Cys Prxs reduced a wide range of hydroperoxides (mainly H2O2), alkyl hydroperoxides, phospholipid hydroperoxides, and peroxynitrites (Rouhier and Jacquot 2005). Altogether, these results suggest that OsTPX, as a 2-Cys Prx family member, can efficiently reduce a wide range of peroxide substrates produced by abiotic stresses through absolutely conserved, activated residues (Cys114 and Thr111) within a strictly conserved active site pocket region, although there is no direct evidence of the physiological roles of these compounds in specific enzymological terms.
To further elucidate the functions of the OsTPX protein, we conducted OsTPX-transformed yeast batch fermentations at the general cultivation temperature (30 °C) and at a high temperature (40 °C) because the most important physical factor influencing the life of yeast is temperature. During fermentation at 30 and 40 °C, the alcohol yields from sugar were 21 % (0.12 g g−1) (Fig. 7a) and 29 % (0.14 g g−1) higher (Fig. 7c) in OsTPX-expressing BY4741 cells (TG cells), respectively, than in BY4741 cells containing empty vector (WT cells). Cell survival was also higher in TG cells than in WT cells during fermentation at 30 °C (Fig. 7b). One of the most common stresses that yeast cells encounter during fermentation is increased ethanol concentration. Under normal fermentation conditions, final ethanol concentrations range from 3 to 6 %, though under high gravity fermentation the alcohol concentration may be >10 % (Gibson et al. 2007), as observed in the present study (Fig. 7). The high toxicity of the ethanol produced endogenously during fermentation causes a reduction in cell viability, growth rate, and fermentation rate (Ding et al. 2009; Gibson et al. 2007). To withstand and/or prevent various types of damage generated by increased ethanol concentrations during fermentation, yeast cells have developed effective mechanisms, including changes in membrane composition, accumulation of L-proline and trehalose, the expression of molecular chaperones, and transcriptome remodeling through the expression of stress-related genes such as zinc finger proteins (Msn2p and Msn4p) and alcohol sensitive ring/PHD finger 1 protein (Asr1p) (Ding et al. 2009; Gibson et al. 2007; Shima and Takagi 2009; Zhao and Bai 2009). However, this approach has the intrinsic limitation that yeast is able to adapt to different metabolic environments such as a high concentration of ethanol during fermentation. The genetically engineered yeast strain (TG), capable of heterologous OsTPX overexpression, displayed improved alcohol yield and cell viability (Fig. 7). Indeed, the final alcohol concentrations in the TG cells from conversion of sugar were 14 % (0.66 g g−1) and 8.5 % (0.40 g g−1), respectively, during fermentation at 30 and 40 °C, while these concentrations were 11 % (0.52 g g−1) and 6 % (0.28 g g−1), respectively, in WT cells under the same fermentation conditions. Considering that the current theoretical yield of ethanol from sugar is 0.50–0.55 g g−1 and in the industrial context 90–95 % of this can generally be achieved by S. cerevisiae (Zhao and Bai 2009), we think that these results are very meaningful and constitute an effective approach to develop stress-tolerant yeast during fermentation. Therefore, in summary, our results indicate that OsTPX-supported fermentation by TG cells could effectively overcome the toxic levels of ethanol generated during fermentation, leading to increased cell viability (Fig. 7b) and fermentation capacity (Fig. 7a, c) when compared to WT cells.
In conclusion, the high expression of OsTPX in transgenic yeast conferred enhanced tolerance to ROS-induced oxidative stress, such as that generated by H2O2 and MD, by maintaining the balance of cellular redox homeostasis and functionally complementing thioredoxin peroxidase activity in ahp1-deficient cells. Further, heterologous OsTPX expression increased the acquired tolerance to high concentrations of ethanol and led to improved fermentation capacity, especially at high temperature. Future studies of the relationship between OsTPX expression and stress tolerance are required to develop better ethanol-tolerant yeast for efficient ethanol production.
References
Ashraf M (2009) Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol Adv 27:84–93
Baier M, Diekz KJ (1999) Protective function of chloroplast 2-Cys peroxiredoxin in photosynthesis: evidence from transgenic Arabidopsis. Plant Physiol 119:1407–1414
Baier M, Noctor G, Foyer CH, Dietz KJ (2000) Antisense suppression of 2-cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzymes associated with ascorbate metabolism but not glutathione metabolism. Plant Physiol 124:823–832
Bhatt I, Tripathi BN (2011) Plant peroxiredoxins: catalytic mechanisms, functional significance and future perspectives. Biotechnol Adv 29:850–859
Chao HF, Yen YF, Ku MS (2009) Characterization of a salt-induced DhAHP, a gene coding for alkyl hydroperoxide reductase, from the extremely halophilic yeast Debaryomyces hansenii. BMC Microbiol 9:182
Dietz KJ (2007) The dual function of plant peroxiredoxins in antioxidant defence and redox signaling. Subcell Biochem 44:267–294
Dietz KJ, Horling F, König J, Baier M (2002) The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J Exp Bot 53:1321–1329
Dinakar C, Abhaypratap V, Yearla SR, Raghavendra AS, Padmasree K (2010) Importance of ROS and antioxidant system during the beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Planta 231:461–474
Ding J, Huang X, Zhang L, Zhao N, Yang D, Zhang K (2009) Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 85:253–263
Fomenko DE, Koc A, Agisheva N, Jacobsen M, Kaya A, Malinouski M, Rutherford JC, Siu KC, Jin DY, Winqe DR, Gladyshev VN (2011) Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc Natl Acad Sci USA 108:2729–2734
Gibson BR, Lawrence SJ, Leclaire JP, Powell CD, Smart KA (2007) Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev 31:535–569
Gietz RD, Woods RA (2001) Genetic transformation of yeast. Biotechniques 30:816–820
González-Párraga P, Sánchez-Fresneda R, Martínez-Esparza M, Argüelles JC (2008) Stress responses in yeasts: what rules apply? Arch Microbiol 189:293–296
Jönsson TJ, Lowther WT (2007) The peroxiredoxin repair proteins. Subcell Biochem 44:115–141
Karplus PA, Hall A (2007) Structural survey of the peroxiredoxins. Subcell Biochem 44:41–60
Kim IS, Jin I, Yoon HS (2011a) Decarbonylated cyclophilin A Cpr1 protein protects Saccharomyces cerevisiae KNU5377Y when exposed to stress induced by menadione. Cell Stress Chaperones 16:1–14
Kim IS, Kim YS, Yoon HS (2012) Rice ASR1 protein with reactive oxygen species scavenging and chaperone-like activities enhances acquired tolerance to abiotic stresses in Saccharomyces cerevisiae. Mol Cells 33:285–293
Kim KH, Alam I, Lee KW, Sharmin SA, Kwak SS, Lee SY, Lee BY (2010) Enhanced tolerance of transgenic tall fescue plants overexpressing 2-Cys peroxiredoxin against methyl viologen and heat stresses. Biotechnol Lett 32:571–576
Kim MD, Kim YH, Kwon SY, Jang BY, Lee SY, Yun DJ, Cho JH, Kwak SS, Lee HS (2011b) Overexpression of 2-cysteine peroxiredoxin enhances tolerance to methyl viologen-mediated oxidative stress and high temperature in potato plants. Plant Physiol Biochem 49:891–897
Kingston-Smith AH, Foyer CH (2000) Overexpression of Mn-superoxide dismutase in maize leaves leads to increased monodehydroascorbate reductase, dehydroascorbate reductase and glutathione reductase activities. J Exp Bot 51:1867–1877
Komatsu S, Yamamoto R, Nanjo Y, Mikami Y, Yunokawa H, Sakata K (2009) A comprehensive analysis of the soybean genes and proteins expressed under flooding stress using transcriptome and proteome techniques. J Proteome Res 8:4766–4778
König J, Lotte K, Plessow R, Brockhinke A, Baier M, Dietz KJ (2003) Reaction mechanism of plant 2-Cys peroxiredoxin. Role of the C terminus and the quaternary structure. J Biol Chem 278:24409–24420
Ma M, Liu ZL (2010) Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 87:829–845
Morgan BA, Veal EA (2007) Functions of typical 2-Cys peroxiredoxins in yeast. Subcell Biochem 44:253–265
Peng L, Kapp EA, McLauchlan D, Jordan TW (2011) Characterization of the Asia Oceania Human Proteome Organisation Membrane Proteomics Initiative Standard using SDS-PAGE shotgun proteomics. Proteomics 11:4376–4384
Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18:2356–2368
Ramagli LS, Rodriguez LV (1985) Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 11:559–563
Rouhier N, Jacquot JP (2005) The plant multigenic family of thiol peroxidases. Free Radic Biol Med 38:1413–1421
Rouhier N, Koh CS, Gelhaye E, Corbier C, Favier F, Didierjean C, Jacquot JP (2008) Redox based anti-oxidant systems in plants: biochemical and structural analyses. Biochim Biophys Acta 1780:1249–1260
Schröder E, Littlechild JA, Lebedev AA, Errington N, Vagin AA, Isupov MN (2000) Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution. Structure 8:605–615
Shima J, Takagi H (2009) Stress-tolerance of baker’s-yeast (Saccharomyces cerevisiae) cells: stress-protective molecules and genes involved in stress tolerance. Biotechnol Appl Biochem 53:155–1564
Sideri TC, Stojanovski K, Tuite MF, Grant CM (2010) Ribosome-associated peroxiredoxins suppress oxidative stress-induced de novo formation of the [PSI+] prion in yeast. Proc Natl Acad Sci USA 107:6394–6399
Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA (2010) The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol 109:13–24
Tripathi BN, Bhatt I, Dietz KJ (2009) Peroxiredoxins: a less studied component of hydrogen peroxide detoxification in photosynthetic organisms. Protoplasma 235:3–15
Wong CM, Siu KL, Jin DY (2004) Peroxiredoxin-null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J Biol Chem 279:23207–23213
Zhao XQ, Bai FW (2009) Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J Biotechnol 144:23–30
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ008115012012), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 94.2 kb)
Rights and permissions
About this article
Cite this article
Kim, IS., Kim, YS. & Yoon, HS. Expression of salt-induced 2-Cys peroxiredoxin from Oryza sativa increases stress tolerance and fermentation capacity in genetically engineered yeast Saccharomyces cerevisiae . Appl Microbiol Biotechnol 97, 3519–3533 (2013). https://doi.org/10.1007/s00253-012-4410-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4410-8