Abstract
Bacterial lipoproteins are crucial antigens for protective immunity against bacterial pathogens. Expression of exogenous lipoproteins in Escherichia coli at high levels is thought to be an extremely difficult endeavor because it frequently results in incomplete or absent lipid modification. Previously, we identified a fusion sequence (D1) from a Neisseria meningitidis lipoprotein that induced a non-lipidated protein, E3 (the domain III of the dengue virus envelope protein), to become lipidated. However, without optimizing the growth conditions, some of the D1-fusion proteins were not lipidated. Here, we report the influence of medium components on the expression of recombinant lipoproteins in E. coli. For high-level expression of mature lipoproteins in the C43 (DE3) strain, M9 medium was better than M63 and the rich medium. Furthermore, we analyzed the influence of other media factors (including nitrogen and carbon sources, phosphate, ferrous ions, calcium, magnesium, and pH) on the levels of lipoprotein expression. The results showed that excess nitrogen sources and phosphate in M9 medium could increase the amount of immature lipoproteins, and glucose was a better carbon source than glycerol for expressing mature lipoproteins. We also found that lipoproteins tended to be completely processed in the alkaline environment, even in the nutrient-rich medium. Additional constructs expressing different immunogens or lipid signal peptides as targets were also utilized, demonstrating that these targets could be expressed as completely mature lipoproteins in the M9 medium but not in the rich medium. Our results provide the useful information for expressing mature exogenous lipoproteins in E. coli.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Post-translational modification of membrane proteins with lipids appears to be ubiquitous in prokaryotic cells. Bacterial lipoproteins share the common structural feature of a consensus sequence known as lipobox, [LVI][ASTVI][GAS]C, at the C region of the signal peptides (Babu et al. 2006). Once they are modified with the lipid moieties at their N termini, they are anchored to the surface and function as structural proteins (e.g., murein lipoprotein) or catalytic proteins (membrane-bound enzyme or transport proteins) (Hayashi and Wu 1990).
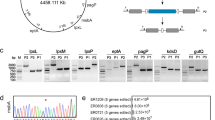
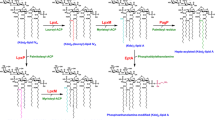
The biosynthesis of lipoproteins in Escherichia coli has been proposed in detail using Braun’s lipoprotein as the model (Braun and Rehn 1969; Sankaran and Wu 1994). The protein is first synthesized as an unmodified prolipoprotein and then modified by the prolipoprotein diacylglyceryl transferase (Lgt). Lgt catalyzes the transfer of the diacylglyceryl moiety from phosphatidylglycerol to the thiol group of the conserved cysteine (Sankaran and Wu 1994). The lipidation signal sequence of diacylglyceryl prolipoprotein is subsequently cleaved at the modified cysteine by prolipoprotein signal peptidase (LspA) (Tokunaga et al. 1982). Phospholipids-apolipoprotein N-acyltransferase (Lnt) catalyzes the final modification by adding a fatty acid at the N-terminal cysteine to form the mature lipoprotein (Jackowski and Rock 1986; Gupta et al. 1993) (Fig. 1a).
Biosynthesis of bacterial lipoproteins and the construct of the recombinant lipoprotein. a The unmodified prolipoprotein contains an N-terminal signal peptide possessing a characteristic consensus sequence of the lipobox. Prolipoprotein diacylglyceryl transferase (Lgt) would catalyze the modification of the cysteine in the lipobox with a diacylglyceryl group attached through a thioether linkage. After lipidation, prolipoprotein signal petidase (Lsp) cleaves the signal peptide. The final modification is the attachment of an amide-linked acyl group to the N-terminal cysteine residue by lipoprotein N-acyl transferase (Lnt). b Schematic layout of the D1E3 fusion construct. c DNA and amino acid sequence of D1 and P3 fragments. The lipobox are framed
Because most of the lipoproteins are located on the bacterial cell surface, they are readily exposed to the host’s immune system. The cysteine-linked diacyl lipid moiety of bacterial lipoprotein is recognized as a danger signal by the immune system to induce antimicrobial activity and triggers host defense mechanisms through Toll-like receptor stimulation (Brightbill et al. 1999; Thoma-Uszynski et al. 2001; Schenk et al. 2009). Lipoproteins are thus critical antigens for protective immunity against bacterial diseases (Hanson and Hansen 1991; Stoll et al. 2005; Bubeck et al. 2006; Bastian et al. 2008; Luo et al. 2009). For example, E. coli lipoproteins stimulate lymphocyte proliferation and immunoglobulin secretion (Melchers et al. 1975; Bessler et al. 1977). Mycobacterium tuberculosis lipoproteins, LpqH, LprG, and LprA, could induce either immunosuppressive responses and/or humoral and cellular responses via Toll-like receptors (Pecora et al. 2006; Drage et al. 2009; Lancioni et al. 2011). OmpL1 and LipL41, lipoproteins of Leptospira species, provide protection against leptospirosis challenge in the hamster model (Haake et al. 1999). Several lipoproteins of Neisseria meningitidis also have been identified as vaccine candidates (Pizza et al. 2000; Pajon et al. 2009; Serruto et al. 2010; Sung et al. 2010). Furthermore, animals immunized with the lipidated outer surface protein A (OspA) were protected against Borrelia burgdorferi challenge, while vaccination with non-lipidated OspA could not provide protective immunity (Fikrig et al. 1990; Bockenstedt et al. 1993; Johnson et al. 1995; Philipp et al. 1997). Importantly, the N-terminal fatty acid-containing region of lipoproteins appears to be responsible for the immunogenic activity (Bessler and Ottenbreit 1976; Lugade et al. 2011).
Application of recombinant lipoproteins as vaccine candidates was limited by the expression level, lipid modification, and downstream process. Different lipid signal peptide sequences have been fused with the target gene, but heterologous expression of recombinant lipoproteins in E. coli often results in incomplete or absent modification (Hansson et al. 1995; Shang et al. 1996). Based on a lipid signal peptide, Lpp, a plasmid, pDUMP, was designed for recombinant lipoprotein expression (Cullen et al. 2003). However, the efficiency of lipidation among the three tested clones varied considerably. Recently, we discovered that the fusion protein of D1, the N-terminal 40 residues of Ag473 (a lipoprotein from N. meningitidis), and E3 (the domain III of the dengue virus envelope protein) could be expressed at high levels with lipid modification in the E. coli C43 (DE3) strain (Chen et al. 2009). The recombinant rlipo-D1E3 was able to elicit stronger anti-E3 and virus-neutralizing antibody responses in animal studies. However, fusion of the D1 fragment with other proteins was not always successful. Some of the recombinant proteins were not lipidated or were incompletely lipidated in rich medium. Therefore, we used the recombinant D1E3 as a model to characterize the effect of individual medium composition on the expression of recombinant lipoproteins. We first utilized various nutrient-rich and nutrient-deficient media to express the recombinant D1E3 and found that M9 medium was favorable for the expression of mature rlipo-D1E3. According to the differences between nutrient-rich and nutrient-deficient media, we then analyzed the influence of various media factors, including nitrogen and carbon sources, phosphate, ferrous ions, calcium, magnesium, and pH, on the levels of lipoprotein expression. In addition, our results also demonstrated that M9 medium was suitable not only for the expression of mature lipidated D1E3 but also for other lipoproteins.
Materials and methods
Chemicals
All chemicals were from Sigma (St. Louis, MO, USA) and Merck (Darmstadt, Germany). Yeast extract, tryptone and casamino acids were obtained from Difco Laboratories (Detroit, MI, USA). Enzymes for molecular cloning were from New England Biolabs, Inc. (Beverly, MA, USA).
Bacterial strains and growth media
E. coli strain C43 (DE3) (Lucigen, Middleton, WI, USA) was used for expressing lipoproteins. The compositions of the media used, including Luria–Bertani (LB) broth, Super broth (SB), Terrific broth (TB), 2xYT, M9 medium, and M63 medium are listed in Table 1. All media were supplemented with 50 μg/ml ampicillin. When applicable, tryptone, yeast extract, casamino acids, or ammonium chloride were used as the nitrogen source of M9 medium. To evaluate the effect of carbon source on the expression of lipoprotein, various amounts of glucose or glycerol were added to M9 medium lacking a carbon source (CM9). Five conditions were tested: CM9/0.08% glucose, CM9/2% glucose, CM9/0.08% glycerol, CM9/0.4% glycerol, and CM9/2% glycerol. In order to assess the suitable conditions for lipoprotein expression, several additional parameters were also manipulated (Table 2). Growth was monitored spectrophotometrically by measuring the culture’s OD600 nm periodically.
Construction and expression of recombinant lipoproteins
The D1E3-expressing plasmid was obtained before (Chen et al. 2009). Briefly, the D1 fragment was cloned into the pET-22b (+) vector using NdeI and BamHI sites. The E3 gene was subsequently cloned into the plasmid using BamHI and XhoI sites to form a D1E3-expressing plasmid (Fig. 1b). P3E3-expressing plasmid was cloned by changing the D1 sequence to the P3 sequence (lipid signal peptide of E. coli acriflavine-resistance protein E precursor). The amino acid sequences of D1 and P3 fragment were shown in Fig. 1c. D1Plyt-expressing plasmid was engineered by replacing the E3 with a truncated pneumolysin (Plyt) sequence. The expression plasmid was transformed into the E. coli C43 (DE3) strain. The transformed cells were grown aerobically overnight in BD Falcon round-bottom tubes containing 5 ml LB medium at 37 °C. Before inoculating into the M9 medium or fresh LB medium, 100 μl of overnight culture were harvested by centrifugation. Bacteria were then resuspended in 5 ml fresh media and grown aerobically in the shaking incubator (200 rev/min) at 37 °C. After the bacteria reached late log-phase (OD600 nnm ~0.7), protein expression was induced by addition of 1 mM isopropylthio-β-d-galactoside (IPTG). It took 2–2.5 h for the bacteria grown in LB medium to reach the IPTG induction point, whereas the bacteria cultured in the M9 medium needed 5–6 h. Cultures were allowed to incubate for additional 3 h, and 500 μl of the cultures were centrifuged. The cells collected were resuspended in water to an absorbance at 600 nm equal to 10. Five microliters of the suspension was then withdrawn and analyzed by tricine SDS-PAGE.
Purification of recombinant lipoproteins
Recombinant lipoproteins were expressed in M9 medium as mentioned above. After induction, 2 l of cell culture were collected and the pellets were resuspended in 100 ml of homogenization buffer (20 mM Tris–HCl, 500 mM NaCl, 10% glycerol, 50 mM sucrose, pH 8.0). After disruption of the cells in a French Press (Constant Systems, Daventry, UK) at 27 kpsi, 0.5% Triton X-100 was added to the homogenate and the cell lysate was then clarified by centrifugation (80,000 × g for 60 min). The supernatant was loaded onto the Ni-NTA resin (Qiagen, San Diego, CA, USA). The column was first washed with the homogenization buffer followed by the same buffer containing 40 mM imidazole. The lipoproteins were then eluted with the homogenization buffer containing 500 mM imidazole.
Tricine SDS-PAGE and immunoblot analysis
For SDS gel electrophoresis, cell suspensions were mixed with an equal volume of sample buffer (63 mM Tris–HCl, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.002% bromophenol blue, pH 6.8) and were boiled for 3 min. Proteins were separated with a Bio-Rad Mini-PROTEAN 3 system (Bio-Rad Laboratories, Hercules, CA, USA) using 16% tricine SDS-PAGE and then stained with Coomassie Brilliant Blue R-250. For immunoblot analysis, proteins were electrophoretically transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% nonfat milk in Tris-buffered saline (20 mM Tris–HCl, 150 mM NaCl, pH 7.5) containing 0.1% Tween-20 (TBST) and then incubated at room temperature for 1 h with mouse anti-(His)6 antibodies (Amersham Biosciences, New Territories, HK) at a 1:15,000 dilution. After washing with TBST, the blot was incubated at room temperature for 1 h with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:15,000) (Bethyl Laboratories, Montgomery, TX, USA). The blot was developed using Immobilon Western HRP Substrate (Millipore) and was exposed to X-ray film. For detection of the lipid signal peptide, the blot was first incubated with the anti-MKKL polyclonal antibodies (LTK BioLaboratories, Taoyuan, Taiwan) (1:2,500) and then reacted with HRP-conjugated goat anti-rabbit IgG (1:5,000). The anti-MKKL polyclonal antibodies were derived from rabbits immunized with ovalbumin-conjugated lipid signal peptide (MKKLLIAAMMAAALAACS).
N-terminal amino acid sequencing
Proteins expressed in E. coli were electrophoretically transferred to a PVDF membrane after separation by 16% tricine SDS-PAGE. The blot was stained with Coomassie Brilliant Blue R-250 for 1 min and washed three times with 50% methanol/1% acetic acid. The protein band was excised from the blot and was subjected to four cycles of Edman degradation using an Applied Biosystems Model 494 Protein Sequencer (Mission Biotech Co. Ltd., Taipei, Taiwan).
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry
Purified lipoproteins were dialyzed against 100 mM ammonium bicarbonate. Dialyzed samples were digested by trypsin at a 50:1 ratio (w/w) in 300 mM ammonium bicarbonate. The digestion was repeated three times to make sure those proteins were fully digested. After adding formic acid to stop the reaction, the digested lipopeptide were purified using the ZipTip Pipette Tips (Millipore). The non-lipidated peptide fragments could be removed by washing the ZipTip Pipette Tips with 70% acetonitrile/0.1% trifluoroacetic acid. The lipid-modified peptide fragments remained on the ZipTip Pipette Tips until they were eluted using isopropanol. The sample was mixed with an equal volume of the saturated solution of α-cyano-4-hydroxycinnamic acid (CHCA) (Sigma) in 70% acetonitrile/0.1% trifluoroacetic acid. The mixture was deposited on the target plate for MALDI-TOF (Waters, Milford, MA, USA) analysis.
Calculation of the specific yields and I/M ratio
Total proteins of the collected cells were measured using Bradford protein assay kit (Bio-Rad Laboratories). The expression levels of immature and mature lipoproteins were estimated by analyzing the corresponding band density on SDS gels using densitometry and were quantified with ImageJ software from two independent experiments. The specific yields of immature and mature lipoproteins were calculated using the equation: \( \left[ {\left( {{\text{Amount}}\;{\text{of}}\;{\text{total}}\;{\text{protein}}} \right) \times \left( {{\text{Expression}}\;{\text{ratio}}\;{\text{of}}\;{\text{immature}}\;{\text{or}}\;{\text{mature}}\;{\text{lipoproteins}}} \right)} \right]/\left( {{\text{Collected}}\;{\text{cell}}\;{\text{weight}}} \right) \). The ratio of immature to mature lipoproteins (I/M ratio) was the division result of the expression ratio of immature and mature lipoproteins.
Results
Recombinant lipoprotein could be expressed homogeneously at high levels in M9 medium
Several media were evaluated for the expression of recombinant lipoproteins. The testing media included four nutrient-rich media (LB, TB, SB and 2xYT) and two nutrient-deficient media (M9 and M63). In all nutrient-rich media, two induced bands were observed by tricine SDS-PAGE, and both bands contained a (His)6 tag at their C termini (Fig. 2a and b). As determined by Edman degradation, the N-terminal sequence of the upper band was MKKL. This result suggested that the upper band had an incomplete lipid modification or was without a lipid moiety. Furthermore, we confirmed that the induced upper band still contains the signal peptide that could be recognized by anti-MKKL antibodies (Fig. 2c). On the other hand, only one induced band corresponding to the position of the lower band was observed after IPTG induction in the M9 medium (Fig. 2a, lane 10). Edman degradation results showed that its N terminus was blocked, and immunoblotting demonstrated that it did not contain the D1 N-terminal sequence (Fig. 2c, lane 10). Since the lipid modification in mature lipoprotein is a N-acyl-S-diacylglyceryl-cystine structure, the N-terminal acylation of proteins would result in N-terminal blockage (Cullen et al. 2003; Chen et al. 2009). These data suggested that the upper band could be an immature lipoprotein, and the lower band was a mature lipoprotein. The different migration on SDS-PAGE could be due to the processed lipid signal peptide. Moreover, the single band expressed in M9 medium was further purified, and analyzed by mass spectrometry. Figure 3a demonstrated that the extracted lipopeptide from the tryptic fragments of rlipo-D1E3 could be separated into two groups (I and II) (Fig. 3a). The peaks in each group differ in mass by 14 atomic mass units (amu). This difference suggested that these signals originated from the N-terminal fragments which were modified by lipids with different fatty acids. The masses of these peaks and the pattern of 14 amu difference were identical to those obtained from rAg473 (Chen et al. 2009). The similar pattern was also observed in the lipid-modified fragments of cyanobacterial lipoprotein PsbQ (Ujihara et al. 2008). All results indicated that the expressed protein in M9 medium was a mature rlipo-D1E3 lipoprotein. Under our culture conditions, the specific yield of mature rlipo-D1E3 in M9 medium was 3.6 mg/g cells, which was higher than in all nutrient-rich media (Fig. 4).
Recombinant D1E3 expression in different media. a Recombinant D1E3 was expressed in LB (lanes 1 and 2), SB (lanes 3 and 4), TB (lanes 5 and 6), 2xYT (lanes 7 and 8), M9 (lanes 9 and 10), and M63 (lanes 11 and 12) media. Expressed proteins were induced and analyzed by tricine SDS-PAGE with Coomassie Brilliant Blue staining. b Immunoblot with anti-(His)6 and c anti-MKKL antibodies. Lanes 1, 3, 5, 7, 9, 11: protein expression in the absence of IPTG induction; lanes 2, 4, 6, 8, 10, 12: protein expression after IPTG induction. Arrows indicate the location of induced proteins on gels. The solid line shows the fully processed lipoproteins, and the dotted line points to the position of unprocessed lipoproteins
MALDI-TOF MS spectrum of the N-terminal fragments of recombinant lipoproteins. N-terminal fragments of lipoproteins were identified following fully digestion by trypsin and eluted from the ZipTip Pipette Tips. a N-terminal fragments of rlipo-D1E3. Four peaks in group I (1,452.07, 1,466.09, 1,480.13, and 1,494.10 m/z) and two peaks in group II (1,492.11 and 1,506.15 m/z) are indicated. b N-terminal fragments of rlipo-P3E3. Four signals with m/z values of 1,295.99, 1,310.01, 1,324.03, and 1,338.06 in group I were indicated. Two signals with value of m/z 1,331.99 and 1,346.01 in group II are shown
Specific yields of mature and immature lipoproteins in various media. The specific yields were calculated as described in “Materials and methods.” The x-axis shows the used medium and the y-axis indicate the specific yields (milligram per gram of cells). The black bar represents the immature lipoproteins and the open bar denotes the mature lipoproteins
Based on these results, it appeared that the nutrient-deficient medium might be better for the expression of mature lipoproteins. We then verified if this was generally true by using another nutrient-deficient medium, M63. Surprisingly, two forms of recombinant proteins were induced. Just like those in the rich medium, the upper band expressed in M63 medium reacted with anti-MKKL antibodies (Fig. 2c, lane 12) and showed MKKL sequence by N-terminal sequencing, whereas the N-terminal sequence of the lower band (Fig. 2a, lane 12, arrow with solid line) could not be detected by Edman degradation. The ratio of immature to mature lipoproteins (I/M ratio) in M63 medium was higher than that in M9 medium (Table 2). These results suggested that the nutrient-rich media were not favorable for lipoprotein expression. Additionally, not all nutrient-deficient media were perfect for lipoprotein expression. Therefore, we selected M9 medium for subsequent studies and tried to identify the ingredient affecting lipoprotein expression.
Addition of nitrogen sources to M9 medium increased the level of immature lipoproteins
One of the differences between rich media and minimal media is the nitrogen source. We added different nitrogen sources to M9 medium to investigate their influence on lipoprotein expression. Tryptone, yeast extracts, or casamino acids were used as the organic nitrogen source, and ammonium chloride was utilized as the inorganic nitrogen source. Expression of lipoproteins in M9 medium supplemented with different amounts of these nitrogen sources was examined. Although the doubling times of bacteria in all the media supplemented with organic nitrogen were shorter than that in M9 medium alone (Table 2), the level of immature lipoproteins were increased significantly (Fig. 5a, lanes 3–9, arrow with dotted line). Indeed, the I/M ratio increased with increasing amount of yeast extract (0.5% to 2%), tryptone (1% to 4%) and casamino acids (0.02% to 4%) (Table 2). Addition of NH4Cl to M9 did not have any significant effect, except that some immature lipoproteins were detected by immunoblot using anti-MKKL antibodies in the presence of 1.5% NH4Cl (Fig. 5b, lane 11). Excess nitrogen sources in media would decrease the specific yield of mature lipoproteins but increase the specific yield of immature lipoproteins (Fig. 4). Our result suggested that the level of immature lipoprotein could be affected by the amount of nitrogen sources in M9 medium.
The effect of nitrogen and carbon sources on the expression of D1E3 in M9 medium. a Recombinant D1E3 was expressed in the M9 medium supplemented with additional nitrogen. IPTG-induced proteins were analyzed by tricine SDS-PAGE with Coomassie Brilliant Blue staining and b by immunoblot with anti-MKKL antibodies. Lane 1, protein expression in the absence of IPTG induction in M9 medium; lane 2, protein expression after IPTG induction in M9 medium; lanes 3 and 4, protein induction in M9 medium supplemented with 0.5% and 2% yeast extract, respectively; lanes 5 and 6, protein induction in M9 medium with extra tryptone added (1% and 4%, respectively); lanes 7–9, protein induction in M9 medium with additional casamino acids (0.02%, 1%, and 4%, respectively); lanes 10 and 11, protein induction in M9 medium with additional ammonium chloride (0.5% and 1.5%, respectively). c D1E3 was expressed in the modified M9 medium containing different carbon sources and analyzed by tricine SDS-PAGE with Coomassie Brilliant Blue staining and d by immunoblot with anti-MKKL antibodies. Lane 1, protein expression in the absence of IPTG induction in M9 medium; lanes 2–4, IPTG-induced protein expression sequentially in the CM9/0.08% glucose, M9, and CM9/2% glucose medium; lanes 5–7, protein induction serially in CM9/0.08% glycerol, CM9/0.4% glycerol, and CM9/2% glycerol medium. Arrows indicate the location of induced proteins on gels. The solid line shows the fully processed lipoproteins, and the dotted line points to the position of unprocessed lipoproteins
Glucose is a better carbon source than glycerol for lipoprotein expression
The level of immature lipoprotein in M63 medium was higher than in M9 medium. Because the major difference between M9 medium and M63 medium is the carbon source, we therefore investigated the effect of the carbon source on lipoprotein expression. Different amounts of glucose or glycerol were added to M9 medium lacking a carbon source (CM9), and the expression of lipoprotein was examined. The concentration of glucose in M9 medium was modified from 0.4% to 0.08% or 2%. Different amount of glycerol (0.08%, 0.4% or 2%) were also added to M9 minimal medium to replace the glucose. The results demonstrated that the doubling times in cM9 with glucose were shorter than that in cM9 with glycerol (Table 2). In M9 medium with glucose, only mature lipoproteins were detected (Fig. 5c and d, lanes 2–4). However, in M9 medium with glycerol, immature lipoproteins were observed using anti-MKKL antibodies (Fig. 5d, lanes 5–7). These results suggested that glucose is a better carbon source than glycerol for lipoprotein expression.
Media pH affects lipoprotein maturation
pH homeostasis in E. coli is relatively good with external pH ranging from 5.0 to 8.5 (Slonczewski et al. 1981; Zilberstein et al. 1984). However, gene expression profiles differ when E. coli are exposed to external acidification or alkalization (Heyde et al. 1991; Tucker et al. 2002). In this study, we explored whether or not medium pH ranging from 5.5 to 8.5 would affect lipoprotein maturation. The results showed that the expression level of rlipo-D1E3 was relatively low in acidic medium (pH 5.5 and pH 6.0) (Fig. 6a and b, lanes 2 and 3), and immature lipoproteins could be detected in M9 medium with pH 5.5 (Fig. 6b, lane 2). The expression level of lipoproteins was similar with pH values ranging from 7.0 to 8.5 (Fig. 6a and b, lanes 4–6). Moreover, the I/M ratio of recombinant lipoproteins expressed in LB medium buffered at pH 5.5 and pH 7.0 were comparable (Table 2). Interestingly, the I/M ratios were reduced if the pH value of LB medium increased to 8.5 (Table 2; Fig. 6a and b, lane 9). The specific yield of mature lipoproteins was also increased when the alkaline LB medium was utilized (Fig. 4). These observations implied that the M9 medium with higher pH values is preferable for expressing mature lipoprotein.
Influence of pH, phosphate, calcium, magnesium, and ferrous ions on lipoprotein expression. a Recombinant D1E3 was expressed in LB or M9 medium with different pH values. Protein expression was observed via tricine SDS-PAGE with Coomassie Brilliant Blue staining and b analyzed by immunoblot with anti-MKKL antibodies. Lane 1, protein expression in the absence of IPTG induction in M9 medium; lanes 2, 3, 5, and 6, protein expression after IPTG induction in modified M9 medium with pH 5.5, 6.0, 8.0, and 8.5, respectively. Lane 4, protein induction in M9 medium (pH 7.0); lane 7, protein induction in LB medium buffered with 100 mM MES at pH 5.5; lane 8, protein expression after IPTG induction in LB medium buffered with 100 mM HEPES at pH 7.0; lane 9, protein induction in LB medium buffered with 100 mM TAPS at pH 8.5. c The concentration of phosphate in M9 medium was reduced by half and increased by adding 25 or 100 mM sodium phosphate or potassium phosphate. Recombinant D1E3 was over-expressed in these cultures and analyzed by tricine SDS-PAGE with Coomassie Brilliant Blue staining and d by immunoblot with anti-MKKL antibodies. Lane 1, protein expression in the absence of IPTG induction in M9 medium; lane 2, protein expression after IPTG induction in M9 medium; lane 3, protein induction in PM9/35 mM phosphate medium; lanes 4 and 5, protein induction in M9 medium supplemented with 25 or 100 mM sodium phosphate, respectively; lanes 6 and 7, protein expression after IPTG induction in M9 medium with additional 25 or 100 mM potassium phosphate, respectively. e The concentration of CaCl2 and MgSO4 were increased ten-fold or decreased to one-tenth of the original concentration in M9 medium. The amounts of FeSO4 added in M9 medium were 0.002 or 0.05 mM. The induced D1E3 protein was analyzed by tricine SDS-PAGE with Coomassie Brilliant Blue staining and f by immunoblot with anti-MKKL antibodies. Lane 1, protein expression in the absence of IPTG induction in M9 medium; lane 2, protein expression after IPTG induction in M9 medium; lane 3, protein induction in MgM9/0.2 mM MgSO4 medium; lane 4, protein induction in MgM9/20 mM MgSO4 medium; lane 5, protein expression after IPTG induction in CaM9/0.01 mM CaCl2 medium; lane 6, protein expression after IPTG induction in CaM9/1 mM CaCl2 medium; lane 7, protein induction in M9 medium with additional 0.002 mM FeSO4; lane 8, protein induction in M9 medium supplemented with 0.05 mM FeSO4. The solid line shows the fully processed lipoproteins, and the dotted line points to the position of unprocessed lipoproteins
Addition of phosphate and ferrous ions might disturb the lipidation process
Because phosphate in medium can impair the growth of E. coli (Malouin et al. 1991), we next investigated whether phosphate might affect lipoprotein expression. Mixtures of monobasic dihydrogen phosphate and dibasic monohydrogen phosphate were used here in order to maintain the medium at pH 7.0. If the rlipo-D1E3 were expressed in M9 medium supplemented with 25 mM phosphate, immature lipoprotein was observed (Fig. 6d, lanes 4 and 6). With an additional 100 mM phosphate, immature lipoproteins became more evident via SDS-PAGE stained with Coomassie blue (Fig. 6c, lanes 5 and 7), and the I/M ratio increased to approximately 0.2 (Table 2). The specific yield results also showed an increasing specific yield of immature lipoproteins when the phosphate amounts in media were increased (Fig. 4). In contrast, reducing the amount of phosphate in M9 medium by 50% had no effect on lipoprotein maturation (Fig. 6c, lane 3). We also investigated the influence of other ingredients in M9 or M63 medium, including CaCl2, MgSO4 and FeSO4. Under our experimental conditions, the maturation of lipoprotein was not affected by MgSO4 and CaCl2 (Table 2; Fig. 6e and 6f, lanes 3–6). However, addition of FeSO4 to M9 medium resulted in an increase of immature lipoproteins as determined by anti-MKKL antibodies (Table 2; Fig. 6e and f, lanes 7 and 8). These results demonstrated that adding phosphate and ferrous ions in the M9 medium might result in defective lipidation.
High levels of mature lipoproteins could also be expressed in M9 medium via other constructs
The last question considered in this study is whether M9 medium is also profitable for expressing other recombinant lipoproteins. So far, our results clearly demonstrated that the M9 medium was better for the expression of rlipo-D1E3. We therefore changed the immunogen or the lipid signal peptide and then examined the expression of these recombinant lipoproteins in E. coli.
D1 was fused with a truncated pneumolysin (Plyt), which is a non-lipoprotein from Streptococcus pneumoniae, and this new construct was expressed in LB or M9 medium. Two induced bands of recombinant D1Plyt proteins were evident in LB medium (Fig. 7, lane 2; arrow ① and arrow ②), whereas the lower band protein was predominant in M9 medium (Fig. 7, lane 3; arrow ②). The upper band (Fig. 7, arrow ①) was an immature lipoprotein as determined by its N-terminal sequence. However, the N-terminal sequence of the lower band (Fig. 7, arrow ②) could not be obtained, indicating that it might be a mature lipoprotein. The rlipo-D1Plyt expressed in M9 medium was partially purified, and the lipid modification was confirmed using MALDI-TOF mass spectrometry (data not shown).
Expression of D1Plyt and P3E3 lipoproteins in LB and M9 medium. Two additional recombinant lipoproteins were over-expressed by IPTG induction in LB and M9 medium. a The induced proteins were observed using 16% tricine SDS gel staining with Coomassie Brilliant Blue and b by immunoblot using anti-(His)6 antibodies. Lane 1, bacterial total lysate in the absence of IPTG induction; lane 2, D1Plyt expression after IPTG induction in LB medium; lane 3, D1Plyt expression after IPTG induction in M9 medium; lane 4, P3E3 induction in LB medium; lane 5, P3E3 induction in M9 medium. The arrows with numbers 1 and 2 indicate the unprocessed and processed D1Plyt, respectively. The arrows with numbers 3 and 4 were the immature rlipo-P3E3 and the mature rlipo-P3E3, respectively
Another signal peptide (P3) of the acriflavine-resistance protein E precursor from E. coli was fused with E3, and P3E3 protein was expressed in both LB and M9 media. As shown in Fig. 7, two bands of P3E3 were expressed in LB medium (lane 4, arrow ③ and arrow ④), and only the lower band was induced in M9 medium (Fig. 7, lane 5). Further analysis using N-terminal sequences and MALDI-TOF mass spectrometry confirmed that the upper band was the immature lipoprotein (Fig. 7, arrow ③), and the lower band was the mature lipoprotein (Fig. 7, arrow ④). The mass spectrum showed that the lipopeptide fragments from rlipo-P3E3 had a similar pattern with those from rlipo-D1E3 (Fig. 3b). Altogether, our results clearly showed that M9 medium was advantageous for recombinant lipoprotein expression in the E. coli C43 (DE3) strain.
Discussion
Immunogenic properties of lipoproteins have numerous applications in the vaccine development. To use recombinant lipoproteins as a vaccine candidate, it is important to produce enough quantities of these materials. As shown in this study, the expression of lipid-modified lipoproteins can be enhanced through the optimization of nitrogen source, carbon source, medium pH, and other ions in the culture medium.
When the exported proteins were over-expressed, including lipoproteins, the translocation machinery is a bottleneck because the export machinery was overloaded (van Mellaert and Anne 2001). An approach for solving this problem is to reduce the protein synthesis using minimal medium (Ignatova et al. 2003). In this study, our results clearly showed that M9 medium is superior for expressing fully modified lipoproteins. In the case of D1E3, the specific yield of mature rlipo-D1E3 in M9 medium was significantly better than in LB medium (Fig. 4). The greater specific yield of entirely modified lipoproteins in M9 medium was also observed for the expression of other recombinant lipoproteins, D1Plyt and P3E3. Although the bacterial growth in M63 medium was much slower than in nutrient-rich media and M9 medium (Table 2), some immature lipoproteins were still detected (Fig. 2). Similarly, when glycerol was utilized as the carbon source in M9 medium, the bacteria grew slowly (Table 2), but some unprocessed lipoproteins were induced (Fig. 5d). In addition, we have tried to decrease the growth rate by expressing recombinant lipoproteins in LB medium at lower temperature (20 and 25 °C), but the immature lipoproteins were still obvious (data not shown). Therefore, in addition to bacterial growth rate, the medium components might also play a role in the lipidation process. The mechanism involved needs to be elucidated.
OprI lipoprotein has been expressed with M9 medium supplemented with 0.02% casamino acid in E. coli strain JM109 (Cote-Sierra et al. 1998). In our study, the use of M9 medium with additional 0.02% casamino acid would result in the expression of some unprocessed lipoproteins (Fig. 5b, lane 7). The existence of immature lipoproteins became more evident as increasing amounts of casamino acid were added (Fig. 5a, lanes 7–9). Although JM109 (a K12 derivative) and C43 (DE3) (a B derivative) were derived from different strains, we have used JM109 to express recombinant D1E3 in LB or M9 medium and the results were similar to those using C43 (DE3) as the expression strain, except the bacterial growth was inhibited (data not shown). Other organic nitrogen sources, such as yeast extract and tryptone, also had the similar effect on the expression of mature lipoproteins (Fig. 5a). Additional inorganic nitrogen source also influence the lipoprotein expression. Although bacteria cultivated in M9 medium supplemented with 1.5% NH4Cl showed a slower growth rate (Table 2), some immature lipoproteins were detected after induction (Fig. 5b, lane 10). These results suggest that expression of mature lipoproteins could be limited by the nitrogen source in the medium.
In this study, we have applied glucose and glycerol as different carbon sources to express lipoproteins. Our results showed that using glycerol in the minimal medium resulted in the existence of immature lipoproteins. The different growth rate could not explain this phenomenon because the slower growth in glycerol-based media still resulted in the existence of unprocessed lipoproteins (Fig. 5d). One possible explanation is that the media compositions might alter the amount of lipid for protein modification. This could be studied in advance using various carbon sources and observing the lipid content in bacteria.
E. coli can grow over a relatively wide range of external pH, and the bacterium can rapidly restore its internal pH to 7.4–7.8 (Padan et al. 1976; Zilberstein et al. 1984). Many genes encoding membrane proteins or proteins correlated with cell envelope structure or modifications are upregulated when external pH becomes acidic or alkaline (Heyde et al. 1991; Tucker et al. 2002). Because most lipoproteins are membrane or membrane-associated proteins (Hayashi and Wu 1990), the expression of lipoproteins might also be regulated by external pH. In this study, we discovered that an alkaline external environment facilitates the lipid modification of lipoproteins both in nutrient-rich and nutrient-deficient media (Fig. 6). This result suggested that the bacterial cell structure might be disturbed by alkaline surroundings, and more lipoproteins are needed to maintain cell integrity. The detailed mechanism of alkaline external pH on lipoprotein biosynthesis needs to be investigated further.
We previously demonstrated that the E. coli strain, C43 (DE3), could be used to over-express the recombinant outer-membrane protein in high yield (Chen et al. 2009). In this study, we further showed that M9 medium is favorable for expressing mature lipoproteins in the E. coli strain C43 (DE3). In addition to high yields of wholly modified lipoproteins, M9 medium usage is also beneficial for lipoprotein purification because the existence of large amounts of immature lipoproteins in LB medium would raise the complexity for the downstream purification process. These detailed studies supply practical information for production of recombinant lipoproteins and it could be applied in the field of vaccine development.
References
Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K (2006) A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 188:2761–2773
Bastian M, Braun T, Bruns H, Rollinghoff M, Stenger S (2008) Mycobacterial lipopeptides elicit CD4+ CTLs in Mycobacterium tuberculosis-infected humans. J Immunol 180:3436–3446
Bessler WG, Ottenbreit BP (1976) Studies on the mitogenic principle of the lipoprotein from the outer membrane of Escherichia coli. Biochem Biophys Res Commun 76:239–246
Bessler W, Resch K, Hancock E, Hantke K (1977) Induction of lymphocyte proliferation and membrane changes by lipopeptide derivatives of the lipoprotein from the outer membrane of Escherichia coli. Z Immunitatsforsch Immunobiol 153:11–22
Bockenstedt LK, Fikrig E, Barthold SW, Kantor FS, Flavell RA (1993) Inability of truncated recombinant osp a proteins to elicit protective immunity to Borrelia burgdorferi in mice. J Immunol 151:900–906
Braun V, Rehn K (1969) Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10:426–438
Brightbill HD, Libraty DH, Krutzik SR, Yang RB, Belisle JT, Bleharski JR, Maitland M, Norgard MV, Plevy SE, Smale ST, Brennan PJ, Bloom BR, Godowski PJ, Modlin RL (1999) Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285:732–736
Bubeck WJ, Williams WA, Missiakas D (2006) Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci USA 103:13831–13836
Chen HW, Liu SJ, Liu HH, Kwok Y, Lin CL, Lin LH, Chen MY, Tsai JP, Chang LS, Chiu FF, Lai LW, Lian WC, Yang CY, Hsieh SY, Chong P, Leng CH (2009) A novel technology for the production of a heterologous lipoprotein immunogen in high yield has implications for the field of vaccine design. Vaccine 27:1400–1409
Cote-Sierra J, Jongert E, Bredan A, Gautam DC, Parkhouse M, Cornelis P, De Baetselier P, Revets H (1998) A new membrane-bound opri lipoprotein expression vector. High production of heterologous fusion proteins in gram (−) bacteria and the implications for oral vaccination. Gene 221:25–34
Cullen PA, Lo M, Bulach DM, Cordwell SJ, Adler B (2003) Construction and evaluation of a plasmid vector for the expression of recombinant lipoproteins in Escherichia coli. Plasmid 49:18–29
Drage MG, Pecora ND, Hise AG, Febbraio M, Silverstein RL, Golenbock DT, Boom WH, Harding CV (2009) TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell Immunol 258:29–37
Fikrig E, Barthold SW, Kantor FS, Flavell RA (1990) Protection of mice against the lyme disease agent by immunizing with recombinant OspA. Science 250:553–556
Gupta SD, Gan K, Schmid MB, Wu HC (1993) Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J Biol Chem 268:16551–16556
Haake DA, Mazel MK, McCoy AM, Milward F, Chao G, Matsunaga J, Wagar EA (1999) Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect Immun 67:6572–6582
Hanson MS, Hansen EJ (1991) Molecular cloning, partial purification, and characterization of a haemin-binding lipoprotein from Haemophilus influenzae type b. Mol Microbiol 5:267–278
Hansson L, Noppa L, Nilsson AK, Stromqvist M, Bergstrom S (1995) Expression of truncated and full-length forms of the Lyme disease Borrelia outer surface protein a in Escherichia coli. Protein Expr Purif 6:15–24
Hayashi S, Wu HC (1990) Lipoproteins in bacteria. J Bioenerg Biomembr 22:451–471
Heyde M, Coll JL, Portalier R (1991) Identification of Escherichia coli genes whose expression increases as a function of external pH. Mol Gen Genet 229:197–205
Ignatova Z, Mahsunah A, Georgieva M, Kasche V (2003) Improvement of posttranslational bottlenecks in the production of penicillin amidase in recombinant Escherichia coli strains. Appl Environ Microbiol 69:1237–1245
Jackowski S, Rock CO (1986) Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J Biol Chem 261:11328–11333
Johnson BJ, Sviat SL, Happ CM, Dunn JJ, Frantz JC, Mayer LW, Piesman J (1995) Incomplete protection of hamsters vaccinated with unlipidated OspA from Borrelia burgdorferi infection is associated with low levels of antibody to an epitope defined by mAb LA-2. Vaccine 13:1086–1094
Lancioni CL, Li Q, Thomas JJ, Ding X, Thiel B, Drage MG, Pecora ND, Ziady AG, Shank S, Harding CV, Boom WH, Rojas RE (2011) Mycobacterium tuberculosis lipoproteins directly regulate human memory CD4(+) T cell activation via Toll-like receptors 1 and 2. Infect Immun 79:663–673
Lugade AA, Bianchi-Smiraglia A, Pradhan V, Elkin G, Murphy TF, Thanavala Y (2011) Lipid motif of a bacterial antigen mediates immune responses via TLR2 signaling. PLoS One 6:e19781
Luo D, Xue F, Ojcius DM, Zhao J, Mao Y, Li L, Lin X, Yan J (2009) Protein typing of major outer membrane lipoproteins from Chinese pathogenic Leptospira spp. and characterization of their immunogenicity. Vaccine 28:243–255
Malouin F, Chamberland S, Brochu N, Parr TR Jr (1991) Influence of growth media on Escherichia coli cell composition and ceftazidime susceptibility. Antimicrob Agents Chemother 35:477–483
Melchers F, Braun V, Galanos C (1975) The lipoprotein of the outer membrane of Escherichia coli: a B-lymphocyte mitogen. J Exp Med 142:473–482
Padan E, Zilberstein D, Rottenberg H (1976) The proton electrochemical gradient in Escherichia coli cells. Eur J Biochem 63:533–541
Pajon R, Yero D, Niebla O, Climent Y, Sardinas G, Garcia D, Perera Y, Llanes A, Delgado M, Cobas K, Caballero E, Taylor S, Brookes C, Gorringe A (2009) Identification of new meningococcal serogroup B surface antigens through a systematic analysis of neisserial genomes. Vaccine 28:532–541
Pecora ND, Gehring AJ, Canaday DH, Boom WH, Harding CV (2006) Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol 177:422–429
Philipp MT, Lobet Y, Bohm RP Jr, Roberts ED, Dennis VA, Gu Y, Lowrie RC Jr, Desmons P, Duray PH, England JD, Hauser P, Piesman J, Xu K (1997) The outer surface protein A (OspA) vaccine against Lyme disease: efficacy in the rhesus monkey. Vaccine 15:1872–1887
Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R (2000) Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816–1820
Sankaran K, Wu HC (1994) Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem 269:19701–19706
Schenk M, Belisle JT, Modlin RL (2009) TLR2 looks at lipoproteins. Immunity 31:847–849
Serruto D, Spadafina T, Ciucchi L, Lewis LA, Ram S, Tontini M, Santini L, Biolchi A, Seib KL, Giuliani MM, Donnelly JJ, Berti F, Savino S, Scarselli M, Costantino P, Kroll JS, O’Dwyer C, Qiu J, Plaut AG, Moxon R, Rappuoli R, Pizza M, Aricò B (2010) Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci USA 107:3770–3775
Shang ES, Summers TA, Haake DA (1996) Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun 64:2322–2330
Slonczewski JL, Rosen BP, Alger JR, Macnab RM (1981) pH homeostasis in Escherichia coli: Measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci USA 78:6271–6275
Stoll H, Dengjel J, Nerz C, Gotz F (2005) Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect Immun 73:2411–2423
Sung JW, Hsieh SY, Lin CL, Leng CH, Liu SJ, Chou AH, Lai LW, Lin LH, Kwok Y, Yang CY, Chong P (2010) Biochemical characterizations of Escherichia coli-expressed protective antigen Ag473 of Neisseria meningitides group B. Vaccine 28:8175–8182
Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, Sieling PA, Barnes PF, Rollinghoff M, Bolcskei PL, Wagner M, Akira S, Norgard MV, Belisle JT, Godowski PJ, Bloom BR, Modlin RL (2001) Induction of direct antimicrobial activity through mammalian Toll-like receptors. Science 291:1544–1547
Tokunaga M, Tokunaga H, Wu HC (1982) Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci USA 79:2255–2259
Tucker DL, Tucker N, Conway T (2002) Gene expression profiling of the pH response in Escherichia coli. J Bacteriol 184:6551–6558
Ujihara T, Sakurai I, Mizusawa N, Wada H (2008) A method for analyzing lipid-modified proteins with mass spectrometry. Anal Biochem 374:429–431
van Mellaert L, Anne J (2001) Gram-positive bacteria as host cells for heterologous production of biopharmaceuticals. In: van Broekhoven A, Shapior F, Anne J (eds) Novel frontiers in the production of compounds for biomedical use. Kluwer Academic Publishers, Dordrecht, pp 277–300
Zilberstein D, Agmon V, Schuldiner S, Padan E (1984) Escherichia coli intracellular pH, membrane potential, and cell growth. J Bacteriol 158:246–252
Acknowledgements
We thank Chiung-Yi Huang for preparing the anti-MKKL antibodies. The authors are grateful to Dr. Chung Wang (Institute of Molecular Biology, Academia Sinica, Taiwan) for comments on the manuscript. This work was supported by National Science Council (NSC 98-2320-B-400 -010 -MY3 to C.-H. L.) and Research Investigator Award from the National Health Research Institutes (99A1-VCPP06-014 to C.-H. L.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tseng, CL., Leng, CH. Influence of medium components on the expression of recombinant lipoproteins in Escherichia coli . Appl Microbiol Biotechnol 93, 1539–1552 (2012). https://doi.org/10.1007/s00253-011-3516-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3516-8