Abstract
The sponge-associated actinomycetes were isolated from the marine sponge Dendrilla nigra, collected from the southwest coast of India. Eleven actinomycetes were isolated depending upon the heterogeneity and stability in subculturing. Among these, Nocardiopsis dassonvillei MAD08 showed 100% activity against the multidrug resistant pathogens tested. The culture conditions of N. dassonvillei MAD08 was optimized under submerged fermentation conditions for enhanced antimicrobial production. The unique feature of MAD08 includes extracellular amylase, cellulase, lipase, and protease production. These enzymes ultimately increase the scope of optimization using broad range of raw materials which might be efficiently utilized. The extraction of the cell free supernatant with ethyl acetate yielded bioactive crude extract that displayed activity against a panel of pathogens tested. Analysis of the active thin layer chromatography fraction by Fourier transform infrared and gas chromatography-mass spectrometry evidenced 11 compounds with antimicrobial activity. The ammonium sulfate precipitation of the culture supernatant at 80% saturation yielded an anticandidal protein of molecular weight 87.12 kDa. This is the first strain that produces both organic solvent and water soluble antimicrobial compounds. The active extract was non-hemolytic and showed surface active property envisaging its probable role in inhibiting the attachment of pathogens to host tissues, thus, blocking host–pathogen interaction at an earlier stage of pathogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infectious diseases are leading health problems with high morbidity and mortality in the developing countries (Black et al. 1982; Walsh and Warren 1974). The development of resistance to multiple drugs is a major problem in the treatment of these infectious diseases caused by pathogenic microorganisms. This multidrug resistance is presently an urgent focus of research and new bioactive compounds are necessary to combat these multidrug resistance pathogens. On this view point, attempts have been made to develop novel drugs against infectious diseases for the mitigation of suffering of the vast masses of humanity. The first systemic search for antibiotics made by Gratia and Dath in 1924 resulted in the discovery of actinomycetin from actinomycetes. The actinomycetes and particularly the genus Streptomyces is widely reported for the production of various antibiotics which are used therapeutically (Argoudelis et al. 1987; Dienstag and Neu 1972; Waksman and Woodruff 1940). About 70% of all known drugs have been isolated from actinomycetes, of which 75% and 60% are used in medicine and agriculture, respectively. The rate of discovery of new compounds from terrestrial actinomycetes has decreased, whereas the rate of reisolation of known compounds has increased (Fenical et al. 1999). Thus, it is crucial that new groups of actinomycetes from pristine habitats need to be explored as sources of novel bioactive secondary metabolites.
Marine microorganisms have been the topic for the investigation of number of natural products. As marine environmental conditions are extremely different from terrestrial ones, it is surmised that marine actinomycetes have different characteristics from those of terrestrial counterparts and, therefore, might produce different types of bioactive compounds. It has been proposed that these antimicrobials are used in competition between microorganisms, offering an advantage to the producer strains (Dienstag and Nue 1972). Thus, marine actinomycetes in particular have received increased attention as potential sources of biologically active metabolites. In this background, the present study was initiated to explore sponge-associated actinomycetes as a sustainable source for the production of novel antibiotics, the mechanism of antibiosis, and the optimization of culture conditions for the production of these antibiotics. In this study, we report the antimicrobial activity of actinomycetes isolated from the sponge Dendrilla nigra collected off from the southwest coast of India.
Materials and methods
Collection of sponges
The marine sponge D. nigra was collected off from the southwest coast of India by SCUBA diving at 10–12 m depth. Samples were surface cleaned with aged sterile seawater and surface sterilized with 70% alcohol. Then, the samples were kept in a sterile incubator oven for 1 h at 40 °C to dry the surface and frozen (−20 °C) immediately in sterile sip-lap bags. Voucher specimens, photographs, and videographs of the specimen and habitat were retained in the laboratory for future reference. The collected specimens were identified and reconfirmed by P.A. Thomas, an eminent sponge taxonomist in India.
Isolation of actinomycetes
A measured area of sponge tissue (1 cm2) was excised using a sterile scalpel from the internal mesohyl area. The tissue was then homogenized in phosphate-buffered saline (PBS) using a sterile mortar and pestle. The resultant homogenate was serially diluted with PBS and preincubated at 40 °C for 1 h for the activation of dormant cells. The aliquot was plated on various isolation media including marine sponge agar (Selvin et al. 2004) and standard media (Himedia®) such as Emerson agar, actinomycetes agar, actinomycetes isolation agar, and marine agar. The inoculated plates were incubated at 27 °C for 14 days in dark aerobic conditions until visible colonies appeared. The morphologically distinct colonies were reisolated and inoculated using the 16-streak method on successive plates until pure cultures were achieved as estimated by colony homogeneity and Gram stain cell morphology criteria. The separated colonies were maintained on actinomycetes isolation agar at 4 °C.
Screening for potential antagonistic producer
Determination of antimicrobial activities of pure actinomycetes cultures were performed by cross-streak method. Muller–Hinton agar plates were prepared and inoculated with actinomycetes cultures by a single streak of inoculum in the centre of the petridish and incubated at 27 °C for 4 days. The plates were seeded with test organisms by a perpendicular streak at a 90° angle to the line of actinomycetes growth. Antagonism was observed based on the inhibitory interaction between the actinomycetes and test strains. The potential antagonistic producer was selected by screening the actinomycetes isolates against a panel of pathogens including 13 gram-positive and nine gram-negative bacteria. The panel of pathogens used in the present study is presented in Table 1. The clinical pathogens (PC1–PC8) were established as multidrug resistant pathogens and deposited in Biomedical Diagnostic Laboratory, Bharathidasan University. The resistance patterns of these isolates were confirmed using selective antibiotics that are routinely used to combat human pathogenic diseases. Preliminary experiments confirmed that the clinical isolates were resistant against chloramphenicol, streptomycin, oxytetracycline, ampicillin, and erythromycin. All the bacterial strains were maintained on nutrient agar slants at 4 °C. A total of 30 Candida species were used to study the anticandidal activity of MAD08. Of the 30 isolates, 28 were obtained from the oral cavity of tuberculosis patients and named as TC1–TC28, one Candida albicans FC1 a secondary pathogen of HIV infection and one type culture C. albicans MTCC 227. All the clinical Candida isolates (TC1–TC28 and FC1) used for the study were resistant against ceftriaxone, vancomycin, and amphotericin. All the Candida species were maintained on SDA slants at 4 °C.
Characterization of antagonistic producer MAD08
Biochemical, morphological, and physiological characteristics of the potential antimicrobial producer (MAD08) was determined by adopting standard methods (Lechevalier 1989). Cultural characteristics of MAD08 were determined based on the color comparisons with standard reference strains (Lechevalier 1989). The genomic DNA of actinobacterium was isolated by CTAB/NaCl method. Universal 16S rRNA eubacterial primer (5′-GAGTTTGATCCTGGCTCAG-3′; 5′-AGAAAGGAGGTGATCCAGCC-3′) was used for the amplification of DNA. The polymerase chain reaction (PCR) was carried out on a thermal cycler (Eppendorf) in a 50-μl reaction mix. The reaction mix contained 10× amplification buffer, 1.5 mM MgCl2, 10 mM forward primer, 10 mM reverse primer, 1 μl dNTP, and 0.25 μl Taq polymerase. After initial denaturation at 95 °C for 1 min, amplification was performed with 35 cycles of 35 s at 94 °C, 40 s at 55 °C, 2 min at 72 °C followed by a final extension at 72 °C for 8 min. The 16S rRNA gene sequence obtained from the isolate MAD08 was compared with other bacterial sequences by using NCBI BLASTn (Altschul et al. 1990, 1997) for their pair wise identities. Multiple alignments of these sequences were carried out by Clustal W 1.83 version of EBI (http://www.ebi.ac.uk/cgi-bin/clustalw/) with 0.5 transition weight. Phylogenetic trees were constructed in MEGA 4.0 version (http://www.megasoftware.net) using neighbor joining (NJ), minimum evolution (ME) and unweighted pair group method with arithmetic mean (UPGMA) algorithms. NJ, ME, and UPGMA bootstrapping were performed with 1,000 replicates with pseudorandom number generators. The sequence used in the analysis was deposited in GenBank, EMBL in Europe, and the DNA Data Bank of Japan with an accession number EU939309.
Optimization of factors affecting antimicrobial production
One loopful of cells from single colony of actinomycetes were inoculated into actinomycetes broth and incubated at 30 °C with shaking at 220 rpm for 120 h. The cell free supernatant (CFS) was obtained by centrifugation at 8,000×g for 20 min (Eppendorf). Factors like different carbon, nitrogen, pH, temperature, and salt concentration affecting the secretion of antimicrobial compound by MAD08 were optimized by adopting search technique varying parameters one at a time. Each of the carbon sources like glucose, sucrose, maltose, mannose, lactose, glycerol, galactose, starch, fructose, and mannitol at 1% (w/v) concentration were added separately to the production media in order to study its effect on the production of antimicrobial compound. Malt extract, yeast extract, beef extract, peptone, ammonium sulfate, sodium nitrate, oat meal, corn meal, urea, and casein at 1% (w/v) concentration was supplemented to the production medium as different nitrogen sources. The influence of various physiological conditions on the production of antimicrobial compound by MAD08 was evaluated by maintaining the production medium at different temperature ranges (4, 26, 30, 40, 50 °C), pH ranged from 5 to 12 (with increments of one unit) and salt concentrations, 0.5% to 3% (with increments of 0.5 units). MAD08 was inoculated in production medium and incubated for 120 h. After incubation, the cells were removed by cold centrifugation at 8,000×g for 20 min. The CFS was analyzed for antimicrobial activity and production of extracellular enzymes.
The extracellular production of amylase, cellulase, lipase, and protease were determined as factors required for the utilization of wider range of nutritional sources including raw materials. Amylase and cellulase activities of the CFS were determined by detecting the amount of reducing sugar (glucose) liberated (Bernfield 1955). The reducing sugar released was measured at 540 nm in a UV/Vis double beam scanning spectrophotometer (Thermospectronic; Miller 1959). One unit of amylase/cellulase activity was defined as microgram of reducing sugar released by 1 ml of enzyme/min, under assay conditions and expressed as unit per milligram protein of the culture broth. Estimation of lipase activity was carried out by titrating the fatty acids liberated from olive oil with an alkali (Ohnishi et al. 1994). The liberated fatty acids were titrated with 1 N NaOH solution. Simultaneously a blank titration was performed by omitting the incubation time and titrating the incubation mixture at the very beginning. The protease activity was measured in terms of tyrosine liberated from casein. One unit (U) was expressed as the µM of tyrosine released by 1 ml of enzyme/min under the assay condition. Tyrosine was estimated by Folin–Ciocalteau’s reagent (Manachini et al. 1988).
Antibacterial assay
The antibacterial assay was carried out as per Selvin et al. (2004). Briefly, the base layer was prepared with 10 ml (1.5%, w/v) of Muller Hinton agar (Himedia). Five numbers of sterile porcelain beads were placed on the base layer at 60° (angle) apart. The overlaid seed layer was prepared by pouring 15 ml of media containing 0.2 ml of prepared inoculum (∼0.2 OD at 630 nm). The porcelain beads were removed carefully with sterile forceps. The resultant wells in triplicate were filled with 100 μl of the CFS. The remaining well was filled with reference drugs (nystatin 30 μg/ml and nalidixic acid, 50 μg/ml). After 24 h of incubation at 37 °C, the area of inhibition around the wells was determined as average of triplicates. The antibiogram of pathogen was analyzed for skewness using SPSS 11.0 software.
Anticandidal assay
Sabouraud dextrose agar was used for bioactivity screening and routine propagation of Candida strains. For plate assay, a yeast suspension of 106 CFU/ml in sterile PBS was prepared (Forbes et al. 1990). The CFS was filled up to the brim of each well. Parallel reference standard (clotimazole 1% topical solution) was used to validate the inferences. The plates were incubated at 30 °C for 48 h. After incubation, the bioactivity was determined by measuring the area of inhibition zone.
Extraction of the antimicrobial compound
Spores at 107/ml of MAD08 strain were inoculated into 1,000-ml Erlenmeyer flask containing 300 ml of actinomycetes broth with 1% oat meal. After incubation at 30 °C for 120 h in an orbital incubator with shaking at 250 rpm, the culture broth was filtered to separate mycelium. The filtrate was subjected to centrifugation at 8,000×g for 20 min. The supernatant was extracted twice with ethyl acetate. The crude extract was concentrated to solvent free by evaporation in a rotary vacuum evaporator (Yamato). The residues obtained were dried in a vacuum desiccator and dissolved in ethyl acetate.
Characterization of the active ethyl acetate extract
The crude extract was subjected for silica gel thin layer chromatography (TLC). The TLC fractions were extracted from the scraped spots and checked for antimicrobial activity against the panel of pathogens presented in Table 1. The infrared spectra of active TLC fraction were recorded on Shimadzu IR-470 model. The spectra were scanned on 400 to 4,000 cm−1 range. The spectra were obtained using potassium bromide pellet technique. Potassium bromide (AR grade) was dried under vacuum at 100 °C for 48 h and 100 mg of KBr with 1 mg of sample was taken to prepare KBr pellet. The spectra were plotted as intensity versus wave number. Then the active ethyl acetate extract was subjected to gas chromatography-mass spectrometry (GC-MS) analysis. An Agilent GC-MS system equipped with a fused silica capillary tube was used to analyze the components in this active fraction. The data was processed by GC-MSD Chemstation column condition was programmed as column oven temperature 150 °C (4 min)–4 °C/min, temperature of inject port 250 °C and detector port 280 °C (Roy et al. 2006). The peaks of the gas chromatography were subjected to mass-spectral analysis. The spectra were analyzed from the available library data. NIST MS search (version 2.0; included with NIST’02 mass spectral library, Agilent p/n G1033A)
Purification of the anticandidal protein from CFS of MAD08 by ammonium sulfate precipitation
The anticandidal protein from the CFS was precipitated by ammonium sulfate at 80% saturation at 4 °C. The protein precipitated was collected by centrifugation at 8,000×g for 20 min at 4 °C (Eppendorf) and dissolved in phosphate buffer of pH 7.0. This aliquot was dialyzed against the same buffer overnight under refrigerated conditions. The anticandidal activity of the protein precipitated was determined as previously described.
Determination of minimal inhibitory concentration and minimal bactericidal concentrations
The minimal inhibitory concentration (MIC) was determined by the broth dilution method. The multidrug resistant Escherichia coli PC1 and Staphylococcus epidermidis PC8 were used for the determination of MIC and minimal bactericidal concentrations (MBC). The 96-well microtiter plates were filled with 0.1 ml of active ethyl acetate extract prepared in Mueller–Hinton broth. The microtiter plates were incubated at 37 °C for 24 h. In every microtiter plate, one row was set for control (without active compound) and standard (nystatin and nalidixic acid). After incubation, the OD was read at 610 nm using an enzyme-linked immunosorbent assay reader. MICs were recorded as the lowest concentrations inhibiting visible growth. To measure the MBC, the MIC cultures were plated on Mueller–Hinton agar with 5% lysed horse blood and incubated them for 24 h at 37 °C. A reduction of at least 90% of the colonies, compared with the culture of the initial inoculum of the strain, was regarded as evidence of bactericidal activity. When the ratio of MBC/MIC is ≤2, the active extract was considered as bactericidal otherwise as bacteriostatic. If the ratio is ≥16, the fractions were considered as ineffective.
Hemolytic property of ethyl acetate extract
Hemolytic activity was detected using blood agar plates with 5% (v/v) human blood. Plates were examined for hemolysis after incubation at 37 °C for 24 h. The plates were inspected for zone of clearance around the colony.
Emulsifying property of ethyl acetate extract
Emulsification activity was measured according to the method of Cooper and Goldenberg (1987). To 4 ml of the ethyl acetate extract, 4 ml of n-hexadecane was added and vortexed at a high speed for 2 min, and the resultant mixture was allowed to stand for 24 h. the E24 was defined as the height of the emulsion layer divided by the total height and expressed as percentage.
Results
Isolation of actinomycetes
Based on the colony morphology and stability in subculturing, 11 heterotrophic actinomycetes were isolated from the marine sponge D. nigra on different culture media. Among these 11 isolates, four were obtained on marine sponge agar, followed by actinomycetes isolation agar (two), starch casein agar (two), marine agar (two), and Emerson agar (one). The actinomycetes isolates were named as Marine Actinomycetes Depository (MAD01–MAD011) and deposited in Marine Bioprospecting Laboratory, Department of Microbiology, Bharathidasan University (India).
Antagonistic potential of actinomycetes isolates
Of the 11 actinomycetes obtained from the sponge D. nigra, four isolates (27%) showed potential antagonistic activity against the panel of clinical pathogens mentioned in Table 1. Figure 1 depicts the antagonistic potential of the actinomycetes strains. MAD08 needs special citation as the isolate showed 100% activity against all the pathogenic microbial strains tested. Moreover, MAD08 showed anticandidal activity as well against 26 Candida strains tested.
Characterization of antagonistic producer MAD08
Based on the biochemical and physiological characteristics, the isolate MAD08 was identified as Nocardiopsis dassonvillei. MAD08 was a gram-negative filamentous bacterium that was found to be catalase, oxidase, methyl red, and Voges–Proskauer negative. The strain did not utilize citrate. The strain was characterized as an eminent producer of urease. The strain grew well at various concentration of NaCl ranging from 1% to 5% (w/v) with optimum growth at 2% NaCl. The strain grew well between temperatures of 27 and 50 °C, with an optimum at 30 °C. The cultural characteristics of the isolate was determined using various media including peptone yeast iron agar, glucose asparagine agar, glycerol asparagine agar, oat meal agar, starch casein agar, sucrose nitrate agar, nutrient agar, and yeast malt agar. The isolate was grown on the media used except sucrose nitrate agar and yeast malt agar. The substrate mycelium was characterized as spreading, translucent, dull brownish yellow, and dark yellow. The aerial mycelium was characterized as dense, muddy, creamy, white, and cotton white.
Molecular characterization
The genomic DNA isolated and subjected to 16S rRNA amplification for the identification of the potent antagonistic producer. PCR product of length 1,200 bp was purified using the GeneiPure™ quick PCR purification kit and sequenced by Sigma–Aldrich. Taxonomic affiliation of the 16S rRNA sequences of the isolate MAD08 was retrieved from classifier program of Ribosomal Database Project II release 10.0 (http://rdp.cme.msu.edu/). The 16S rRNA sequence of the isolate was blasted using megablast tool of GenBank (http://www.ncbi.nlm.nih.gov/). This revealed that the isolate was a N. dassonvillei. Representative of maximum homologous (98–99%) sequences of each isolate were obtained from seqmatch programme of RDPII and were used for the construction of phylogenetic affiliation (Fig. 2). The isolate showed a unique cluster with N. dassonvillei JPL-3. Among the 30 closest neighboring strains of NCBI-BLASTn used in the phyogenetic analysis, 24 strains were Nocardiopsis (Fig. 3). In particular, four strains were N. dassonvillei with >98% similarity. Therefore, the strain MAD08 might have the characteristics of N. dassonvillei. It revealed that “N. dassonvillei MAD08” has probable nestering with many number of these Nocardiopsis groups.
The UPGMA tree of 20 closest type cultures retrieved from SeqMatch program of Ribosomal Database Project II. The optimal tree with the sum of branch length = 5.633 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option)
The evolutionary history was inferred using the NJ-bootstrap method. The optimal tree with the sum of branch length = 5.53 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 828 positions in the final dataset
Antimicrobial activity of MAD08
The CFS of MAD08 showed highest activity against five gram-positive (Staphylococcus aureus MTCC 2940, S. aureus MTCC 96, Micrococcus luteus MTCC 106, Rhodococcus rhodochrous MTCC 265, Streptomyces MTCC 5) and six gram-negative bacteria (Vibrio cholerae PC6, Pseudomonas aeuroginosa PC3, E. coli PC1, P. aeuroginosa MTCC 2453, E. coli MTCC 2939, E. coli MTCC 739). The antibiogram showed positive skewness towards gram-negative bacteria. The CFS of MAD08 showed highest activity against C. albicans FC1 and C. albicans MTCC 227. Simultaneously the CFS showed moderate activity against 24 Candida strains (TC1–TC9, TC12–TC14, and TC17–TC28). These results envisaged that a potent anticandidal compound can be isolated and purified from the CFS of MAD08.
Optimization of culture conditions and quantification of antimicrobial activities in liquid media
The production of antimicrobial compound was correlated with the growth curve of MAD08. The maximum production was observed during the late log phase (onset of stationary phase). The antimicrobial compound production appeared after 72 h of growth in 168 h batch fermentation. This production remained to be stable between 96 and 144 h and then decreased drastically after 144 h of incubation with maximum antimicrobial compound production at 120 h. It was found that supplementation of carbon sources had no direct effect on the antimicrobial production by Nocardiopsis sp MAD08 when media. Among the ten nitrogen sources used, the presence of oat meal induced antimicrobial compound production by MAD08 (Fig. 4). The supplementation of oat meal to the production medium resulted in antimicrobial activity against 89%, 90%, and 88% of gram-negative, gram-positive, and Candida sp. respectively. Under submerged fermentation conditions, maximum antimicrobial production was attained at 30 °C, pH 6.0, and with 2% NaCl supplementation. At optimum conditions for antimicrobial production, the CFS showed amylase, protease, lipase, and cellulase activity of 120, 93, 13, and 50 U/mg protein, respectively. These enzymes are nutritional enzymes required for the growth of MAD08. These hydrolytic enzymes convert complex carbon and nitrogen sources into simpler and easily available forms for the isolate MAD08 and, thus, facilitating its optimal growth and antimicrobial production.
Effect of nitrogen source (Oat meal) on the production of antimicrobials in Nocardiopsis sp MAD08. Different nitrogen sources including beef extract (BE), meat extract (ME), peptone(PE), ammonium sulfate (AS), sodium nitrate (SN), oat meal extract (OAT), corn meal (CORN), UREA, casein were used in the optimization process. Among these, maximum antimicrobial protein production was observed in medium supplemented with the oat meal. The antimicrobial activity was presented as area of halo in mm2 produced by the antimicrobial compound
Purification of antibacterial fraction
The extraction of antimicrobial compound from the supernatant with equal volumes of ethyl acetate resulted in antimicrobial activity against 90% and 95% of gram-positive and gram-negative organisms, respectively. The crude extract obtained by organic solvent extraction showed no activity against any of the Candida isolates used in the present study. The TLC pattern of ethyl acetate extract showed a mixture of compounds. The FT-IR spectrum exhibited absorption bands at 2,924 and 2,854, which indicated a  and at 1,741.88–1,377.47 cm–1 indicating a double bond. The compounds in the active crude extract were analyzed by GC-MS. A total of 26 peaks were observed with retention times as mentioned in Table 2. The retention time, molecular weight and the bioactivity of the compounds corresponding to the 26 peaks are presented in Table 2. Among the 26 peaks, 11 compounds including acetic acid butyl ester (116 g/mol), ethanol,2-(octyloxy−) (174 g/mol), oxalic acid allyl nonyl ester (256 g/mol), butylated hydroxytoluene (220 g/mol), 2-isopropyl-5-methyl-1-heptanol (172 g/mol), cyclohexanecarboxylic acid hexyl ester (212 g/mol), diethyl phthalate (222 g/mol), pentadecanal (226 g/mol), 1-tridecanol (200 g/mol), 9-octadecenal (266 g/mol), 9-octadecenamide, (Z) (281 g/mol) were determined to have antimicrobial activity.
and at 1,741.88–1,377.47 cm–1 indicating a double bond. The compounds in the active crude extract were analyzed by GC-MS. A total of 26 peaks were observed with retention times as mentioned in Table 2. The retention time, molecular weight and the bioactivity of the compounds corresponding to the 26 peaks are presented in Table 2. Among the 26 peaks, 11 compounds including acetic acid butyl ester (116 g/mol), ethanol,2-(octyloxy−) (174 g/mol), oxalic acid allyl nonyl ester (256 g/mol), butylated hydroxytoluene (220 g/mol), 2-isopropyl-5-methyl-1-heptanol (172 g/mol), cyclohexanecarboxylic acid hexyl ester (212 g/mol), diethyl phthalate (222 g/mol), pentadecanal (226 g/mol), 1-tridecanol (200 g/mol), 9-octadecenal (266 g/mol), 9-octadecenamide, (Z) (281 g/mol) were determined to have antimicrobial activity.
Purification of anticandidal protein
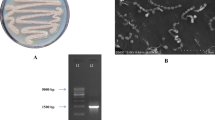
Although the CFS showed anticandidal activity, the extraction with different organic solvents did not yielded anticandidal bioactive principle. Considering the possibility that the bioactivity could be due to an extracellular protein, ammonium sulfate precipitation was carried out serially using different concentration of ammonium sulfate starting from 20% to 90% saturation. As predicted and expected, an anticandidal bioactive protein was obtained at 80% saturation. The bioactive protein was found to be active against 90% of Candida strains tested. A prominent activity was observed against C. albicans FC1. The molecular weight of the partially purified bioactive protein was 87.12 KDa as determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and activity staining (Fig. 5).
Mechanism of antibiosis
The active antimicrobial compound of MAD08 was tested against E. coli PC1 and S. epidermidis PC8 in order to calculate the minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC). The MIC and MBC values are presented in Table 3. The antibiotic compound from MAD08 exhibited antimicrobial activity that was comparable with the activity of chloramphenicol (positive control) against E. coli PC1 and S. epidermidis PC8. Among the organisms tested both E. coli PC1 and S. epidermidis PC8 was highly sensitive to the metabolites of MAD08. MIC values of the active extract in the range of 300–600 µg/ml were observed against the tested bacteria. The bioactive compound was found to be bactericidal for E. coli and S. aureus (MBC/MIC ≤2). The positive control, chloromphenicol showed MIC values ranging from 600–1200 μg/ml.
It was hypothesized that promotion of active compounds from MAD08 for use in clinical trials, the compounds needs to be non-hemolytic. This consideration was confirmed by incubating the bioactive extract on blood agar plates for 18 h at 27 °C. Since, the bioactive compound was non-hemolytic, the compound can be used for the development of antimicrobials for human use. The probable mechanism of action of active ethyl acetate extract is through the formation of surface active compounds known as biosurfactants. The bioactive ethyl acetate extract showed an emulsification activity (E24) of 50% envisaging that the antimicrobial activity is due to surface active compounds.
Discussion
It has already been envisaged that marine sponges are microbial fermenters, which provide exciting new avenues in marine microbiology and biotechnology (Hentschel et al. 2006). Considering the diversity of sponge associated marine microorganisms, the sponges are elucidated as depositories of marine microbial diversity. These microbes are being evaluated as sustainable source of bioactives, biosynthetic genes, and novel structural leads (Hentschel et al. 2003, 2006; Fieseler et al. 2004). Sponge-associated bacteria showed potential antagonistic activity against fouling bacteria, which ultimately envisaged the possible epibiotic defense of host sponge (Thakur et al. 2004; Kanagasabhapathy et al. 2005). In the present study, actinomycetes isolates from the marine sponge D. nigra was screened for inhibitory interactions with pathogenic strains. Among the isolates, MAD08 showed broad spectrum of antagonistic potential against all the pathogens including Candida strains tested. It is a known fact that gram-negative bacteria are the most prominent pathogens compared to gram-positive bacteria. Thus, the present study brings out a new insight towards the development of antimicrobials against gram-negative bacteria from N. dassonvillei MAD08.
The antimicrobial production under submerged fermentation conditions of a batch culture was optimized for enhanced production. In the batch culture, the antimicrobial production was decreased drastically after 144 h of incubation with maximum antimicrobial compound production at 120 h. This drastic loss in antimicrobial activity after 144 h may be due to the degradation of the compound (Gross and Morell 1971) or due to the adsorption of the compound on the surface of the producer (De Vuyst and Vandamme 1992). It was reported that different carbon sources have an influence on the production of secondary metabolites. But controversially, there was no production of antimicrobial compound by Nocardiopsis sp. MAD08 when media was supplemented with different carbon sources at a concentration of 1% (w/v). In contrast with the present findings, maximal production of antimicrobial compound by Streptomyces strain US80 and Streptomyces sp. TN 97 strain was obtained in a medium supplemented with glucose and glycerol or fructose, respectively (Fguira et al. 2005; Mehdi et al. 2006). In the previous work, the addition of sucrose to the production medium favored the production of nisin antibiotic by Lactobacillus lactis (Vessoni penna and Moraes 2002). The advantage of the isolate MAD08 over the other antimicrobial producers is that it does not require any additional carbon sources for its growth and/or antimicrobial compound production, which envisaged the usefulness of the strain as a cost effective one for the production of antimicrobial compound in industrial level.
The supplementation of oat meal (nitrogen source) to the production medium resulted in enhanced antimicrobial activity. The production of antimicrobial compound in the media supplemented with oat meal can be correlated to the presence of maximum growth in the supplemented media over the non-supplemented media. Thus, the supplementation of nitrogen sources to the production media was found to be an essential factor for the production of antimicrobial compounds. Similar results on nisin production were obtained in a complex medium supplemented with cotton seed meal as nitrogen source (De Vuyst and Vandamme 1993). The maximum bacteriocin like substance by Bacillus sp. strain P34 was obtained in a medium supplemented with soybean protein, but no antimicrobial activity was obtained by cultivation on fish meal, feather meal, whey, and grape waste (Motta and Brandelli 2008). Thus, the production of antibiotics in a nitrogen source supplemented media depends and varies along with the requirement of nitrogen source by the organism for its growth and production. It was found that the CFS of MAD08 showed amylase, protease, lipase, and cellulase activities. These enzymes are nutritional enzymes required for the growth of MAD08. It is likely that these hydrolytic enzymes provide a favorable source of nutrients for bacterial growth in addition to provide a suitable environment for replication and survival. Organisms capable of producing all these extracellular enzymes are infrequent in the environment (Dixit and Pant 2000).
The extraction of antimicrobial compound from the CFS with equal volumes of ethyl acetate resulted in antimicrobial activity against bacteria but no activity against Candida isolates. These findings envisaged that the extraction method had definite effect on the isolation of bioactive principles. It has also been reported that organic solvents always provides a higher efficiency in extracting compounds for antimicrobial activities compared to water based methods (Masuda et al. 1997; Lima-Filho et al. 2002). In the present study, the TLC fraction of ethyl acetate extract showed a mixture of compounds in Fourier transform infrared and GC-MS analysis. Claeson and Sunnesson (2005) analyzed many volatile compounds including benzyl alcohol and phenyl ethyl alcohol from S. albidoflavus grown on tryptone glucose medium by GC-MS. A bioactive compound dibutyl phthalate has been reported in culture broth of S. albidoflavus by GC-MS (Roy et al. 2006). Narayana et al. (2008) analyzed benzyl alcohol, phenyl ethyl alcohol, and 2H-1, 4 benzoxazin-3(4H)-one from Streptomyces sp. ANU6277 by GC-MS analysis. In the present study, the presence of (Z)-9-octadecenamide and 9,12-octadecadienoic acid methyl ester, the two prominent antimicrobial compounds, was observed by GC-MS analysis. Antimicrobial extract of Dendrilla rosea showed the presence of three main components including oxacycloheptadec-8-en-2-one (Ambrettolide), a polyketide; 9,12-octadecadienoic acid methyl ester, a diterpene and (Z)-9-octadecenamide, the amide of oleic acid, which is a fatty acid derivative (Haan et al., in press).
In the present study, possible mechanism of antibiosis including bacteriostatic or bactericidal, hemolytic activity, and surfactin production were explored. It was found that the active compounds were non-hemolytic, bactericidal, and surfactive molecules. Considering the surfactive property, the bioactive compound may interact with the interfaces and affect the adhesion of pathogenic bacteria to host tissues. Unless a pathogen is introduced directly into the tissues, the first step in initiation of infection is usually adherence or attachment of the pathogen to some surfaces of the host. Due to surfactant property of the active ethyl acetate fraction, the first step of infection was suggested to be blocked by bioactive compound, thus, proving the active compound to be as an efficient candidate for the treatment of infectious diseases at earlier stages. Gottenbos et al. (2001) demonstrated that positively charged biomaterial surfaces exert an antimicrobial effect on adhering gram-negative bacteria but not on gram-positive bacteria. Several biosurfactants have shown antimicrobial action against bacteria, fungi, algae, and viruses. Potentially broad classes of antimicrobials that target adhesion are called “pilicides.” Bicyclic 2-pyridones and N-substituted amino acid derivatives have been shown to competitively inhibit binding of chaperones to pilin subunits by surface plasmon resonance (Svensson 2001). In vitro, bicyclic 2-pyridones have also been shown to inhibit hemagglutination and biofilm formation in laboratory and clinical E. coli strains, and ex vivo, they have been shown to inhibit adhesion of the bacteria to bladder carcinoma cells by 90% (Pinkner 2006).
In conclusion, the strain Nocardiopsis MAD08 showed a broad range of antibacterial and anticandidal activity. The antibacterial activity was assigned to the presence of 11 compounds and the anticandidal activity to a single protein. Thus, to our knowledge, this is the first strain that express both organic solvent (antibacterial) and water soluble (antifungal) antimicrobial compounds. A judicial and effective combination of these bioactive strategies might, in the future, lead the way towards large-scale profitable production of antimicrobials from the marine actinomycetes N. dassonvillei MAD08.
References
Altschul SF, Gise W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment tool. J Mol Biol 215:403–410
Altschul SF, Thomas LM, Alejandro AS, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Argoudelis A, Baczyneky DL, Duo MT, Laborde AI, Sabek OK, Truesdel SK, Shillidag FB (1987) In vitro studies of arginomycin and its biodegradation products. J Antibio 40:750–760
Bernfield P (1955) Amylase, alpha and beta. Methods of Enzymol 1:149–158
Black RE, Brown KD, Becker S, Yunus M (1982) Longitudinal Studies of infectious diseases and physical growth of children in rural Bangladesh. Am J Epidemiol 115:305–314
Claeson AS, Sunnesson AL (2005) Identification using versatile sampling and analytical methods of volatile compounds from S. albidoflavus grown on four humid building materials and one synthetic medium. Indoor Air 15:41–47
Cooper DG, Goldenberg BG (1987) Surface active agents from two Bacillus species. Appl Environ Microbiol 53:224–229
De Vuyst L, Vandamme EJ (1992) Influence of the carbon source on nisin production in Lactococcus lactis subsp. lactis batch fermentations. J Gen Microbiol 138:571–578
De Vuyst L, Vandamme EJ (1993) Influence of the phosphorus and nitrogen source on nisin production in Lactococcus lactis subsp. lactis batch fermentations using a complex medium. Appl Microbiol Biotechnol 40:17–22
Dienstag J, Nue HC (1972) In vitro studies of tobramycin and aminoglycoside antibiotic. J Antimicrob Ag Chem 1:41–45
Dixit VS, Pant A (2000) Hydrocarbon degradation and protease production by Nocardiopsis sp. NCIM 5124. Lett Appl Microbiol 30:67–69
Fenical W, Baden D, Burg M, de Goyet CV, Grimes JD, Katz M, Marcus NH, Pomponi S, Rhines P, Tester P, Vena J (1999) Marine-derived pharmaceuticals and related bioactive compounds. In: Fenical W (ed) From Monsoons to Microbes:Understanding the Ocean’s Role in Human Health. National Academic Press, Washington, DC
Fguira LFB, Fotso S, Mehdi RBA, Mellouli L, Laatsch H (2005) Purification and structure elucidation of antifungal and antibacterial activities of newly isolated Streptomyces sp. strain US80. Res Microbiol 156:341–347
Fieseler L, Horn M, Wagner M, Hentschel U (2004) Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl Environ Microbiol 70:3724–3732
Forbes BA, Sahm DF, Weissfeld AS, Trevino EA (1990) Methods for testing antimicrobial effectiveness. In: Baron EJ et al. (eds) Bailey and Scott’ Diagnostic Microbiology. Mosby Co, St Louis, Missouri
Gottenbos B, Grijpma DW, Van der Mei HC, Jan Feijen, Busscher HJ (2001) Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 48:7–13
Gross E, Morell JL (1971) The structure of nisin. J Am Chem Soc 93:4634–4635
Haan R, Heng RNY, Poynter S (in press) Extraction and analysis of bioactive agents from Tasmanian marine organisms. Journal of undergraduate science and Technology
Hentschel U, Fieseler L, Wehrl M, Gernert C, Steinert M, Hacker J, Horn M (2003) Microbial diversity of marine sponges. Prog Mol Subcell Biol 37:59–88
Hentschel U, Usher KM, Taylor MW (2006) Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55(2):167–77
Kanagasabhapathy M, Sasaki H, Nakajima K, Nagata K, Nagata S (2005) Inhibitory Activities of surface associated bacteria isolated from the marine sponge Pseudoceratina purpurea. Microbes Environ 20:178–185
Lechevalier HA (1989) The actinomycetes III. A practical guide to generic identification of actinomycetes. In: Williams ST et al. (eds) Bergey’s Manual Systematic Bacteriology. Volume 4. Williams & Wilkins, pp. 2344–2347
Lima-Filho JVM, Carvalho AFFU, Freitas SM (2002) Antibacterial activity of extracts of six macroalgae from the Northeastern Brazillian coast. Brazilian J Microbiol 33:311–333
Manachini PL, Fartina MG, Parini C (1988) Thermostable alkaline protease produced by Bacillus thermoruber a new species of Bacillus. Appl Microbiol Biotechnol 28:409–413
Masuda M, Abe T, Sato S (1997) Diversity of halogenated secondary metabolites in the red algae Laurencia nipponica (Rhodomelaceae Ceramiales). J Phycol 33:196–208
Mehdi RBA, Sioud S, Fguira LFB, Bejar S, Mellouli L (2006) Purification and structure determination of four newly isolated Streptomyces sp. TN97 strain. Process Biochem 41:1506–1513
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem 31:426–428
Motta AS, Brandelli A (2008) Evaluation of environmental conditions for production of bacteriocin-like substance by Bacillus sp. strain P34. World J Microbiol Biotechnol 24:641–646
Narayana JP, Prabhakar P, Vijayalakshmi M, Venkateswarlu Y, Krishna SJ (2008) Study of bioactive compounds from Streptomyces sp. ANU 6277. Pol J Microbiol 57:35–39
Ohnishi K, Yoshida Y, Sekiguchi J (1994) Lipase production of Aspergillus oryzae. J Ferment Bioeng 77:490–495
Pinkner JS (2006) Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc Natl Acad Sci U S A 103:17897–17902
Roy RN, Laskar S, Sen SK (2006) Dibutyl phthalate the bioactive compound produced by S. albidoflavus 321.2. Microbiol Res 161:121–126
Selvin J, Soniya J, Asha KRT, Manjusha WA, Sangeetha VS, Jayaseema DM, Antony MC, Vinitha DAJ (2004) Antibacterial potential of antagonistic Streptomyces sp. isolated from the marine sponge Dendrilla nigra. FEMS Microbiol Ecol 50:117–122
Svensson A (2001) Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli. ChemBioChem 2:915–918
Thakur NL, Anil AC, Müller WEG (2004) Culturable epibacteria of the marine sponge Ircinia fusca: temporal variations and their possible role in the epibacterial defense of the host. Aqua Microb Ecol 37:295–304
Vessonipenna TC, Moraes DA (2002) Optimisation of nisin prodcution by Lactococcus lactis. Applied Biochem Biotech 98–100:775–789
Waksman SA, Woodruff HB (1940) Bacteriostatic and bactericidal substance produced by a soil Actinomyces. Proc Soc Ex Biol Med 45:609–614
Walsh JA, Warren KS (1974) Selective primary healthcare: an interim strategy for disease control in developing countries. N Engl J Med 301:367–370
Acknowledgements
RG is thankful to Council of Scienctific and Industrial Research (CSIR) for Senior Research Fellowship. SS and TRR are thankful to Ministry of Earth Sciences (MoES) for the award of Junior Research Fellowship in MoES funded research project. JS is thankful to CSIR, New Delhi and MoES, New Delhi for research grants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Selvin, J., Shanmughapriya, S., Gandhimathi, R. et al. Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetes Nocardiopsis dassonvillei MAD08. Appl Microbiol Biotechnol 83, 435–445 (2009). https://doi.org/10.1007/s00253-009-1878-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-1878-y