Abstract
Seeds are known to harbor diverse microorganisms offering protective effects on them with the prospects of quick root colonization at germination, selective recruitment as endophytes, and possible vertical transmission. The study was undertaken to assess the gross seed-internal bacterial community in tomato and to confirm if spore-forming Firmicutes constituted major seed endophytes adopting cultivation versus molecular approach on surface-sterilized seeds. Testing the initial seed wash solutions of “Arka Vikas” and “Arka Abha” cultivars showed > 1000 bacterial cfu per dry seed, largely Bacillus spp. Tissue homogenates from surface-disinfected seeds did not show any cultivable bacteria on enriched media for 1–2 weeks, while 16S rRNA V3-V4 taxonomic profiling revealed a huge bacterial diversity (10–16 phyla per cultivar). Proteobacteria formed the dominant phylum (65.7–69.6% OTUs) followed by Firmicutes, Actinobacteria, Bacteroidetes, and a notable share of Euryarchaeota (1.1–3.1%). Five more phyla appeared common to both cultivars in minor shares (Acidobacteria, Planctomycetes, Chloroflexi, Spirochaetes, Verrucomicrobia) with the ten phyla together constituting 99.6–99.9% OTUs. Class level and family level, the cultivars displayed elevated bacterial diversity, but similar taxonomic profiles. Arka Vikas and Arka Abha showed 114 and 107 genera, respectively, with 63 common genera constituting 96–97% OTUs. Psychrobacter formed the dominant genus. Bacillus and related genera constituted only negligible OTU share (0.16–0.28%). KEGG functional analysis showed metabolism as the major bacterial community role. One-month-old in vitro seedlings showed the activation of some originally uncultivable bacteria uninfluenced by the OTU share. The study reveals a high diversity of cultivation-recalcitrant endophytic bacteria prevailing in tomato seeds with possible vertical transmission and significant roles in plant biology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endophytic bacteria are generally known to be recruited by plants from the surrounding environment, primarily from soil through roots [17, 38]. Seed-associated bacteria often formed a topic of considerable interest for plant biologists on account of their protective effects on seeds and the prospects of quick plant colonization at germination [37, 54]. At variance from the previous understanding about the plant acquisition of endophytic bacteria from soil or the atmosphere, seed transmission of endophytic bacteria is now considered an important entry route [4, 12, 49]. Germinating seeds release a body of exudates to the immediate surroundings, the spermosphere, which stimulate seed surface microflora and the rhizospheric organisms with their build-up and potential seedling internal colonization [36, 49]. Further, vertical transmission of endophytes from one generation to the next is a major topic of interest among plant biologists [40, 52, 54]. Past studies on seed-associated endophytes relied mostly on cultivation employing surface-sterilized seeds which often showed Proteobacteria as the commonest phylum followed by Firmicutes, Actinobacteria, and occasional Bacteriodetes [37, 49]. Genus level, members of Proteobacteria such as Burkholderia, Enterobacter, Agrobacterium, Pantoea, Pseudomonas, and Firmicutes including Bacillus and Paenibacillus were often reported as seed-associated bacteria [12, 36, 37].
Tomato is a major vegetable crop world over for fresh market as well as for processing [5]. Seed-associated bacteria and in particular endophytic bacteria in tomato have been topics of significance to researchers towards their exploitation in agriculture. Xu et al. [55] studied Bacillus sp. associated with tomato seeds and documented five species, some with growth promotion ability. Cultivation-based studies on seedling root-associated endophytic bacteria in different Ralstonia solanacearum-resistant and susceptible tomato cultivars displayed more diverse organisms in resistant cultivars with higher biocontrol potential suggesting the possibility of seed transmission of beneficial endophytes [50]. Cultivation versus 16S rRNA gene-mediated taxonomic profiling on two tomato cultivars brought out more diversity with the latter approach exhibiting a dominance of Proteobacteria, while cultivation-based study showed Firmicutes (Bacillus and Paenibacillus spp.) as common associates [29]. Assessing the seed transmission of bacteria in two successive generations of tomato through cultivation and molecular profiling, Bergna et al. [5] documented vertical transmission of different bacteria including B. aryabhattai and B. nakamurai.
Cultivation-reliant studies on seed endophytes often showed Bacillus spp. and other related genera of spore formers such as Paenibacillus as common seed associates including rice [32], wheat [18], maize [7, 19], cucurbits [24], alfalfa [30], quinoa [39], and even tomato [5, 29, 55]. In most plant species, including tomato, the embryo is firmly attached to the seed coat and it is hard to exclude the seed coat tissues during direct seed studies. Our recent study employing dry seeds versus surface-sterilized seeds of two tomato cultivars showed Bacillus spp. as the major seed external-associates while the tissue homogenates from surface-sterilized seeds, in vitro germinating seeds, or 10-day-old in vitro grown seedlings on sucrose-minus MS medium did not show any cultivable bacteria [41]. On the other hand, 1-month-old in vitro raised seedlings where the seed coat could be excluded post seed germination exhibited bacterial association in a notable share of seedlings. The associated organisms included mainly Proteobacteria followed by Actinobacteria and Firmicutes, which appeared to arise from the activation of embryo-associated cultivation-recalcitrant endophytic bacteria (CREB) during the course of seedling development. Only one spore-former was isolated in this study which proved to be an unusual Gram-negative Bacillus sp. showing high 16S rRNA gene sequence homology to B. phocheonensis [41].
A major share of endophytic bacteria is now known to be non-amenable to cultivation. Next-generation sequencing (NGS)-based studies on roots [17, 28] and more recently on shoot tissues [43, 47] have brought out a huge diversity of bacterial endophytes. In these studies, Proteobacteria formed the commonest phylum followed by Actinobacteria, Firmicutes, and Bacteriodetes along with minor shares of several phyla including some candidate phyla with no cultured relatives and a low share of Archaea. Cultivation-independent approaches to study the seed-associated/seed endophytic bacteria based on NGS such as 16S rRNA gene amplicon profiling and metagenomics [1, 5, 56], or alternate molecular methods such as 16S rDNA clone library sequencing and PhyloChip analysis [10], have shown considerable bacterial diversity. An NGS-based study would be the way out to unravel the CREB community in tomato and to avoid the chances of wrong depiction of bacteria that possibly escape surface sterilization treatment as seed-transmitted organisms. The present study was undertaken to get an estimate of the gross seed-internal bacterial diversity in tomato and to assess if spore-forming Firmicutes constituted major seed endophytes adopting cultivation versus 16S rRNA gene profiling.
Materials and Methods
Seed Material and Surface Sterilization with Bacterial Monitoring
Seeds of tomato (Solanum lycopersicum L.) cultivars “Arka Vikas” and “Arka Abha” procured from the vegetable seed store of ICAR-Indian Institute of Horticultural Research, Bengaluru, stored in polythene bags or aluminum lined seed storage bags at 16 °C or ambient temperature were employed. Post procurement, the seed lots were stored at 4 °C. The initial study employed a freshly procured lot (2018 batch) in aluminum-lined bags to assess the external bacterial load and the effectiveness of surface sterilization, done essentially as per Shaik and Thomas [41]. This involved six steps of washes in autoclaved and filter-sterilized (0.2 μm) distilled water (FDW) with one step wash in FDW containing 0.01% Tween-20 (Tw-FDW). In the first treatment (T1), 100 seeds were vortexed at top speed in 10 ml Tw-FDW for 5 min in a 15-ml centrifuge tube followed by five repeated vortex-washings in FDW. In the second treatment (T2), the first three washes were made in FDW with one Tw-FDW wash (fourth step) followed by two ADW washes to assess if Tween-20 had any adverse effect on the bacterial cfu. Each wash solution was monitored for bacterial cfu by spotting 10 μl lots on nutrient agar (NA) and the total surface bacteria removed was assessed through the SP-SDS [48] of the pooled wash solutions employing four replications. The seeds were next treated with 70% ethanol (1 min), rinsed once in FDW, and agitated in NaOCl (4% available chlorine) for 10 min followed by six rinses in FDW with wash solution monitoring through spotting (10 μl) on NA. Effectiveness of surface sterilization was assessed by plating up to 400 μl last wash solution through spotting-and-tilt-spreading (SATS) [46]. After 10 min treatment with 2% Na2S2O3 to remove any residual chloramines, the seeds were imprinted on NA for 10 min. The NA plates were sealed in sterile polypropylene (PP) bags pre- and post-plating to avoid external organisms. Bacterial cfu was assessed after 1–7 days of incubation (first day at 36 °C and thereafter at room temperature to provide a trigger to the initial growth). The major colony types from the two cultivars in the seed wash solutions were taken up for identification through 16S rRNA sequence megablast homology analysis to the type strains of cultured organisms at the National Centre for Biotechnological Information (NCBI) as described by Upreti and Thomas [50]. Based on the outcome, additional seed lots were tested for the bacterial load and the effectiveness of surface sterilization as above.
Cultivation-Based Testing of Surface-Sterilized Seeds
Surface-sterilized seeds (100 nos. from 2018 lot) with the monitoring for effective disinfection were homogenized aseptically in a mortar which warranted an extended grinding for 8–10 min. The seed tissue homogenate (STH; 100 μl FDW per seed) was tested on NA and trypticase soy agar (TSA) through SP-SDS and 400 μl original STH through SATS. Plates were observed for 7 days (37 °C for one night and thereafter 30 °C) for any colony growths.
In the next trial, three batches of Arka Vikas and Arka Abha seeds (2016, 2017, and 2018 lots stored at 4 °C) were taken through surface sterilization with the monitoring for external bacteria. STH from 100 seeds dispersed in 10 ml FDW was tested through SATS on NA employing three replications at 100 to 103 dilutions, and through SP-SDS of 100 to 105 dilutions on TSA for each sample. The remaining STH was stored at − 20 °C.
DNA Isolation fromSeed Tissue Homogenate (STH)
STH from the three seed lots of Arka Vikas and Arka Abha stored at − 20 °C were employed for DNA isolation after 1 week by which time no cultivable bacteria were detected on NA/ TSA. The thawed STH was mixed well and after 1 h static incubation at 4 °C in 15 ml centrifuge tubes, the upper 4 ml clearer portion was used for bacterial DNA isolation. The sample was spun down at high speed (18,000×g) for 2 min. The supernatant part was spun down again, and the pooled pellet from the two rounds of spinning was used for DNA isolation employing the PowerFood® (PF) microbial DNA isolation kit (MO BIO Laboratories Inc., CA) adopting the extended lysis protocol (10 min at 70 °C) with two replicate columns based on the earlier observations with banana tissues [43]. DNA was eluted in 60 μl buffer per column and subjected to quality assessments. The three DNA samples were pooled in equal amounts for each cultivar, and the consolidated sample was employed for further studies.
PCR-Based Testing of STH DNA for Uncultivable Bacteria
Since no bacterial growth was observed from the STH on NA/TSA, a PCR was undertaken targeting uncultivable bacteria using 16S rRNA gene primers 27F and 1492R on the pooled DNA from the three seed lots as per Thomas et al. [47]. Considering that the above primers could amplify plant plastid and mitochondrial 16S rRNA gene, PCR employing bacterial class-specific primers targeting α, β, and γ-Proteobacteria, Actinobacteria, and Firmicutes was undertaken as per [42, 47]. PCR reactions were set up in 20 μl volume and 10 μl samples were tested in 1% agarose gel.
16S rRNA V3-V4 Taxonomic Profiling of Seed Endophytic Bacterial Biome
The pooled DNA sample from the three seed lots was employed for the cultivation-independent assessment of seed internal bacteria after monitoring the STH applied plates for 2 weeks with no cultivable bacterial detection on NA/TSA. The DNA samples after the quality checks were submitted to M/s Eurofins Genomics India Pvt. Ltd., Bengaluru (www.eurofinsgenomics.co.in) for the 16S rRNA metagene taxonomic profiling targeting the V3–V4 hypervariable region, undertaken as per the standard Illumina protocol. This involved the standard Illumina library preparation with 16S rRNA F (5′-GCCTACGGGNGGCWGCAG-3′) and 16S rRNA R (5′-ACTACHVGGTATCTAATCC-3′) primers and 2 × 300 MiSeq sequencing, PE read stitching, and quality filtration steps as described elsewhere [43, 45]. QIIME-based OTU picking and taxonomy assignments were made with the Greengenes as the reference database. Since the majority of reads matched to the plastid and mitochondrial OTUs, a second round QIIME analysis was adopted excluding the reads that corresponded to plant OTUs [43, 47].
Activation of CREB from In Vitro Seedlings and Their Identification
Based on the documentation of high taxonomic diversity in the two tomato cultivars and the previous observation on the possibility of activation of CREB in aseptically grown tomato seedlings [41], Arka Vikas and Arka Abha seedlings (50 each) were raised singly in glass culture tubes (150 × 25 mm) on Murashige and Skoog [35] basal medium (without sucrose) employing the surface-sterilized seeds from the 2018 lot with wash solution monitoring as described above. Post-germination, the detached seed coat was removed to exclude bacteria associated with the seed-external tissues [41]. Seedlings were observed for any visible microbial growth, and after 4 weeks, the apparently clean seedlings were indexed for any covert bacteria by transferring a small bit of culture medium to NA [41]. The collar region from 1-month-old seedlings showing bacterial association was pooled, homogenized in a mortar, and applied on NA as per SP-SDS and SATS. Distinct colony types were selected, subjected to three rounds of single-colony purification, and identified as described earlier.
Culturing the Surface-Sterilized Seeds on Enriched Medium
Considering that the application of 100 μl original STH corresponded to the monitoring of only one seed, direct planting of surface-disinfected seeds on enriched bacteriological medium (NA) was undertaken. About 100 seeds were taken through surface disinfection with wash solution monitoring and seed imprinting, and 96 seeds of each cultivar were cultured directly on NA (16 seeds per plate). After one night at 36 °C, the plates were observed for 1 week at room temperature for any bacterial growth from the seeds.
Functional Annotation of Seed Whole Endophytic Bacterial Biome
In order to get an insight on the functional role of CREB community in tomato seeds, PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) software [27] version 1.1.0 was employed on 16S rRNA amplicon sequence OTUs derived after filtering out the plant sequences as outlined previously [45]. Greengenes picked OTUs were used to generate metagenomic data with PICRUSt application. The resultant Biological Observation Matrix (BIOM) table was used to derive the relative Kyoto Encyclopedia of Genes and Genomes (KEGG) abundance information [23].
Sequence Data
The NGS data generated in this study have been deposited with NCBI SRA with the BioProject ID PRJNA552773 (Tomato seed tissue endophytic microbiome analysis; TaxID: 1297885) with the BioSample accession nos. SAMN12217409 for EG-MG01 Arka Vikas seed endophytic microbiome and SAMN12217410 for EG.MG02 Arka Abha seed endophytic microbiome. 16S rRNA sequence data of the bacterial strains isolated from the in vitro seedling cultures have been deposited with the NCBI GenBank (acc. nos. MN134002 to MN134011 for the seed wash isolates and MN134014 to MN134029 for the in vitro seedling-derived isolates).
Results
Seed Surface Sterilization and External Bacteria
Fresh seeds of 2018 lot showed abundant external bacteria which came down with repeated seed washings, particularly employing Tw-FDW in the first washing step (T1) than the fourth step (T2) (Fig. 1a). The wash solutions showed predominantly Bacillus spp. with no obvious differences in population structure between the two treatments but for the better bacterial release in T1. SP-SDS on the pooled wash solutions (6×) showed 425 and 162.5 bacterial cfu removed per seed for Arka Vikas in T1 and T2, respectively, and 1262 and 1125 cfu, respectively, for Arka Abha seeds. These numbers are expected to be higher considering the fast-spreading Bacillus spp. colonies impairing with the cfu counts beyond day 1. Following the ethanol step, a few Bacillus colonies were observed, but none appeared after the NaOCl treatment. Two predominant colony types were present in both Arka Vikas and Arka Abha which were identified as Bacillus safensis and B. subtilis with occasional B. circulans and B. megaterium in Arka Vikas and B. cereus, B. megaterium, B. aerius, and Paenibacillus favisporus in Arka Abha (Table 1).
Monitoring of six serial seed wash solutions of raw tomato seeds for external bacteria through spotting 10 μl solution per row on NA (a), testing the pooled 6× wash solution for gross bacterial cfu through SP-SDS of three dilutions (b), with both a and b displaying abundant Bacillus colonies, and testing the tissue homogenate from surface-sterilized seeds for cultivable bacteria through SP-SDS of five serial dilutions (c). Av and Ab indicate cultivar “Arka Vikas” and “Arka Abha,” respectively; T1, and T2: Tw-FDW in the first and the fourth washing step, respectively. (Note: the white spots in c correspond to micro-tip marks on the nutrient medium)
Trials employing different seed lots displayed varying amounts of initial bacterial load. The surface sterilization approach proved mostly effective, but for occasional bacterial cfu observed, linked to the seed lot, initial bacterial load, or reduction in the efficacy of NaOCl subsequent to the initial container opening. Therefore, monitoring the wash solutions and seed imprinting appeared essential in critical experiments to ensure the freedom from all external organisms. Invariably, Bacillus sp. formed the common seed external associate.
Cultivation-Based Testing of STH
The original STH from surface-sterilized 2018 seed lot or its serial dilutions did not show any bacterial cfu on NA or TSA for 1 week but for some grainy spots at the original sample applied spots (Fig. 1b). Re-streaking of these raised spots to NA or TSA did not elicit any new colony growths, indicating the absence of cultivable bacteria with the effectively surface-sterilized seeds.
The three seed lots employed in the next trial displayed some differences in the external bacterial load as per the wash solution monitoring (Supplementary Fig. S1). Here again, testing the wash solutions post-ethanol treatment displayed a few Bacillus colonies but none after the NaOCl step. The STH from these three seed lots also did not show any active bacterial colony growths on NA/TSA in both the cultivars during the 2-week period of observation.
DNA Isolation and PCR-Based Testing of STH-DNA for Uncultivable Bacteria
STH from the three seed lots (2016, 2017, and 2018) yielded 4.6, 5.9, and 10.8 ng DNA μl−1 in Arka Vikas while Arka Abha seed lots gave 4.3, 12.9, and 6.8 DNA μl−1, respectively. The pooled DNA from the three samples yielded a good amplicon with the bacterial primers 27F and 1492R (Fig. 2a). Class-specific PCR showed a prominent amplicon against γ-Proteobacteria and Firmicutes followed by β-Proteobacteria (Fig. 2b). Relatively less intense bands were observed with the Actinobacteria and α-Proteobacteria primers. The results suggested the presence of different bacterial classes in the STH in an uncultivable form.
PCR screening for uncultivable bacteria employing 16S rRNA gene primers 27F and 1492R showing the amplicon of ~1500 bp (a), and class-specific PCR (b) targeting α-Proteobacteria (lanes 1, 2 for Av and Ab), β-Proteobacteria (3, 4), γ-Proteobacteria (5, 6), Actinobacteria (7, 8,) and Firmicutes (9, 10) with the expected amplicon sizes of 674, 342, 496, 640, and 483 bp, respectively
16S rRNA V3-V4 Taxonomic Profiling of Seed Endophytic Bacterial Biome
The pooled DNA from the three seed lots gave rise to good amplicon libraries with a mean fragment size of 574 and 583 bp for Arka Vikas and Arka Abha, respectively (Supplementary Table S1). QIIME round-1 analysis showed 43.1 and 30.3% reads in Arka Vikas matching to chloroplast and mitochondria, respectively. The corresponding values for Arka Abha were 44.2 and 30.3%, which indicated a high masking effect due to the plant sequences.
QIIME round-2 analysis excluding the reads corresponding to plant OTUs revealed high amounts of endophytic bacterial taxonomic diversity in both the cultivars. Phylum level, Arka Vikas showed 12 constituents with the dominance of Proteobacteria (69.6%) followed by Firmicutes, Actinobacteria, and Bacteroidetes with minor shares of eight other phyla, while Arka Abha displayed altogether 16 constituents with the four major phyla accounting for 96.7% OTUs (Table 2). A notable share of phylum Euryarchaeota was associated with the surface-sterilized seeds (1.06 and 3.06%, respectively). The two cultivars shared ten common phyla including Acidobacteria, Planctomycetes, Chloroflexi, Spirochaetes, and Verrucomicrobia comprising 99.9% OTUs in Arka Vikas and 99.6% OTUs in Arka Abha. Arka Vikas displayed two genotype-specific phyla while Arka Abha showed six distinct phyla.
Class level, γ-Proteobacteria formed the dominant constituent in both the cultivars accounting for 98.1 and 97.4% of Proteobacteria (Table 2). Bacilli constituted the next major class followed by Actinobacteria, Flavobacteriia and Sphingobacteriia under Eubacteria, and Methanobacteria under Euryarchaeota. Order level, Pseudomonadales formed the major component in both the cultivars (66.9 and 62.5%, respectively) followed by Bacillales, Actinomycetales, and Flavobacteriales (data not shown).
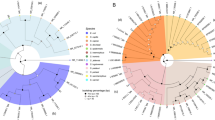
Family level distribution showed high taxonomic diversity in both the cultivars with similar taxonomic profiles with respect to the major constituents (Fig. 3). Arka Vikas displayed altogether 84 families and Arka Abha 78 families with 50 families shared between them, which in turn constituted 97.8 and 97.9% of the OTUs, respectively. Moraxellaceae (Gammaproteobacteria) formed the single largest constituent with 66.7 and 62.3% OTUs in Arka Vikas and Arka Abha, respectively. Other major families under Eubacteria included Planococcaceae (Bacilli), Nocardiaceae (Actinobacteria), Flavobacteriaceae (Bacteroidetes), and Methanobacteriaceae under Archaea (Methanobacteria).
Genus level too, the two cultivars displayed very identical taxonomic profiles considering the OTUs assigned to the dominant genera (Fig. 4). Psychrobacter (Moraxellaceae; Gammaproteobacteria) formed the single dominant constituent (52.2% in Arka Vikas and 49.3% in Arka Abha). Other notable genera (≥ 1.0% OTUs) included unclassified Moraxellaceae, unclassified Enterobacteriaceae (Class Gammaproteobacteria), Planococcus and Planomicrobium (Bacilli), Arthrobacter, Rhodococcus (Actinobacteria), Gillisia (Flavobacteriia), Pedobacter (Sphingobacteriia), and most significantly, Methanobrevibacter, a Methanobacterium under the phylum Euryarchaeota, all of them present in both the cultivars. Altogether, Arka Vikas showed 114 genera and Arka Abha 107 genera (gross 160 genera) and 63 common genera shared between them which constituted 97.0% of the OTUs in Arka Vikas and 96.9% OTUs in Arka Abha (Supplementary Dataset 1). It was significant to note that Bacillus spp. which appeared as the commonest seed external organism formed only a minor share (0.164% OTUs in Arka Vikas and 0.163% in Arka Abha). Other spore-forming Firmicutes included Lactobacillus (0.081 and 0.401% OTUs, respectively) and Brevibacillus in Arka Vkias (0.123%).
A few organisms could be defined at the species level based on the V3-V4 data. Among these, Psychrobacter pulmonis (52.2 and 49.3% OTUs in Arka Vikas and Arka Abha, respectively), formed the major constituent. Other notable ones present in both the cultivars included Planococcus pelagicus (9.1–9.4%), Rhodococcus fascians (0.69–0.98%), Arthrobacter nitroguajacolicus (0.041–0.41%), Paracoccus marcusii (0.20–0.29%), Prevotella copri (0.20–0.37), and Acinetobacter lwoffii (0.12–0.16%). Arka Vikas showed a Shannon Alpha diversity index of 3.41 (193 Observed OTUs) and Arka Abha 3.62 (199 OTUs). The rarefaction curve (Fig. 5) indicated that a large share of the species diversity remained uncovered. The Krona for the two seed samples showed identical bacterial profiles (Supplementary Fig. S2).
Activation of CREB to Cultivation from In Vitro Seedlings
A small share of seedling cultures on sucrose-minus MS medium (12 and 10%, in Arka Vikas and Arka Abha, respectively) showed fungal colony growth after seed germination despite the extensive surface sterilization treatments. The remaining seedlings appeared clean. Culture-indexing of 4-week-old seedlings showed 6/44 cultures of Arka Vikas and 7/45 cultures of Arka Abha as index-positive on NA displaying bacterial growth. Tissue homogenates from the pooled collar region of these 1-month-old index-positive seedlings showed 105 cfu g−1 tissue in Arka Vikas and 106 cfu g−1 in Arka Abha. Nine distinct organisms were isolated from Arka Vikas (Table 3) which included the genera Kosakonia, Pseudomonas, Acinetobacter (γ-Proteobacteria), Sphingomonas, Methylobacterium (α- Proteobacteria), Staphylococcus, Bacillus (Firmicutes), Micrococcus (Actinobacteria), and Elizabethkingia (Flavobacteriia; Bacteroidetes). Six strains were obtained from Arka Abha which included Enterobacter, Pantoea (γ-Proteobacteria), Paracoccus (α-Proteobacteria), Rhodococcus, Micrococcus (Actinobacteria), and Staphylococcus (Firmicutes).
Culturing the Surface-Sterilized Seeds on Enriched Medium
About 7% seeds of Arka Vikas and 10% from Arka Abha from the 2018 seed lot displayed bacterial colony outgrowth from the seeds cultured on NA after 2–4 days (Fig. 6). In all the instances, the organism appeared to be spore-forming Bacillus sp. (morphologically B. safensis and B. subtilis) or related genera as per colony characteristics, Gram reaction, and spore formation. The number of seeds displaying bacterial growth varied with different seed lots and batches, low in some instances and more in others, particularly for the newer seed lots.
Functional Annotation of Seed Endophytic Bacterial Biome
Considering the prevalence of diverse bacteria in dry seeds in an uncultivable state, it was only feasible to get the information on the gross functional roles of endophytic bacterial community based on the known functions of organisms as per the relative OTUs. The abundance of different KEGG pathways as per the PICRUSt analysis showed very similar profiles for both the cultivars at K01 level (Fig. 7). Metabolism (63% abundance) formed the major functional role for the endophytic bacterial biome in both the cultivars followed by genetic information processing (20%), environmental information processing (13%), cellular processes, and organismal systems.
Discussion
This study deciphers the high diversity of endophytic bacteria associated with the surface sterilized seeds in two tomato cultivars through cultivation and the cultivation-independent approach of 16S rRNA gene V3–V4 amplicon profiling. While the former approach showed Bacillus spp. as the common seed externally associated organism, no cultivable bacteria were detected in the seed tissue homogenates after the surface-disinfection treatments. The cultivation-independent approach, on the other hand, brought to light the prevalence of a high diversity of CREB in line with the observations on aseptically grown seedlings of Miscanthus sp. [10] and the NGS approach applied on the seeds of melon, tomato, and rice [1, 5, 56]. This study has brought out several rare eubacterial phyla (Acidobacteria, Planctomycetes, Chloroflexi, Spirochaetes, Verrucomicrobia, TM7, Cyanobacteria, Thermotogae, [Thermi], Tenericutes, Synergistetes, Fusobacteria) and archaeal phyla (Euryarchaeota, Crenarchaeota) as seed internally associated bacteria in tomato, in addition to the four phyla of Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes commonly documented as endophytes. Bergna et al. [5] documented seed-transmissible Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes Planctomycetes, Acidobacteria, Chloroflexi, Cyanobacteria, and some other minor OTUs in tomato seeds. In this study, we laid the emphasis on whole bacterial diversity including very low abundant constituents to a get a holistic picture of seed endophytic bacterial biome. Although the 16S rRNA gene profiling was confined to just two DNA samples, they were derived from three different seed lots and thus a proper representative of the two cultivars.
The NGS-based observations in this study, together with the isolation of different bacteria from 4-week-old in vitro seedlings endorsed the prevalence of CREB in tomato seeds and the possibility of their activation to cultivable form with the seedling growth [41], additionally illuminating their high taxonomic diversity. The seedling growth conditions in vitro enabled to study the seed-activated endophytic bacteria protected from external organisms unlike in field studies. It was significant to document a high share of genera under Proteobacteria (Kosakonia, Sphingomonas, Pseudomonas, Acinetobacter, Methylobacterium, Enterobacter, Paracoccus, Pantoea) among the seedling-activated cultivable organisms besides some members of Actinobacteria (Micrococcus, Rhodococcus), Firmicutes (Bacillus, Staphylococcus), and Bacteroidetes (Elizabethkingia). All the activated organisms in this study except for Kosakonia, Enterobacter, and Elizabethkingia spp. were documented in the molecular profiling study. Endorsed by the rarefaction curve, this suggested that a greater diversity of seed endophytes than that presented here remain to be uncovered. The activated organisms in this study and in the earlier study [41] were not those with high OTU abundance but that with relatively low share of reads. Further, the activated organisms in the two studies did not share much commonality except for a few bacterial species such as Kosakonia oryzendophytica, Sphingomonas paucimobilis, and Micrococcus aloeverae indicating that the activation is perhaps a random event [42, 44]. In this study, the terms uncultivable bacteria and CREB have been used between them; there is some difference. Uncultivable means that the organisms are not culturable with the routine cultivation-based approaches. Cultivation-recalcitrant implies that the organisms are not readily cultured or their cultivation requirements are not understood, and they become cultivable under suitable conditions for their growth.
Past seed microbiology studies relied mainly on cultivation where Bacillus spp. and the related genera of spore-formers were documented as common seed associated bacteria/seed endophytes based on their isolation from surface-sterilized seeds or so-derived seedlings [10, 22, 32, 36, 37]. On the other hand, the documentation of a high share of Bacillus spp. in seed wash solutions, low OTU abundance of Bacillus during V3–V4 profiling, and the low share of spore-formers among the seedling-activated bacteria in this study suggested that Bacillus spp. are major seed external colonizers and less abundant embryo-colonizers. On the other hand, Bacillus spp. has been documented microscopically inside the seeds of grape [8] and melon [15] through 16S rRNA gene fluorescent in situ hybridization. Bergna et al. [5] isolated B. aryabhattai and B. nakamurai from tomato seeds in successive generations suggesting their vertical transmission. The frequent isolation of Bacillus and Paenibacillus spp. in cultivation-based studies of surface-sterilized seeds could be attributable to the hardy spores that possibly escape the surface disinfection as observed with the seeds directly cultured on NA. It would warrant the use of substantial amounts of tissue homogenates from seeds to capture such low abundant organisms.
One limitation in the studies on seed endophytes is the difficulty in separating the seed coat from the embryo to assess the true vertical transmission as is the case with maze, brassica, Miscanthus sp. etc. The seed coat constituted an integral component in such cultivation-based studies with Bacillus spp. forming the major isolates [7, 10, 18, 24, 29, 30]. In some instances where the seed external tissues were removed aseptically, the spore-formers formed a small or less common constituent [16, 53]. Even the cultivation-independent approach on merely surface-washed seeds showed mainly Proteobacteria with very low shares of Bacillus spp. [1]. The present study does not exclude spore-forming bacteria as a vertically transmissible organism but identifies it as a relatively minor constituent among the wide range of seed endophytic bacterial flora. Alternatively, the spore-formers may be present in a form which is recalcitrant to DNA recovery. Spore-formation is a possible mechanism by which the organisms remain dormant and survive within the desiccated seeds under nutrient-limiting conditions. This may also explain the failure to culture the organisms from dry seeds as also observed with Miscanthus sp. [10] and tomato [41]. Both the cultivation-independent approach and the cultivation-based observations on in vitro seedlings showed Proteobacteria as the dominant phylum inside the seeds. Although Firmicutes formed a significant phylum in the 16S rRNA profiling, this was contributed by the non-spore formers such as Planococcus and Planomicrobium.
Molecular profiling approach has shown Psychrobacter (Moraxellaceae) as the commonest seed internal bacterial genus in both tomato cultivars with the maximum homology to P. pulmonis which was first isolated from the lungs of lambs associated with respiratory distress [51]. This suggests the ability of an organism to be in different ecosystems with possible strain differences. Psychrobacter is a genus of Gram-negative bacteria found in a variety of marine and terrestrial environments besides a common food spoilage organism [6]. The genus also comprises of psychrophilic or psychrotolerant organisms with the ability to grow between − 10 and 42 °C [25]. The three seed lots employed in the 16S rRNA gene amplicon profiling were refrigeration stored (4 °C) for 6–24 months. It is not clear whether this formed a contributing factor for the high abundance of this genus. Psychrobacter has been documented as a dominant operational phylogenetic unit in cultivation-independent study covering roots, stems, and the rhizosphere in the halophyte Arthrocnemum [34]. Psychrobacter has also been documented as a common root endophyte of Pennisetum sinese in cultivation-independent studies [11]. The other dominant genera as per 16S rRNA profiling included unclassified Moraxellaceae and Planococcus sp. (Bacilli). Planococcus sp. also constitutes psychrophilic organisms such as P. maitriensis from Antarctica [2]. The endophytic bacterial population could be dynamic over the lifetime of the plant, seed stages, or depending on the environmental condition [10].
For the organisms to be transmitted vertically from one generation to the next, colonization of embryo, or the endosperm is a requisite. Considering that the NGS approach in this study involved the microflora from both the embryo and the seed coat, it was not feasible to make a clear assessment of the embryo-transmissible organisms. Observations employing in vitro grown seedlings of tomato here and in the earlier study [41] where the seed coat part could be segregated post-seed germination indicated embryo colonization and vertical transmission of different bacterial genera. Preliminary observations with watermelon seeds, where the embryo could be studied distinctly from the seed-coat tissues, endorse the observations of Bacillus as a dominant external organism and the high diversity of embryo-colonizing organisms as CREB (Thomas, unpublished data). Currently, the knowledge about the transmission of endophytes and seed microbiome assembly remains incomplete [38]. Association of diverse bacteria with the pollen of different plant species [31] and bacterial introduction inside seeds through stigmatic bacterial application [33] have been demonstrated. Microscopic documentation of bacterial colonization in the seed embryos and the aseptically germinated seeds [8, 10, 15] suggest vertical transmission of bacterial endophytes. It is not clear as of now whether the seed bacteria are transmitted continuously from generation to generation without exiting the plant [5, 12].
The composition and structure of seed microbiota of different plant species are yet to be characterized [9, 38]. Very few investigations have addressed the origins and routes of colonization of seeds by bacteria. Bacterial endophytes can possibly get into the seeds either from the vegetative parts through the vascular connections, through gametes, through the apical meristem getting converted to reproductive structures or the direct transfer from mature fruits to seeds [37, 49]. Several endophytic bacteria associated with flowers are considered to colonize the developing ovules and ultimately end up in fruits and seeds [37]. Bacillus and Pseudomonas spp. are particularly prevalent in cucurbit fruits, especially within the seed cavity from where they could possibly enter the seeds [13, 14, 24]. Once established within the endophytic seed microbiome, these bacteria can transfer naturally to seedlings during germination and promote the seedling growth [5, 33, 52]. The seed external organisms like Bacillus spp. also have the potential for easy colonization of seedlings at germination [22, 37] which applies to the normal farmers’ practice of sowing seeds without surface sterilization.
A number of studies have shown substantial amounts of variability existing in the seed microbiome with different plant species, geographical locations, and soils [3, 20, 26] and between genotypes within a species [19]. In contrast, a conserved core seed microbiome within a plant species is known to exist [10, 20, 56]. This study on two tomato cultivars which were grown in the same locality and soil conditions displayed very identical taxonomic diversity and functional profiles. Whether this is a reflection of the bacterial population acquired from the soil cannot be discounted now [10]. Both seed and soil apparently contribute to the endophyte population of new seedling [21]. The population structure could vary depending on the seedling growth phase and further plant development. It calls for more targeted research on understanding the dynamics of seed-associated bacteria and their integration to the developing plant versus the soil community interactions. Further, studies specifically targeting the embryo tissues are needed to make a clear assessment of the vertical transmission of endophytic bacteria from one generation to the next which is envisaged in the future. The presence of diverse bacteria and their prevalence in an uncultivable form came in the way of deciphering the functional roles of different organisms. As of now, the functional information on the uncultivable bacterial community as a whole was only feasible which indicated the role of the community in metabolic pathways. There is also scope for exploiting the activated endophytic bacteria as seed inoculants in agriculture. It warrants more research to reach a proper conclusion on such useful organisms.
In summary, the cultivation-independent analysis on surface-sterilized seeds with the rigid monitoring of the efficacy of surface-disinfection treatments in this study helped in unraveling the enormous endophytic bacterial diversity in tomato seeds. The bacterial community inhabits the seeds in a cultivation-recalcitrant state with the chances of their activation to cultivable form with the seedling growth under in vitro conditions which may also apply to the normal seedlings. The seed integrally associated organisms bear the potential for quick seedling colonization at germination with the prospects of vertical transmission to the next generation. Bacillus spp. and related genera of spore-forming bacteria appeared to be dominant seed external colonizers with very limited share detected in molecular analysis on surface sterilized seeds. The high share of seed-associated Bacillus spp. documented in different cultivation-based studies could be attributed to the hardy spores that possibly escape the surface sterilization and their rapid multiplication on enriched medium.
Abbreviations
- CREB:
-
Cultivation-recalcitrant endophytic bacteria
- FDW:
-
Filter-sterilized autoclaved distilled water
- MS medium:
-
Murashige and Skoog medium
- NA:
-
Nutrient agar
- NGS:
-
Next Generation Sequencing
- PP bags:
-
Polypropylene bags
- SATS:
-
Spotting-and-tilt-spreading
- SP-SDS:
-
Single plate-serial dilution spotting
- STH:
-
Seed tissue homogenate
- TSA:
-
Trypticase soy agar
References
Adam E, Bernhart M, Müller H, Winkler J, Berg G (2018) The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil 422:35–49
Alam SI, Singh L, Dube S, Reddy GSN, Shivaji S (2003) Psychrophilic planococcus maitriensis sp. nov. from Antarctica. Syst Appl Microbiol 26:505–510
Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, Hunault G, Simoneau P, Jacques MA (2015) Emergence shapes the structure of the seed microbiota. Appl Environ Microbiol 81:1257–1266
Berg G, Raaijmakers JM (2018) Saving seed microbiomes. ISME J 12:1167–1170
Bergna A, Cernava T, Rändler M, Grosch R, Zachow C, Berg (2019) Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes J 2:183–193
Betts G (2006) Other spoilage bacteria. In: Blackburn CW (ed) Food spoilage microorganisms. Woodhead Publishing Series in Food Science, Technology and Nutrition, Sawston, Cambridge, pp 668–693
Bodhankar S, Grover M, Hemanth S, Reddy G, Rasul S, Yadav SK, Desai S, Mallappa M, Mandapaka M, Srinivasarao C (2017) Maize seed endophytic bacteria: dominance of antagonistic, lytic enzyme-producing Bacillus spp. 3 Biotech 7:232
Compant S, Mitter B, Colli-Mull JG, Gangl H, Sessitsch A (2011) Endophytes of grapevine flowers, berries and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb Ecol 62:188–197
Compant S, Saikkonen K, Mitter B, Campisano A, Mercado-Blanco J (2016) Editorial special issue: soil, plants and endophytes. Plant Soil 405:1–11
Cope-Selby N, Cookson A, Squance M, Donnison I, Flavell R, Farrar K (2017) Endophytic bacteria in Miscanthus seed: implications for germination, vertical inheritance of endophytes, plant evolution and breeding. GCB Bioenergy 9:57–77
Deng ZS, Zhang BC, Qi XY, Sun ZH, He XL, Liu YZ, Li J, Chen KK, Lin ZX (2019) Root-associated endophytic bacterial community composition of Pennisetum sinese from four representative provinces in China. Microorganisms 7:47
Frank AC, Saldierna Guzmán JP, Shay JE (2017) Transmission of bacterial endophytes. Microorganisms 5:70
Fürnkranz M, Lukesch B, Mueller H, Huss H, Grube M, Berg G (2012) Microbial diversity inside pumpkins: microhabitat specific communities display a high antagonistic potential against phytopathogens. Microb Ecol 63:418–428
Glassner H, Zchori-Fein E, Compant S, Sessitsch A, Katzir N, Portnoy V, Yaron S (2015) Characterization of endophytic bacteria from cucurbit fruits with potential benefits to agriculture in melons (Cucumis melo L.). FEMS Microbiol Ecol 91:fiv074
Glassner H, Zchori-Fein E, Yaron S, Sessitsch A, Sauer U, Compant S (2018) Bacterial niches inside seeds of Cucumis melo L. Plant Soil 422:101–113
Hardoim PR, Hardoim CC, van Overbeek LS, van Elsas JD (2012) Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7:e30438
Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320
Herrera SD, Grossi C, Zawoznik M, Groppa MD (2016) Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol Res 186:37–43
Johnston-Monje D, Raizada MN (2011) Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One 6:1–22
Johnston-Monje D, Mousa WK, Lazarovits G, Raizada MN (2014) Impact of swapping soils on the endophytic bacterial communities of pre-domesticated, ancient and modern maize. BMC Plant Biol 14:233
Johnston-Monje D, Lundberg DS, Lazarovits G, Reis VM, Raizada MN (2016) Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 405:337–355
Kandel SL, Joubert PM, Doty SL (2017) Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32(suppl 1):D277–D280
Khalaf EM, Raizada MN (2016) Taxonomic and functional diversity of cultured seed associated microbes of the cucurbit family. BMC Microbiol 16:131
Kim SJ, Shin SC, Hong SG, Lee YM, Choi I-G, Park H (2012) Genome sequence of a novel member of the genus psychrobacter isolated from antarctic soil. J Bacteriol 194:2403
Klaedtke S, Jacques MA, Raggi L, Préveaux A, Bonneau S, Negri V, Chable V, Barret M (2016) Terroir is a key driver of seed-associated microbial assemblages. Environ Microbiol 18:1792–1804
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Liu H, Carvalhais LC, Crawford M, Singh E, Dennis PG, Pieterse CM, Schenk PM (2017) Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front Microbiol 8:2552
López S, Pastorino G, Franco M, Medina R, Lucentini C, Saparrat M, Balatti P (2018a) Microbial endophytes that live within the seeds of two tomato hybrids cultivated in Argentina. Agronomy 8:136
López JL, Alvarez F, Principe A, Salas ME, Lozano MJ, Draghi WO, Jofré E, Lagares A (2018b) Isolation, taxonomic analysis, and phenotypic characterization of bacterial endophytes present in alfalfa (Medicago sativa) seeds. J Biotechnol 267:55–62
Manirajan BA, Ratering S, Rusch V, Schwiertz A, Geissler-Plaum R, Cardinale M, Schnell S (2016) Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ Microbiol 18:5161–5174
Mano H, Tanaka F, Watanabe A, Kaga H, Okunishi S, Morisaki H (2006) Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ 21:86–100
Mitter B, Pfaffenbichler N, Flavell R, Compant S, Antonielli L, Petric A, Berninger T, Naveed M, Sheibani-Tezerji R, von Maltzahn G, Sessitsch A (2017) A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front Microbiol 8:11
Mora-Ruiz MDR, Font-Verdera F, Orfila A, Rita J, Rosselló-Móra R (2016) Endophytic microbial diversity of the halophyte Arthrocnemum macrostachyum across plant compartments. FEMS Microbiol Ecol 92:fiw145
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nelson EB (2004) Microbial dynamics and interactions in the spermosphere. Annu Rev Phytopathol 42:271–309
Nelson EB (2018) The seed microbiome: origins, interactions and impacts. Plant Soil 422:7–34
Nelson EB, Simoneau P, Barret M, Mitter B, Compant S (2018) Editorial special issue: the soil, the seed, the microbes and the plant. Plant Soil 422:1–5
Pitzschke A (2016) Developmental peculiarities and seed-borne endophytes in quinoa: omnipresent, robust bacilli contribute to plant fitness. Front Microbiol 7:2
Shahzad R, Khan AL, Bilal S, Asaf S, Lee IJ (2018) What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Front Plant Sci 9:24
Shaik SP, Thomas P (2019) In vitro activation of seed-transmitted cultivation- recalcitrant endophytic bacteria in tomato and host-endophyte mutualism. Microorganisms 7:132
Thomas P (2011) Intense association of non-culturable endophytic bacteria with antibiotic-cleansed in vitro watermelon and their activation in degenerating cultures. Plant Cell Rep 30:2313–2325
Thomas P, Sekhar AC (2017) Cultivation versus molecular analysis of banana (Musa sp.) shoot-tip tissue reveals enormous diversity of normally uncultivable endophytic bacteria. Microb Ecol 73:885–899
Thomas P, Soly TA (2009) Endophytic bacteria associated with growing shoot tips of banana (Musa sp.) cv. Grand Naine and the affinity of endophytes to the host. Microb Ecol 58:952–964
Thomas P, Agrawal M, Bharathkumar CB (2019) Diverse cellular colonizing endophytic bacteria in field shoots and in vitro cultured papaya with physiological and functional implications. Physiol Plant 166:729–747
Thomas P, Sekhar AC, Mujawar MM (2012) Non-recovery of varying proportions of viable bacteria during spread-plating governed by the extent of spreader usage and proposal for an alternate spotting-spreading approach to maximize the CFU. J Appl Microbiol 113:339–350
Thomas P, Sekhar AC, Pasha SS (2017) High taxonomic diversity of cultivation-recalcitrant endophytic bacteria in grapevine field shoots, their in vitro introduction and unsuspected persistence. Planta 246:879–898
Thomas P, Sekhar AC, Upreti R, Mujawar MM, Pasha SS (2015) Optimization of single plate-serial dilution spotting (SP-SDS) with sample anchoring as an assured method for bacterial and yeast cfu enumeration and single colony isolation from diverse samples. Biotech Rep 8:45–55
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7:40–50
Upreti R, Thomas P (2015) Root-associated bacterial endophytes from Ralstonia solanacearum resistant and susceptible tomato cultivars and their pathogen antagonistic effects. Front Microbiol 6:255
Vela AI, Collins MD, Latre MV, Mateos A, Moreno MA, Hutson R, Dominguez L, Fernandez-Garayzabal JF (2003) Psychrobacter pulmonis sp. nov., isolated from the lungs of lambs. Int J Syst Evol Microbiol 53:415–419
Verma SK, Kharwar RN, White JF (2019) The role of seed-vectored endophytes in seedling development and establishment. Symbiosis 78:107–113
Walitang DI, Kim K, Madhaiyan M, Kim YK, Kang Y, Sa T (2017) Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of rice. BMC Microbiol 17:209
White JF, Kingsley KL, Butterworth S, Brindisi L, Gatei JW, Elmore MT, Verma SK, Yao X, Kowalski KP (2019) Seed-vectored microbes: their roles in improving seedling fitness and competitor plant suppression. In: Verma SK, White Jr JF (eds) Seed endophytes. Springer, Cham, pp 3–20
Xu M, Sheng J, Chen L, Men Y, Gan L, Guo S, Shen L (2014) Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J Microbiol Biotechnol 30:835–845
Zhang J, Zhang C, Yang J, Gao J, Zhao X, Zhao J, Zhao D, Zhang X (2018) Insights into endophytic bacterial community structures of seeds among various Oryza sativa L. rice genotypes. J Plant Growth Regul 38:93–102
Acknowledgments
The NGS and bioinformatics support by M/s Eurofins Genomics India Pvt. Ltd., Bengaluru, is gratefully acknowledged. This study partly formed the component of the Ph.D. thesis of the co-author at the Jain University, Bengaluru, India.
Funding
The study was funded under the ICAR-AMAAS Net-work project “Genomics-mediated taxonomic and functional analysis of endophytic microbiome in horticultural crops and plant-microbe interaction studies” by the ICAR-National Bureau of Agriculturally Important Microorganisms, Mau Nath Bhanjan, Uttar Pradesh, India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Thomas, P., Shaik, S.P. Molecular Profiling on Surface-Disinfected Tomato Seeds Reveals High Diversity of Cultivation-Recalcitrant Endophytic Bacteria with Low Shares of Spore-Forming Firmicutes. Microb Ecol 79, 910–924 (2020). https://doi.org/10.1007/s00248-019-01440-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01440-5