Abstract
The aim of this work was to study the diversity of vaginal lactobacilli in Lebanese women and to evaluate the antagonism, hydrophobicity, and safety characteristics of these strains. This study was performed on samples from 135 women who visited a gynecology clinic in the north of Lebanon, between September 2012 and January 2013. From these samples, 53 different isolates of vaginal lactobacilli were collected from vaginal swabs and identified using biochemical and molecular methods. The use of genotypic Rep-PCR fingerprinting allowed for the organization of these isolates into 23 different groups. Seven of the isolated lactobacilli were antagonistic against the following vaginal pathogens: Gardnerella vaginalis CIP7074T, Staphylococcus aureus ATCC33862, Escherichia coli CIP103982, and Candida albicans ATCC10231. The antagonistic lactobacilli strains were then identified using 16S rDNA sequence. The data of this study show that the antagonistic lactobacilli were non-hemolytic, sensitive to most antibiotic tests, free of plasmid DNA, and exhibited interesting hydrophobicity and autoaggregation properties positioning them as potential candidates for probiotic design.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Probiotics are defined as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” [17]. The safe use of lactobacilli as probiotic agents in the human genitourinary tract dates back to 1915 [36].
The vaginal microbiota has been largely studied as an “ecosystem” whose normal composition helps to protect against invading pathogens responsible of urinary infections or sexually transmitted diseases. Lactobacilli are the predominant species observed in the healthy vaginal ecosystem. Lactobacilli are facultative anaerobic bacteria that convert lactose and other sugars into lactic acid, usually employed in the industry as “starters” in cheese and yogurt making. The associated medical benefits are regularly deciphered and outlined by relevant studies. For the last century, lactobacilli were described as the most important vaginal microorganism of women of childbearing age but their numbers vary considerably dependant on age and among tested populations [43]. In the vaginal microbiota, lactobacilli play a determinant role in the maintenance of a healthy ecosystem through the production of organic acids, bacteriocins, H2O2, and bacteriocin-like inhibitory substances (BLIS). The normal pH of approximately 4.5 is important for maintaining a healthy barrier to the external environment. It has been found that the bactericidal properties exerted by feminine hygiene products may modify the normal vaginal microbiota mentioned before, which is essential for protection against pathogens. The vaginal microbiota comprises Gram negative and Gram positive bacteria, anaerobic bacteria, and uncultured bacteria which play an important role in preterm birth [24]. An aberrant vaginal microbiota is known as the bacterial vaginosis; disease is associated with adverse health conditions and infectious complications. In postpartum women, bacterial vaginosis constitutes a major risk for both the woman’s health and the health of her newborn due to inflammatory complications [27]. Bacterial vaginosis and lower tract infections have shown and association with an increased risk of human immunodeficiency viruses (HIV) acquisition [30, 49]. Treatment of bacterial vaginosis with oral or topically applied antibiotics is often associated with failure and high rates of recurrence. The dominance of lactobacilli in healthy vaginal microbiota and its depletion in bacterial vaginosis has given rise to the concept of oral or vaginal use of probiotic Lactobacillus strains for treatment and prevention of this multispecies infection [32]. Thus, the role of vaginal lactobacilli was established in different reports. Diminution in vaginal lactobacilli count increased transmission of viral infections [10]. Further, more vaginal lactobacilli may prevent premature delivery and illness in newborns [19]. A recent in vitro study established the role of Lactobacillus spp. in suppressing the toxic shock syndrome toxin-1 caused by Staphylococcus aureus [28]. The concept of using lactobacilli as probiotics was even developed and successfully tested in women [32, 44]. The aim of this study was to establish preliminary lactobacilli cartography from a Lebanese cohort, to characterize them at a molecular level and to decipher the principle functions requested for their design as probiotics, especially as antimicrobials continue to fail.
Material and Methods
Target Population
This study was carried out on 135 women visiting a gynecology clinic in the north of Lebanon, between September 2012 and January 2013. A patient history was provided to a gynecologist for each woman. Questions included basic information (e.g., age, sexual activities, urinary or genital symptoms or pathology, drug and antibiotic intake, vaginal intimate washes) as well as questions surrounding pregnancies (i.e., routine pregnancy visits, number of pregnancies, and number of abortions).
Isolation and Identification of Vaginal Lactobacilli
Growth on de Man–Rogosa–Sharpe (MRS) agar was observed only for 43 samples of the 135 analyzed. Isolated colonies were selected and picked for each of the 43 samples. Briefly, the vaginal swab was serial diluted and 1 ml of each dilution was spread onto MRS [12] agar plates (Becton Dickinson-USA), and incubated at 37 °C for 48 h under a 5 % CO2 condition. Morphological and biochemical preliminary tests permitted to select Gram positive bacteria, with a bacilli shape, and without catalase activity. The isolates were then subjected to API and pyrosequencing identifications.
The inoculation of API50 CH strips (Biomérieux-France) was performed as recommended by the manufacturer. After 48 h at 37 °C, the strips were checked for fermentation or non-fermentation of carbohydrates. Species assignment was determined by https://apiweb.biomerieux.com/.
A pyrosequencing of short variable regions such as V1 and V2 regions in the 16S rDNA gene permitted rapid and efficient identification [54]. The pyrosequencing was conducted as described by Tarnberg et al. [54] with some modifications. The V1 and V2 regions of the 16S rDNA were amplified by PCR using primers Bio-pBR5 and pBR-V1, and BiopBR-V2 and RevV2, respectively (Table 1). One of these primers was biotinylated to obtain a biotinylated amplicon. The single-stranded DNA amplicons were prepared semi-automatically with a Vacuum Prep Tool and Vacuum Prep Worktable (Biotage-Sweden). Briefly, a 20-μl aliquot of biotinylated PCR products was immobilized onto 4 μl of streptavidin-coated SepharoseTM High Performance Beads (Amersham Biosciences, UK) with 26 μl of binding buffer, pH 7.6 (10 mM Tris–HCl, 2 M NaCl, 1 mM ethylenediamine tetraacetic acid, 0.1 % Tween 20), and incubated at 65 °C under agitation at 1,400 rpm for at least 5 min. Double-stranded DNA immobilized on Sepharose beads was washed with 70 % ethanol and denatured with 0.2 M NaOH. Unbound single-stranded DNA was washed with 0.1 M Tris–HCl (pH 7.6). The beads carrying single-stranded DNA amplicons were suspended in 38.4 μl of annealing buffer (pH 7.6) (20 mM Tris–acetate, 5 mM Mg-acetate) containing 200 nmol of sequencing primers. The single-stranded DNA was annealed to the sequencing primer at 80 °C for 2 min followed by incubation for 2 min at 20 °C. The single-stranded PCR products were sequenced using the PyroMarkTM Q96 ID System (Biotage, Sweden). To assign the species of these lactobacilli, a blast of the V1 and V2 sequences was performed using the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Genomic Fingerprinting
Total DNA was extracted from each vaginal lactobacilli with the Wizard® Genomic DNA Purification Kit (Promega, USA). Rep-PCR requested only the use of primer 5′- (GTG)5-3′ which is considered the most suitable for clustering lactic acid bacteria and yeasts at the species level [1, 37]. The reaction mixture of 50 μl contain 50 ng of DNA template, 5 μl of primer 5′- (GTG)5- 3 at 20 μM, 1.2 mM of dNTPs, 7 mM of MgCl2, 2.5U of DreamTaq DNA polymerase, and 2× PCR buffer (Thermoscientific, USA). The Rep-PCR program consisted of an initial step of denaturation (4 min at 95 °C), followed up by 29 cycles of denaturation (1 min at 94 °C), annealing (1 min at 40 °C), and extension (8 min at 72 °C). The program was ended by an additional extension step at 72 °C for 16 min. The Rep-PCR products were analyzed on 1 % agarose gel. Electrophoresis was carried out at 100 V for 2 h using 1× Tris–borate–EDTA. The gels were stained with GEL-RED (Biotium, Canada) and visualized by GelDoc (Bio-Rad). The construction of phylogenetic trees was performed using the software DIVERSILAB® (Biomérieux, France).
Inhibition of Pathogens Microorganisms by Vaginal Lactobacilli
The antagonism activity was done by the well diffusion method [53] with some modifications. The vaginal lactobacilli were grown in MRS broth (pH 6.5) inoculated with 1 % of an overnight culture and incubated at 37 °C for 18–24 h. Cells were harvested by centrifugation (8,000×g for 20 min, 4 °C), and the cell-free supernatant was sterilized by filtration through a 0.22-μm Millipore filter. The antimicrobial activity of vaginal lactobacilli was determined against ESBL positive E. coli CIP103982 which was cultured on nutrient broth for 24 h at 37 °C. After this period, 0.1 ml of the bacterial suspension was added to Muller Hinton agar and then kept at 40 °C for 4 h. Then, 6 mm wells were made and filled with 100 μl of supernatant obtained from each isolate. The plates were incubated aerobically for 2 h at 4 °C, and then for 24 h at 37 °C, and finally they were inspected for inhibition zones.
Attempts to characterize the inhibitory substance(s) involved in the aforementioned antagonism were conducted as follows: fresh supernatant was prepared from each putative antagonistic vaginal lactobacilli. The supernatant was (i) heat treated for 10 min at 80 °C, (ii) or treated by different proteases (Sigma-Aldrich, Germany) such as proteinase K (1 mg/ml), trypsin (1 mg/ml), α-chemotrypsin (1 mg/ml), and papain (1 mg/ml), and incubated for 2 h at 37 °C, and (iii) ultimately neutralized with 2 M NaOH (pH 6.5). After each treatment, the samples were assessed against the indicator strain Escherichia coli CIP102982.
The antagonism was extended to an enteric strain of S. aureus (ATCC33862), Gardnerella vaginalis 7074T isolated from vaginal infection and vaginal strain of Candida albicans ATCC10231. Thus, S. aureus ATCC33862 was grown in Brain Heart Infusion (BHI) broth (Bio-Rad, France) for 24 h at 37 °C, under aerobic conditions. One milliliter of 24 h cultured BH broth culture was added to 9 ml of Brain Heart agar (0.8 %), and then the mixture was thoroughly mixed and poured into Petri plates containing a very thick Brain Heart Infusion agar. For C. albicans ATCC10231, the same procedure was used but with the Sabouraud agar (Bio-Rad, France) and broth media instead of BH broth. Notably, G. vaginalis CIP7074T was cultured using Brain Heart broth supplemented with 10 % horse serum and incubated for 48 h at 37 °C under anaerobic using GenBox anaerobic strip (Biomérieux, France).
The inhibition of S. aureus ATCC33862 was followed by epifluorescence microscopy. The growth curve and microscopic observations were performed using a co-culture of S. aureus ATCC33862 and Lactobacillus plantarum CMUL140, which displayed the upmost anti-staphylococcal activity. After inoculation of MRS with L. plantarum CMUL 140, and Brain Heart with S. aureus ATCC33862, the cultures were left for 18 h at 37 °C. After this period, 0.1 ml of each culture was added to 20 ml of Brain Heart containing 5 g/l glucose, allowing a cell concentration of approximately 106 CFU/ml for L. plantarum CMUL140 and 103 CFU/ml for S. aureus ATCC33862. Samples were taken every 2 h during a total incubation period of 10 h at 37 °C. For each sample, the number of bacterial cells with intact cell membranes (viable cells) was determined with a LIVE/DEAD® BacLight Bacterial Viability Kit (Invitrogen, UK). All microscopic observations were made with a Nikon Optiphot-2 epifluorescence microscope equipped with a 100-W mercury lamp and a filter package B2A (Nikon, USA). To assess culturability of S. aureus ATCC33862 and L. plantarum CMUL140, culturable counts were determined throughout these experiments. Samples were serially diluted in physiological water, and dilutions were plated onto Plate Count Agar medium, Baird–Parker agar medium (Becton Dickinson, USA), and de Man Rogosa and Sharpe medium [13]. The counting of colonies was determined after 24 h of incubation at 37 °C.
Molecular Characterization of Antagonistic Vaginal Lactobacilli
The antagonistic strains were molecularly characterized with three independent methods. These methods were PCR-based methods and the primers used in each PCR are indicated in Table 1. The first method consisted in the amplification of a 250-bp Lactobacillus-specific region within the 16S ribosomal gene [15]. The second method was the amplification of 16S-23S region [26]. The third method was the amplification and determination of the 16S rDNA sequence. For this last purpose, the PCR amplicon was excised from 0.6 % agarose gel using MinElute Gel Extraction Kit (Qiagen, Germany) and then cloned into the ρGEM-T Easy vector (Promega, USA). The ligation product was transferred to E. coli JM 109 (Promega, USA) by heat shock treatment. The recombinant plasmids carrying the anticipated DNA amplicons were selected for their resistance to ampicillin (10 mg/ml) (Sigma-Aldrich, Germany). Upon transformation of E. coli JM 109, the competent cells were cultured on Luria–Bertani agar medium (Biokar, France) supplemented with 50 μl of X-Gal at 20 mg/ml, 20 μl of isopropyl β-d-1-thiogalactopyranoside (IPTG) at 100 mM, and ampicillin (10 mg/ml). After 24 h at 37 °C, colonies lacking lactose operon were cultured in Luria–Bertani broth medium containing ampicillin (10 mg/ml). Plasmid extractions were done with GeneJET Plasmid Miniprep Kit (ThermoScientific, USA). A digestion with Eco RI was used to check and validate the genetic construction. The 16S rDNA contained in the recombinant plasmids were sequenced at Eurofins MWG (Germany). The blast of each 16S rDNA sequence was performed in NCBI-Standard Nucleotide BLAST and the Ribosomal Database Project (RDP) databases (rdp.cme.msu.edu/seqmatch). Remarkably, the same procedure and conditions were applied for extraction of plasmids from antagonistic strains.
Antibiotic Susceptibility
The susceptibility of the antagonistic vaginal lactobacilli was assessed against different antibiotics (Masts, UK) such as penicillin G (10 μg), ampicillin (10 μg), cefoxitin (30 μg), cefotaxime (30 μg), cefepime (30 μg), vancomycin (5 μg), ciprofloxacin (5 μg), gentamicin (10 μg), streptomycin (10 μg), kanamycin (10 μg), erythromycin (15 μg), tetracycline (30 μg), cotrimoxazole (30 μg), rifampicin (30 μg), fosfomycin (30 μg), and lincomycin (30 μg). Antibiotic susceptibility assays were carried out on Muller–Hinton Agar (Bio-Rad, France) supplemented with sheep blood (5 %) (Biomérieux, France) and antibiotics were purchased from (MAST, UK). Briefly, 106 CFU/ml of antagonistic vaginal lactobacilli strains was suspended in 0.9 % sodium chloride and the solution was poured into blood agar plates; after 3 min of incubation at room temperature, the excess of solution was removed and the plates were dried for 15 min at 20 °C (room temperature), and then incubated at 37 °C for 24 h, under anaerobic conditions.
Hemolytic Activity
The hemolytic activity of fresh cultures of vaginal lactobacilli was tested by inoculation on blood agar medium containing 5 % (w/v) sheep blood or human blood, 10 μl of 24 h old culture. The plates were incubated for 48 h at 37 °C. After this period, they were examined for hemolytic activity by the observation of clear halos around the colonies.
Autoaggregation
Autoaggregation assays were performed according to Del Re et al. [11]. Bacteria were grown for 18 h at 37 °C in MRS solid or liquid medium. Cells were harvested by centrifugation (5,000×g, 15 min), washed twice with phosphate saline (PBS) buffer (pH = 7.0), and resuspended in their own culture supernatant or in PBS to give a viable counts of 108 CFU/ml. Cell suspensions (4 ml) were mixed by vortexing for 10 s and autoaggregation was determined during 5 h of incubation at room temperature. Every hour, 0.1 ml of the upper suspension was transferred to another tube containing 3.9 ml of PBS and the absorbance (A) was measured at 600 nm. The percentage of autoaggregation was determined using the following formula:
Where A t represents the absorbance at time t = 1, 2, 3, 4, or 5 h and A 0 the absorbance at t = 0. This assay was repeated three independent times.
Hydrophobicity
Microbial adhesion to solvents (MATS) was measured according to Rosenberg et al. [46] with modifications [8]. Briefly, bacteria were harvested in the stationary phase by centrifugation (5,000×g, 15 min), washed twice, and resuspended in phosphate buffer (pH = 7.0) to approximately 108 CFU/ml. The absorbance of the cell suspension was measured at 600 nm (A 0). One milliliter of xylene (Fluka, Germany) was added to 3 ml of cell suspension. After 10 min of preincubation at room temperature, the two-phase system was mixed by vortexing for 2 min. The aqueous phase was removed after 2 h of incubation at room temperature, and its absorbance at 600 nm (A t ) was measured. The percentage of bacterial adhesion to solvent was calculated using the formula given below. This assay was repeated three times.
Where A t represents the absorbance at time t = 1, 2, 3 h and A 0 the absorbance at t = 0. This assay was repeated three times.
Results
Isolation and Identification of Vaginal Lactobacilli
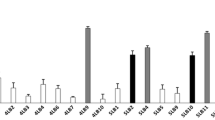
In this study, 53 isolates were obtained from 135 vaginal swab samples of healthy Lebanese women, whose age ranged from 19 to 55. These isolates were obtained by a cultured-based method in MRS medium. Vaginal lactobacilli were phenotypically identified through typical lactobacilli shape, Gram positive staining, and lack of catalase activity. According to the API50 CH test (Table 2), Lactobacillus acidophilus was the most common species in the analyzed samples, followed at equal percentage by L. plantarum and Lactobacillus brevis (Fig. 1a). The API 50 CH method also allowed for the identification of Lactobacillus crispatus, Lactobacillus fermentum, and Lactobacillus jensenii (Table 2).
Molecular identification carried out using pyrosequencing of V1 and V2 variable region in 16S rDNA allowed for the identification of more species. The pyrosequencing analysis revealed that the most common species were Lactobacillus salivarius and L. jensenii, followed at equal percentage by L. acidophilus, L. crispatus, Lactobacillus gasseri, and Lactobacillus helveticus and at lesser extent by Lactobacillus ruminis (n = 2), L. fermentum (n = 1), and L. plantarum (n = 1) (Fig. 1b and Table 2). Further, a model of the program obtained from the V1 pyrosequencing for one Lactobacillus strain is depicted in Fig. 2. The program is clearly indicating that no signal errors was present in the short sequenced fragment (V1–V3). In addition, when necessary, all sequences were manually examined and compared with reference peaks (AATGGC) installed within the dispensation order.
It is noteworthy that the vaginal lactobacilli identification by the routinely API 50CH method was not very accurate as only 13 species were identically identified by the API 50CH and pyrosequencing of 16S rDNA method. After which, all the isolated vaginal lactobacilli were subjected to Rep-PCR (GTG)5 fingerprinting technique for genotypic grouping. Numerical analysis of the (GTG)5-fingerprint band profile resulted in 23 different groups (Fig. 3). A hierarchical clustering tree based on this rep-PCR fingerprint pattern was constructed by the DiversiLab® (Biomérieux-France), the distance cutoff of relatedness was 70 %. Interestingly, the five antagonistic L. gasseri strains were not necessarily contained in the same group except for strains CMUL57 and CMUL99.
Inhibition of Pathogens and Partial Characterization of Inhibitory Substances
The antagonism against E. coli CIP103982 was tested using the well diffusion method [5, 17]. As result, seven isolates designed as CMUL34 (L. gasseri), CMUL54 (L. fermentum), CMUL57 (L. gasseri), CMUL67 (L. acidophilus), CMUL80 (L. gasseri), CMUL99 (L. gasseri), and CMUL140 (L. plantarum) were shown to inhibit the growth of the aforementioned indicator strain. This antagonism was measured by the inhibition zones observed after 24 h at 37 °C. Remarkably, all these antagonistic strains were isolated from healthy women. Furthermore, the supernatants from the aforementioned antagonistic strains were also active against S. aureus ATCC33862, G. vaginalis CIP7074T, and C. albicans ATCC10231 (Fig. 4). To unravel the nature of the inhibitory substance(s) secreted by each antagonistic strain into the growth medium, each cell-free supernatant gathered from each antagonistic strain was submitted to different treatments. Thus, the inhibitory substances were resistant to heat and catalase treatments, but not to pH neutralization.
Interactions Between L. plantarum CMUL140 and S. aureus ATCC33862
The co-culture of S. aureus ATCC33862 and L. plantarum CMUL140 resulted in the inhibition of growth of the pathogen, and in the log reduction of the CFU/milliliter by 1 log after approximately 8 h (Fig. 5a). However, when S. aureus ATCC33862 was grown alone, its concentration has increased from 3.1 log CFU/ml to 6.3 log CFU/ml in 10 h (Fig. 5b). To strengthen this anti-staphylococcal data, epifluorescence microscopy experiments were carried out targeting 8 h aged co-cultures. Thus, the red cells correspond to dead S. aureus ATCC33862 cells, whereas the green correspond to live L. plantarum CMUL140 cells (Fig. 6a). A remarkably bright green fluorescence was observed for the S. aureus ATCC33862 co-cultured with L. plantarum compared with the sample containing only S. aureus ATCC33862 (Fig. 6b).
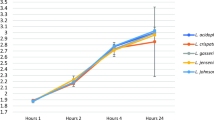
Growth curve of S. aureus ATCC 33862 alone (diamond), S. aureus ATCC 33862 with L. plantarum CMUL140 (square), and L. plantarum CMUL140 in the same co-culture (red) and L. plantarum CMUL140 alone (triangle) over 10 h of incubation. S. aureus ATCC33862 counts were determined on both Plate Count Agar and Baird–Parker agar media and L. plantarum CMUL140 counts were determined on MRS medium at 37 °C
Live/dead fluorescence microscopy of a co-culture between S. aureus ATCC 33862 and L. plantarum CMUL 140 after 8 h of incubation. a Co-cultured, dead cells of S. aureus ATCC 33862, the live cells of L. plantarum CMUL 140, and the bright green fluorescence of live S. aureus ATCC 33862 in co-culture in comparison to the green fluorescence obtained from S. aureus ATCC 33862 cells in monoculture (b)
Molecular Characterization of Antagonistic Vaginal Lactobacilli
The antagonistic strains CMUL34 (L. gasseri), CMUL54 (L. fermentum), CMUL57 (L. gasseri), CMUL67 (L. acidophilus), CMUL80 (L. gasseri), CMUL99 (L. gasseri), and CMUL140 (L. plantarum) were characterized using three independent methods. The first two methods (specific lactobacilli PCR specific and ITS analysis) were used as preliminary diagnostic tools or confirmation of API 50CH identification to achieve the complete 16S rDNA sequencing. The use of specific lactobacilli PCR, as expected, permitted the amplification of 250-bp DNA fragment [18], the 16S-23S polymorphism, gathered two 2 ISR, of about 600 and 800 bp [19], and ultimately the sequencing of the 16S rDNA concluded the molecular identification.
The 16S rDNA sequences obtained from the complete 16S rDNA sequences were deposited into the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) and the accessions numbers are given in Table 3.
Antibiotic Susceptibility and Hemolytic Activity
Fifteen antibiotics from different families were used. The data are indicated in Table 4. All antagonistic vaginal lactobacilli were resistant to fluoroquinolones and fosfomycin, while only two isolates CMUL34 (L. gasseri) and CMUL140 (L. plantarum) were sensitive to SXT. In regard to the resistant phenotype, three isolates CMUL34 (L. gasseri), CMUL54 (L. fermentum), and CMUL67 (L. acidophilus) appeared to be resistant to cefoxitin, and two of them CMUL54 (L. fermentum) and CMUL140 (L plantarum) were resistant to vancomycin. The susceptibility to these antibiotics was determined based on the recommendations of the Antibiogram committee of the French Microbiology Society (http://www.sfm-microbiologie.org/UserFiles/file/CASFM/CASFM_2013.pdf). No hemolytic activity was detected after 48 h for any of these isolates. Importantly, none of these antagonistic strains appeared to contain natural plasmid DNA.
Autoaggregation and Hydrophobicity Assessments
Sedimentation rate was measured over a 5-h period. Three isolates named CMUL57 (L. gasseri), CMUL67 (L. acidophilus), and CMUL140 (L. plantarum) exhibited the highest level of autoaggregation (Table 5). Moreover the MATS method was used to assess the hydrophobic/hydrophilic functions of each antagonistic vaginal lactobacilli. To this end, xylen was used as a hydrophobic solvent to estimate the cell surface properties of the strains. The most hydrophobic strains were CMUL57 (L. gasseri), CMUL67 (L. acidophilus), and CMUL140 (L. plantarum), which interestingly exhibited the upmost level of autoaggregation (Table 5).
Statistical Analysis
All data were the mean of at least three independent experiments. Standard deviation (SD) was calculated and indicated for each test. Statistical comparisons between the different methods were performed by the t test analysis, and the Kappa index was determined with SPSS software version 2011.
Discussion
The aim of this study was to establish the lactobacilli diversity in a Lebanese cohort, to characterize them at the molecular level, and to evaluate the main properties required for making a successful vaginal probiotic, in a time when antimicrobials continue to fail. In this context, the analysis that was performed on swab samples collected from 135 Lebanese women has led to the isolation of 53 vaginal lactobacilli, which were divided up into 23 groups according to the genotyping analysis.
The low number of isolates obtained from the reasonably larger number of samples was due to the high number of women who have genital infections and low Nugent–Krohn–Hillier [38] (NKH) scores observed by microscope from vaginal slides made at the same time of vaginal swabs. Table 6 shows the number of lactobacilli strains isolated from each category of woman, the clinical situation, visit reason, and pathogens isolated from infected woman. As this is the first report dedicated to the vaginal lactobacilli diversity from Lebanese women, the data presented and discussed here could be considered as a preliminary indication for the vaginal lactobacilli cartography in North Lebanon.
The data resulting from our study indicated that L. salivarius and L. jensenii were the most abundant species within the lactobacilli complex, which contains other species such as L. acidophilus, L. crispatus, L. gasseri, L. ruminis, L. fermentum, L. plantarum, and L. helveticus. The presence of L. plantarum in the vaginal environment has already been reported and associated with a decreased risk of bacterial vaginosis [2]. Notably, L. jensenii, L. gasseri, Lactobacillus iners, and L. crispatus were the most frequently identified species from the vagina, although their relative abundance varied widely and appeared dependant on the studied population [3, 56, 60]. Thus, studies focusing the vaginal microbiota from different population worldwide need to be regularly reported [20, 31, 35, 40, 41, 50, 54]. All these studies outlined the importance of lactobacilli in maintaining the balance of the vaginal microbiota. Tarnberg et al. [14] and Pendharkar et al. [32] reported the predominance of L. crispatus, whereas in our study L. salivarius and L. jensenii, respectively, were more representative. The species L. crispatus and L. jensensii are strong H2O2 producers as compared to L. gasseri [51, 57]. Furthermore, the presence of L. salivarius found in the healthy vagina of Indian and Bulgarian women has been shown to produce significant amounts of lactic acid, display optimal adhesion, and eradicate Helicobacter pylori in the gastric mucosa [13, 20, 21, 23, 33]. Taken together, these examples delineate the sights of L. salivarius from human sources as potential probiotic candidate.
The vaginal lactobacilli were grown in contact with G. vaginalis CIP7074T, S. aureus ATCC33862, E. coli CIP103982, and C. albicans ATCC10231 which are potential urogenital infections agents [39]. Justly, the potential of some lactobacilli to co-aggregate may help in the clearance of these pathogens [39]. Indeed, coaggregation is recognized as one of the mechanisms through which lactobacilli can exert their probiotic effects by creating a hostile micro-environment around a pathogen [4, 59]. In accordance, we elucidated by epifluorescence microscopy the capacity of L. plantarum CMUL140 to co-aggregate with S. aureus ATCC33862 leading to a decrease of this pathogen, which is known to afflict women of premenopausal age worldwide [8].
Based on the data presented in Figs. 6 and 7, we hypothesize that the mode of action is likely bactericidal, in regard to the high number of dead S. aureus ATCC33862 cells. Further, the brightly green cells of S. aureus ATCC33862 correspond to the high metabolic activity of cells since the mechanism of Live/Dead probe is targeting the ARNm production, and this could explain that the S. aureus ATCC33862 in co-culture was in stress situation before death. Besides, the coaggregation phenomenon was clearly observed and the S. aureus ATCC33862 cells in this aggregation were either in death or in stress situation (Fig. 7).
Of the 53 vaginal lactobacilli, only seven isolates comprising CMUL 034 (L. gasseri), CMUL 054 (L. fermentum), CMUL 057 (L. gasseri), CMUL 067 (L. acidophilus), CMUL 080 (L. gasseri), CMUL 099 (L. gasseri), and CMUL 140 (L. plantarum) were able to inhibit the growth of the aforementioned microorganisms. Remarkably, none of the isolates corresponding to the most abundant species (L. salivarius and L. jensenii) were able to inhibit the aforementioned pathogens. The antagonism against the panel of pathogens is likely due to the production of BLIS as neutralization of the supernatant or protease treatment have both abrogated the antagonism, while the heat treatment unaffected this activity. It is worth mentioning that the production of BLIS by vaginal lactobacilli with large antagonism has been already reported [47].
The antagonistic vaginal lactobacilli were fully characterized at the molecular level. The 16S rDNA analysis carried out on these seven vaginal lactobacilli confirmed the pyrosequencing data, arguing the perfect reliability of these two methods. The preliminary molecular characterization of the seven strains was done by specific PCR allowing the discrimination of the Lactobacillus genus through the amplification of 250 bp amplicon was obtained (data not shown) while the amplification of 16S-23S region confirmed the presence of two IS typically found for lactobacilli (data not shown). The specific PCR as well as the 16S-23S PCR corroborate the previously reported data obtained on other species of lactobacilli from different sources.
To be qualified as a probiotic, the candidates must satisfy some safety and functional properties such as antimicrobial activity, antibiotic susceptibility, hemolytic activity, and adhesion capabilities. The natural bacterial resistance to antibiotics is not considered as a risk to animal or human health. The antagonistic lactobacilli isolated here were sensitive to β-lactamines, except CMUL34 (L. gasseri), CMUL54 (L. fermentum), and CMUL 67 (L. acidophilus), which exhibited resistance to cefoxitin. Also, all these antagonistic vaginal lactobacilli were sensitive to aminosides, tetracyclines, rifampicin, and macrolides. The genus Lactobacillus is inherently resistant to glycopeptides such as vancomycin [55, 58]. Resistance to vancomycin was observed for CMUL 54 (L. plantarum) and CMUL 140 (L. plantarum). Furthermore, our strains were sensitive to kanamycin; this data is different to that reported by Zhou et al. [61], who showed a high resistance level of certain lactobacilli to kanamycin. The sensitivity of strains to tetracycline and erythromycin is in accordance with results previously reported [7, 14]. The resistance to erythromycin in the human vaginal lactobacilli was attributed to transition mutation in the V region of 23S rRNA codifying gene [6]. Finally, in regard to the hemolytic activity, our strains do not appear to have such activity. These antagonistic strains do not contain a plasmid DNA as indicated in the “Results” section, which bolsters them as safe for use as probiotics.
In addition to antagonism, the adherence capacity to human cells is considered of major importance for the in vitro evaluation of probiotic properties [17]. Clearly, the hydrophobicity is an important mechanism in bacterial adherence [25]. The mechanism of infection requires pathogens adhesion and displacement of indigenous lactobacilli. An alternative therapeutic approach to antimicrobials is to reintroduce and keep lactobacilli in this microbiome by probiotic administration [59]. Highly negatively charged hydrophilic lactobacilli adhered minimally [34], while lactobacilli with a slightly negatively charged, hydrophobic cell surfaces adhered strongly [18]. Adherence of lactobacilli to epithelial cells as well as their aptitudes to form biofilms is due to their autoaggregation and surface hydrophobicity [16]. Furthermore, coaggregation constitutes a barrier which prevents colonization by pathogens [9]. Antagonistic lactobacilli isolated in this study showed some autoaggregation properties, and the percent of autoaggregation appeared to be strain dependant (Table 5). The highest percent of autoaggregation was observed for CMUL57 (L. gasseri) CMUL67 (L. gasseri) and CMUL140 (L. plantarum), respectively (Table 5). The adhesion potency of L. gasseri species was shown to be attributed to an aggregation-promoting factor-like protein, which serves as a bridge between pathogen and human cells [29]. In this context, L. plantarum and L. salivarius strains secreted extracellular proteins with LysM domains known to be as the attachment site of the autolysin AcmA of Lactococcus lactis to peptidoglycan [52]. An extracellular chitin-binding protein from L. plantarum, containing LysM domain, was shown to attach to the cell surface and to bind N-acetylglucosamine containing polymers [48]. In our study, the difference in the level of autoaggregation could be explained by the presence of such specific compounds based on the comparison with the other vaginal lactobacilli CMUL34 (L. gasseri) CMUL54 (L. fermentum), CMUL80 (L. gasseri), and CMUL99 (L. gasseri), which displayed lower levels of autoaggregation. Isolates of CMUL57 (L. gasseri), CMUL67 (L. acidophilus), and CMUL140 (L. plantarum) adhered highly to xylene; an apolar solvent, confirming the hydrophobic properties of these strains. Studies on the microbial cell surfaces revealed the presence of (glyco-) proteinaceous material leading higher hydrophobicity, whereas hydrophilic surfaces were associated with the presence of polysaccharides [18, 22, 34, 42, 45].
Conclusion
This is the first species distribution study of the Lactobacillus genus of vaginal microbiota from women in North Lebanon. Seven antagonistic strains were observed in this study. These lactobacilli exhibited potential probiotic characteristics such as antagonism against vaginal pathogens including G. vaginalis CIP7074T, C. albicans ATCC10231, E. coli CIP103982, and S. aureus ATCC33862. Autoaggregation and hydrophobicity as well as safety characteristics such as hemolytic activity, and antibiotic susceptibility were established. Taken together, these data allowed us to design preliminary probiotic proprieties and therefore consider these strains good candidates for probiotic applications. This study will be pursued mainly through in vivo experiments in order to highlight further probiotic capabilities of these strains.
References
Andrade MJ, Rodriguez M, Sánchez B, Aranda E, Córdoba JJ (2006) DNA typing methods for differentiation of yeasts related to dry-cured meat products. Int J Food Microbiol 107:48–58
Antonio MA, Rabe LK, Hillier S (2005) Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J Infect Dis 192:394–398
Antonio MA, Hawes SE, Hillier SL (1999) The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J Infect Dis 180:1950–1956
Aslim B, Onal D, Beyatli Y (2007) Factors influencing autoaggregation and aggregation of Lactobacillus delbrueckii subsp. bulgaricus isolated from handmade yogurt. J Food Prot 70:223–227
Batdorj B, Dalgalarrondo M, Choiset Y, Pedroche J, Métro F, Prévost H, Chobert JM, Haertlé T (2006) Purification and characterization of two bacteriocins produced by lactic acid bacteria isolated from Mongolian airag. J Appl Microbiol 101:837–848
Begovic J, Huys G, Mayo B, D’Haene K, Florez AB, Lozo J, Kojic M, Strahinic I, Topisirovic L (2009) Human vaginal Lactobacillus rhamnosus harbor mutation in 23S rRNA associated with erythromycin resistance. Res Microbiol 160:421–426
Belletti N, Gatti M, Bottari B, Neviani E, Tabanelli G, Gardini F (2009) Antibiotic resistance of lactobacilli isolated from two Italian hard cheeses. J Food Prot 72:2162–2169
Bellon-fontaine MN, Rault J, van Oss CJ (1996) Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf B: Biointerfaces 7:47–53
Boris S, Suárez JE, Vázquez F, Barbés C (1998) Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect Immun 66:1985–1989
Brotman RM, Erbelding EJ, Jamshidi RM, Klebanoff MA, Zenilman JM, Ghanem KG (2007) Findings associated with recurrence of bacterial vaginosis among adolescents attending sexually transmitted diseases clinics. J Pediatr Adolesc Gynecol 20:225–231
Del Re B, Sgorbati B, Miglioli M, Palenzona D (2000) Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol 31:438–442
de Man JC, Rogosa M, Sharpe ME (1960) A medium for the cultivation of lactobacilli. J Appl Bacteriol 23:130–135
Dimitonova SP, Bakalov BV, Aleksandrova-Georgieva RN, Danova ST (2008) Phenotypic and molecular identification of lactobacilli isolated from vaginal secretions. J Microbiol Immunol Infect 41:469–477
Drago L, Mattina R, Nicola L, Rodighiero V, De Vechi E (2011) Macrolide resistance and in vitro selection of resistance to antibiotics in Lactobacillus isolates. J Microbiol 49:651–656
Dubernet S, Desmasures N, Guéguen M (2002) A PCR-based method for identification of lactobacilli at the genus level. FEMS Microbiol Lett 21:271–275
Dunne C, O’Mahony L, Thornton G, Morrissey D, O’Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, Kiely B, O’Sullivan GC, Shanahan F, Collins JK (2001) In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr 73:386–392
FAO/WHO (2006) Probiotics in food. Health and nutritional properties and guidelines for evaluation. FAO Food and Nutritional paper No. 85 (ISBN 92-5-105513-0)
Felten A, Barreau C, Bizet C, Lagrange PH, Philippon A (1999) Lactobacillus species identification, H2O2 and antibiotic resistance and correlation with human clinical status. J Clin Microbiol 37:729–733
Fichorova RN, Onderdonk AB, Yamamoto H, Delaney ML, Dubois AM, Allred E, Leviton A (2011) Maternal microbe-specific modulation of inflammatory response in extremely low-gestational-age newborns. MBio 2:280–10
Garg KB, Ganguli I, Das R, Talwar GP (2009) Spectrum of Lactobacillus species present in healthy vagina of Indian women. Indian J Med Res 129:652–657
Gil NF, Martinez RC, Gomes BC, Nomizo A, De Martinis EC (2010) Vaginal lactobacilli as potential probiotics against Candida spp. Braz J Microbiol 41:6–14
Greene JD, Klaenhammer TR (1994) Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl Environ Microbiol 60:4487–4494
Hsieh PS, Tsai YC, Chen YC, Teh SF, Ou CM, King VA (2012) Eradication of Helicobacter pylori infection by the probiotic strains Lactobacillus johnsonii MH-68 and L. salivarius ssp. salicinius AP-32. Helicobacter 17:466–477
Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, Vo KC, Caughey AB, Hilton JF, Davis RW, Giudice LC (2013) Diversity of the vaginal microbiome correlates with preterm birth. Reproductives Sciences 28
Juarez Tomas MS, Zonenschain D, Morelli L, Nader Macias ME (2005) Characterization of potentially probiotic vaginal lactobacilli isolated from Argentinian women. Br J Biomed Sci 62:170–174
Kabadjova P, Dousset X, Le Cam V, Prevost H (2002) Differentiation of closely related Carnobacterium food isolates based on 16S-23S ribosomal DNA intergenic spacer region polymorphism. Appl Environ Microbiol 68:5358–5366
Larsson PG, Bergstrom M, Forsum U, Jacobsson B, Strand A, Wolner-Hanssen P (2005) Bacterial vaginosis. Transmission, role in genital tract infection and pregnancy outcome: an enigma. APMIS 113:233–245
MacPhee RA, Miller WL, Gloor GB, McCormick JK, Hammond JA, Burton JP, Reid G (2013) Influence of the vaginal microbiota on toxic shock syndrome toxin 1 production by Staphylococcus aureus. Appl Environ Microbiol 79:1835–1842
Marcotte H, Ferrari S, Cesena C, Hammarstrom L, Morelli L, Pozzi G, Oggioni MR (2004) The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. J Appl Microbiol 97:749–756
Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinyaachola JO, Bwayo, Kreiss J (1999) Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis 180:1863–1868
Martirosian G, Radosz-Komoniewska H, Pietrzak B, Ekiel A, Kaminski P, Aptekorz M, Dolezych H, Samulska E, Jozwiak J (2012) Characterization of vaginal lactobacilli after kidney transplantation. Anaerobe 18:209–213
Mastromarino P, Vitali B, Mosca L (2013) Bacterial vaginosis: a review on clinical trials with probiotics. New Microbiol 36:229–238
Mastromarino P, Brigidi P, Macchia S, Maggi L, Pirovano F, Trinchieri V, Conte U, Matteuzzi D (2002) Characterization and selection of vaginal Lactobacillus strains for the preparation of vaginal tablets. J Appl Microbiol 93:884–893
Millsap K, Reid G, van der Mei C, Busscher H (1996) Cluster analysis of genotypically characterized Lactobacillus species based on physicochemical cell surfaces properties and their relationship with adhesion to hexdecane. Can J Microbiol 43:284–291
Motevaseli E, Shirzad M, Raoofian R, Hasheminasab SM, Hatami M, Dianatpour M, Modarressi MH (2013) Differences in vaginal lactobacilli composition of Iranian healthy and bacterial vaginosis infected women: a comparative analysis of their cytotoxic effects with commercial vaginal probiotics. Iran Red Crescent Med J 15:199–206
Newman D (1915) The treatment of cystitis by intravesical injections of lactic bacillus cultures. Lancet 330
Nielsen DS, Schillinger U, Franz CM, Bresciani J, Amoa-Awua W, Holzapfel WH, Jakobsen M (2007) Lactobacillus ghanensis sp. nov., a motile lactic acid bacterium isolated from Ghanaian cocoa fermentations. Int J Syst Evol Microbiol 57:1468–1472
Nugent RP, Krohn MA, Hillier SL (1991) Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol 29:297–301
Osset J, Bartolomé RM, García E (2001) Assessment of the capacity of Lactobacillus to inhibit the growth of uropathogens and block their adhesion to vaginal epithelial cells. J Infect Dis 183:485–491
Pavlova SI, Kilic AO, Kilic SS, So JS, Nader-Macias ME, Simoes JA, Tao L (2002) Genetic diversity of vaginal lactobacilli from women in different countries based on 16S rRNA gene sequences. J Appl Microbiol 92:451–459
Pendharkar S, Magopane T, Larsson PG, de Bruyn G, Gray GE, Hammarström L, Marcotte H (2013) Identification and characterisation of vaginal lactobacilli from South African women. BMC Infect Dis 13:43
Pelletier C, Bouley C, Cayuela C, Bouttier S, Bourlioux P, Bellon-Fontaine MN (1997) Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp. paracasei, and Lactobacillus rhamnosus strains. Appl Environ Microbiol 63:1725–1731
Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S (2013) Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol Rev 7:762–792
Reid G (2012) Probiotic and prebiotic applications for vaginal health. J AOAC Int 295:31–34
Rojas M, Conway PL (1996) Colonization by lactobacilli of piglet small intestinal mucus. J Appl Microbiol 81:474–480
Rosenberg M, Gutnick D, Rosenberg E (1980) Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9:29–33
Ruiz FO, Gerbaldo G, Garcia MJ, Giordano W, Pascual L, Barberis L (2012) Synergistic effect between two bacteriocin-like substances produced by lactobacilli strains with inhibitory activity for Streptococcus agalactiae. Curr Microbiol 64:349–356
Sánchez B, González-Tejedo C, Ruas-Madiedo P, Urdaci MC, Margolles A (2010) Lactobacillus plantarum extracellular chitin-binding protein and its role in the interaction between chitin, Caco-2 cells, and mucin. Appl Environ Microbiol 77:1123–1126
Sewankambo N, Gray RH, Wawer MJ, Paxton L, McNaim D, Wabwire-Mangen F, Serwadda D, Li C, Kiwanuka N, Hillier SL, Rabe L, Gaydos CA, Quinn TC, Konde-Lule J (1997) HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet 23:546–550, Erratum in: Lancet 1997, 350:1036
Sivester ME, Dicks LM (2003) identification of lactic acid bacteria from human vaginal secretions. Antonie Van Leeuwenhoek 83:117–123
Song YL, Kato N, Matsumiya Y, Liu CX, Kato H, Watanabe K (1999) Identification of and hydrogen peroxide production by fecal and vaginal lactobacilli isolated from Japanese women and newborn infants. J Clin Microbiol 37:3062–3064
Steen A, Buist G, Leenhouts KJ, El Khattabi M, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J (2003) Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem 278:23874–23881
Tagg JR, McGiven AR (1971) Assay system for bacteriocins. Appl Microbiol 21:943
Tarnberg M, Jakobsson T, Jonasson J, Forsum U (2002) Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rRNA variable V1 and V3 regions. Acta Pathol Microbiol Immunol Scand 110:802–810
Tulini FL, Winkelströter LK, De Martinis ECP (2013) Identification and evaluation of the probiotic potential of Lactobacillus paraplantarum FT259, a bacteriocinogenic strain isolated from Brazilian semi-hard artisanal cheese. Anaerobe 22:57–63
Vásquez A, Jakobsson T, Ahrné S, Forsum U, Molin G (2002) Vaginal Lactobacillus flora of healthy Swedish women. J Clin Microbiol 40:2746–2749
Wilks M, Wiggins R, Whiley A, Hennessy E, Warwick S, Porter H, Corfield A, Millar M (2004) Identification and H2O2 production of vaginal lactobacilli from pregnant women at high risk of preterm birth and relation with outcome. J Clin Microbiol 42:713–717
Yuksekdag ZN, Beyatli Y (2008) Antimicrobial activity and antibiotic susceptibility of some lactic acid bacteria isolated from different dairy products. Arch Leb 59:216–220
Younes JA, van der Mai HC, van den Heuvel E, Busscher HJ, Reid G (2012) Adhesion forces and coaggregation between vaginal staphylococcal and lactobacilli. PLoS ONE 7:e36917
Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, Foster JA, Forney LJ (2007) Differences in the composition of vaginal microbial communities found in healthy Caucasian and Black women. ISME J 1:121–133
Zhou JS, Pillidge CJ, Gopal PK, Gill HS (2005) Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int J Food Microbiol 98:211–217
Acknowledgments
This work was possible through the financial support of Lebanese University and Lille1 University. The authors would like to thank Prof. Mike Chikindas (Rutgers University) and Delphine Caly (Lille 1 University) for the critical reading of the manuscript. The authors are indebted to Mr. Taha ABDOU and Mrs Mariam YEHIA for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Al Kassaa, I., Hamze, M., Hober, D. et al. Identification of Vaginal Lactobacilli with Potential Probiotic Properties Isolated from Women in North Lebanon. Microb Ecol 67, 722–734 (2014). https://doi.org/10.1007/s00248-014-0384-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0384-7