Abstract
Freshwater aquifers in granitic rocks are widespread microbial habitats in the terrestrial subsurface. Microbial populations in deep granitic groundwater from two recently drilled (1 and 2 years) and two old boreholes (14 and 25 years) were compared. The 16S rRNA gene sequences related to “Candidatus Magnetobacterium bavaricum”, Thermodesulfovibrio spp. of Nitrospirae (90.5–93.1 % similarity) and a novel candidate division with <90 % similarity to known cultivated species were dominant in all boreholes. Most of the environmental clones closely related to the novel lineages in Nitrospirae, which have been detected exclusively in deep groundwater samples. In contrast, betaproteobacterial sequences related to the family Rhodocyclaceae were obtained only from the recently drilled boreholes, which had higher total cell numbers. Catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) analysis supported the result from clone library analysis; betaproteobacterial cells were dominantly detected in recently drilled boreholes. These results suggest that while indigenous microbial populations represented by the novel phylotypes persisted in the boreholes for 25 years, betaproteobacterial species disappeared after 2 years owing to the change of substrate availability.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Granite is one of the major rock types and widely distributed in the terrestrial upper crust [35]. Based on an average continental geothermal gradient of 19 °C/km, a mean surface temperature of 15 °C [37], and an upper growth limit for life of 122 °C, the biosphere harbored in granite potentially extends downward to ∼6 km [40]. Although microbiological studies of deep granitic aquifers associated with residual seawater [11–13, 31, 34] and mineral ore deposits [26, 27, 36] have been conducted, the majority of granitic aquifers simply represented by the percolation of recharged meteoric water through granitic rocks remain unexplored [3, 17].
Although it is important to determine the biomass and community structure of subsurface microorganisms that thrive in granitic aquifers, their habitats within fracture networks are vulnerable to the intrusion of drilling fluid [5, 30, 33]. Indeed, it is likely that microbial contaminants thrive in boreholes by utilizing compounds artificially introduced during drilling [5]. Therefore, long-term monitoring of population dynamics after drilling is necessary to distinguish microbial populations that depend on nutrient fluxes originally available in the deep aquifer. It is therefore critical to examine the levels of electron donors and acceptors introduced during drilling and hydrogeochemically supplied for the activities of subsurface microorganisms [2, 12, 30].
The Grimsel Test Site (GTS), which is an underground laboratory of the Swiss National Cooperative for the Disposal of Radioactive Waste (NAGRA), has been in operation since 1984 [1, 21]. The GTS lies at a depth of approximately 400–500 m within the Aare Massif granitic rocks. For microbiological investigations, four horizontal boreholes drilled between 1985 and 2009 were available. This unique opportunity enables us to collect different groundwater samples as a function of time after drilling during one sampling campaign rather than multiple sampling over a long period of time. The main objective of this study is to evaluate whether boreholes that span a wide range of drilling age are useful to distinguish microbial populations stably colonizing under borehole conditions from those flourished by drilling-induced disturbance. It is expected that the former might represent the deep life widely distributed in the terrestrial subsurface. For this objective, we coupled molecular phylogenetic analyses of microbial populations based on 16S rRNA gene sequence to geochemical analyses of groundwater mainly for biologically produced or consumed substrates.
Materials and Methods
The Grimsel Test Site (GTS)
The underground laboratory at the GTS extends about 1 km horizontally in a north–south direction. The northern and southern parts of the laboratory lie about 400 and 500 m below the ground surface, respectively (Fig. 1). The surface vegetation is poor owing to the high altitude (1,730 m), which suggests that the groundwater in the region is recharged with only a small amount of organic matter. In September 2010, groundwater samples from four horizontal boreholes were collected: US85.02 and JGP09.002 from the northern part and BOADUS96.001 and LCS08.002 from the southern part (Table 1). As horizontal drilling with oxygenated tap water without mud can alter indigenous microbial populations, one recently drilled borehole and one old borehole were compared in both parts of the aquifer. Although the lengths of the borehole intervals varied, the variation in their outflow rates allowed flushing at least one interval volume before sampling (Table 1). It is noted that the borehole intervals were separated by a Solexperts rubber multi-packer system, and that the hydraulic conductivities of borehole intervals ranged from 3.3 × 10−8 to 5.1 × 10−9 m/s (Table 1).
Topographic and geological features of the Grimsel Test Site (GTS). Upper: photograph of a mountainous area in which the GTS is located; middle: a cross-section diagram around the GTS tunnel system; bottom: layout of a horizontal-section of an underground laboratory with the investigated horizontal boreholes
Geochemical Analyses
From the multi-packer system, Tygon tubing was extended and directly connected to a flow-cell measuring system (Knick Stratos, Berlin, Germany) for on-site measurements of the physicochemical parameters (pH, Eh, and temperature). Dissolved oxygen (DO) was also measured on site using a DO sensor (HQ40d; Hach, Loveland, CO, USA) inserted into a flow cell. The concentration of aqueous Fe2+ was measured on site with a spectrophotometer (DR2400; Hach, Loveland, CO, USA) using the 1,10-phenanthroline method. To measure the concentration and isotopic composition of HS−, 1 L of groundwater was fixed with 5 mM Zinc acetate and 2.5 mM NaOH on site and then measured by spectrophotometry using the methylene blue method. For HS− measurement, 10 ml of the fixed sample was used without being concentrated.
Following geochemical analyses were conducted in Japan for the groundwater samples, which were kept at 4 or −20 °C during 1-week transportation mainly by air. The alkalinity of the unfiltered groundwater was determined by potentiometric titration with 0.1 N HCl solution using the Gran plot method [9]. A subsample was filtered through a 0.22-μm pore size filter (type GVWP; Millipore, Billerica, MA, USA) with a polypropylene housing for the following analyses. Cationic species including Na+, Mg2+, K+, Ca2+, and NH4 +, and anionic species including F−, Cl−, NO2 −, NO3 −, and SO4 2− were measured by ion chromatography (Prominence; Shimadzu Corp., Tokyo, Japan). NO3 −, and NO2 − were analyzed with a high sensitivity UV detector (SPD-20A; Shimadzu Corp., Tokyo, Japan), while the other ionic species were analyzed with a conductivity detector (CDD-10Avp; Shimadzu Corp., Tokyo, Japan). Dissolved organic carbon (DOC) was measured by thermocatalytic oxidation and MC-NDIR detection (multi N/C 3100; Analytik Jena Japan, Kanagawa, Japan). Organic acids including acetate, fumarate, formate, lactate, succinate, malate, pyruvate, and citrate were analyzed by high-performance liquid chromatography (Prominence; Shimadzu Corp., Tokyo, Japan) according to the manufacturer’s instructions. Subsamples for DOC and organic acids were stored at −20 °C.
For measurements of the concentrations of TIC, CH4, and C2H6 and carbon isotopic compositions of TIC (δ 13CTIC) and CH4 (δ 13CCH4), groundwater was collected into evacuated 67-mL glass vials and stored in the dark at 4 °C until analysis. The TIC is the sum of CO2, HCO3 −, and CO3 2−. The dominant species is HCO3 − because the GTS groundwater was slightly alkaline. The concentrations and carbon isotopic compositions were measured using a Finnigan MAT 252 isotope-ratio monitoring mass spectrometer (Thermo Finnigan, Bremen, Germany), as previously described [15, 41]. The carbon isotopic composition was expressed as a delta-notation; δ 13C = (13C/12C)sample/(13C/12C)VPDB − 1, where VPDB refers to the Vienna Peedee Belemnite isotopic standard.
To analyze the sulfur isotopic compositions of sulfate (δ 34SSO4) and sulfide (δ 34SHS), the remaining of 1 L groundwater fixed with 5 mM Zinc acetate and 2.5 mM NaOH was filtered a glass fiber filter (GF/F filter, Whatman, Springfield Mill, UK) for the recovery of precipitated ZnS. Next, the filtrate was amended with 0.1 M BaCl2 to precipitate BaSO4 for sulfate analysis. The BaSO4 was collected by filtration using a glass fiber filter. The filters were subsequently freeze-dried overnight, after which they were subjected to combustion using an elemental analyzer (Flash EA1110; Thermo Fisher Scientific, Waltham, MA, USA) to transform the ZnS or BaSO4 into SO2. The sulfur isotopic ratio of SO2 was measured using an isotope-ratio monitoring mass spectrometer (DELTAplus Advantage ConFlo III System, Thermo Fisher Scientific, Waltham, MA, USA). The sulfur isotopic composition was expressed as a delta-notation; δ 34S = (34S/32S)sample/(34S/32S)VCDT − 1, where VCDT refers to the Vienna Canyon Diablo Troilite isotopic standard.
The precision of the geochemical analyses is described in the Supplementary Information.
Microscopic Observations of Groundwater Samples
The total cell numbers of the groundwater samples were determined by the direct count method using SYBR Green I (Takara, Tokyo, Japan). Briefly, groundwater samples were fixed with 3.7 % formaldehyde at neutral pH, after which they were filtered through a 0.22-μm pore size, 25-mm diameter black polycarbonate filter (Advantec, Tokyo, Japan). In Japan, the filter was stained wtih 1× SYBR Green I in 1× TAE buffer for 5 min at 25 °C. The stained filter was then rinsed with 10 mL of DDW and examined under epifluorescence using an Olympus BX51 microscope equipped with a DP70 digital microscope camera (Olympus, Tokyo, Japan).
DNA Extraction, Library Construction and Sequencing for Phylogenetic Analysis
Groundwater was collected in a sterile Tedlar sampling bag, which was evacuated and flushed with Ar gas three times, and passed through a 0.22-μm pore size filter (type GVWP; Millipore, Billerica, MA, USA) with a sterilized polypropylene housing on site immediately after sampling. Filter samples were kept frozen until DNA was extracted from the cells on the filter using a Soil DNA Kit (Mo Bio Lab. Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions.
Using a universal bacteria primer set of Bac27F and Uni1492R [20] and a universal archaeal primer set of Arch21F and Uni1492R [8], the near-full region of the 16S rRNA gene was amplified by polymerase chain reaction (PCR) using LA Taq polymerase (TaKaRa, Tokyo, Japan). Each oligonucleotide primer (0.1 μM) and DNA template (1 μL) was prepared in a reaction mixture. The reaction mixture was then subjected to the following conditions using a GeneAmp 9700 thermocycler (PE Applied Biosystems, Foster City, CA, USA): 35 cycles of denaturation at 96 °C for 20 s, annealing at 50 °C (for PCR with archaeal primer set) or 53 °C (for PCR with bacterial primer set) for 45 s, and extension at 72 °C for 120 s. Whereas PCR amplification with the bacterial primer set was successful for all groundwater samples, no PCR-amplified products were obtained with the archaeal primer set. Bacterial PCR-amplified products with the expected sizes were purified using a PCR purification kit (Mo Bio Lab. Inc., Carlsbad, CA, USA). The purified PCR product was inserted into the pCR2.1 vector (Invitrogen, Carlsbad, CA, USA) using a DNA Ligation Kit (Ver. 2.1, TaKaRa, Tokyo, Japan). Ligation was conducted at 16 °C for 6 h, after which the inserted vectors were cloned using an original TA cloning kit.
The inserted nucleotide sequences were amplified directly by PCR from randomly selected colonies using M13 primers treated with exonuclease I and shrimp alkaline phosphatase (Amersham Pharmacia Biotech, Buckinghamshire, UK). The nucleotides were sequenced with a 3130xl genetic analyzer (PE Applied Biosystems) by the dideoxynucleotide chain-termination method using a BigDye sequencing kit (PE Applied Biosystems) according to the manufacturer’s recommendations. The Bac27F primer was used in the partial sequencing analysis. Single-strand sequences more than 500 nucleotides in length were analyzed.
A single phylogenetic clone type (phylotype) was obtained from the clone type analysis (those with >97 % similarity were grouped). The closest cultivated species, closest environmental clones and sequence similarities were determined using the BLAST program in conjunction with the DNASIS Taxon software (Hitachi Software, Tokyo, Japan). The partial sequence was extended to the near full length of 16S rRNA gene with lengths of ∼1.4 kbp and manually aligned according to the secondary structures using ARB (a software environment for sequence data) [24]. Chimeric sequences were also checked by comparing the phylogenetic affiliations of the 5′ and 3′ halves of each sequence. Sequences were reduced to unambiguously alignable positions using the ARB FastAligner utility, and phylogenetic analysis of alignments was performed employing distance methods with ARB. The DNA sequences determined in this study are available through DDBJ (AB746199 and AB746487–AB746521).
Catalyzed Reporter Deposition-Fluorescence In Situ Hybridization Analysis
For catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) analysis, cells were fixed with formaldehyde immediately after collection, washed twice with PBS and then stored them in 1:1 ethanol/PBS at −20 °C on site. In Japan, bacterial cell wall were permeabilized with lysozyme solution as previously described [29]. Filters were soaked in lysozyme solution (10 mg/ml of lysozyme, Wako Pure Chemical Industries, Ltd., Osaka, Japan), 50 mM EDTA (SHOWA CHEMICAL, Tokyo, Japan) and 0.1 M Tris–HCl (Nakalai tesque, Kyoto, Japan), and incubated at 37 °C for 1 h, washed with MilliQ water for 1 min. The filters were subsequently treated with achromopeptidase (Wako Pure Chemical Industries, Ltd., Osaka Japan) as previously described [38]. For inactivation of EPX, the sample filters were soaked in 0.15 % H2O2 in methanol for 30 min at room temperature, washed with 99.5 % ethanol, and dried. The sample filters were stored at −20 °C until further processing.
CARD-FISH were performed as previously described [29, 38] with some modification. Each sample filer was soaked in 10 μl of hybridization buffer [29] including 2 % (w/v) blocking reagent (Roche Molecular Biochemicals, Mannheim, Germany). Probes used were HRP-labeled EUB(I–III) (mixture of following 3 sequences; 5′-GCT GCC TCC CGT AGG AGT-3′, 5′-GCA GCC ACC CGT AGG TGT-3′, and 5′-GCT GCC ACC CGT AGG TGT-3′) for bacteria detection [6], and HRP-labeled Bet42a (5′-GCC TTC CCA CTT CGT TT-3′) mixed with non-labeled competitor (5′-GCC TTC CCA CAT CGT TT-3′) for β-Proteobacteria detection [25]. Formamide concentration to adjust probe stringency of the hybridization buffer was 35 % (v/v) for both probes. Filters were hybridized in a humid chamber for at least 2 h at 46 °C, then washed in 15 ml of prewarmed washing buffer at 48 °C for 15 min. Prior to tyramide signal amplification, the sample filters were equilibrated with PBS for 15 min at room temperature. The sample filters were soaked in the substrate mix, consisting of PBS containing 0.1 % blocking reagent, 0.0015 % H2O2, 2 M NaCl, 10 μg/ml p-iodophenylboronic acid (KANTO CHEMICAL CO., INC., Tokyo, Japan), and 1/500 parts of fluorescein-labeled tyramide, and incubated for 30 min at 37 °C. After the signal amplification, the sample filters were washed with PBS for 15 min at room temperature and with sterile MilliQ water for 1 min. They were dehydrated in 99.5 % ethanol and finally dried. DAPI (4′,6-diamidino-2-phenylindole) staining were performed as previously described [29]. Microscopic observation with 123 to 131 fields and photograph were taken with fluorescent microscope (BX51, OLYMPUS, Tokyo, Japan) and Digital camera (DP71, OLYMPUS, Tokyo, Japan).
Results and Discussion
Geochemical Characteristics of the GTS Groundwater
The geochemical parameters of groundwater samples from four boreholes were analyzed to investigate the levels of biologically utilized compounds and their correlation with the abundance and diversity of microbial populations (Table 2). Although pH and Eh measurements were unsuccessful for BOADUS96.001due to analytical errors, the pH was ca. 9.6 in good agreement with data from Degueldre et al. [7] for the rest of boreholes. The Eh value ranged from −100 to 19 mV. Variations in DO appeared to be roughly correlated with the flow rates of the boreholes (Tables 1 and 2). The DO and the flow rate of LCS08.002 were likely correlated because the low flow rate leads to oxygen penetrating the plastic tubing that connects the borehole interval to the DO flow cell. Consequently, the Eh value of LCS08.002 was positive.
The salinity of the GTS groundwater is low and classified as a type of Na-Ca-HCO3-F [7]. The concentrations of Na+, Mg2+, K+, Ca+, F−, and Cl− were similar, regardless of borehole age and location (north or south) (Table 2). Biological compounds utilized as electron acceptors (NO3 − and NO2 −) and electron donors (NH4 +, Fe2+, HS−, acetate, formate, lactate, succinate, malate, pyruvate, propionate, citrate, CH4, and C2H6) were below our detection limits or detected at very low concentrations. The concentrations of SO4 2−, DOC and TIC ranged from 51.6 to 176.8, 24.7 to 82.7, and 294 to 383 μM, respectively.
Stable isotopes can be used to trace biological activities in subsurface environments [14, 23]. The carbon isotopic compositions of CH4 and TIC ranged from −45.6 to −79.5‰ vs. VPDB and −11.6 to −13.4‰ vs. VPDB, respectively, which is indicative of biogenic CH4 [43]. However, the in situ microbial production of methane in the study area was likely negligible because sulfate generally must be depleted for methanogenesis to be thermodynamically favorable. The sulfur isotopic compositions of sulfate and sulfide are also known to indicate microbial sulfate reduction [4, 19]. Because the isotopic fractionation between sulfate (−0.4 to +11.5‰ vs. VCDT) and sulfide (−9.4 to +11.5‰ vs. VCDT) was smaller than that typically encountered for microbial sulfate reduction, ongoing sulfate reduction also appears unlikely. As a result, the ongoing redox reactions might be not active in the groundwater from the GTS.
Microbial Abundance and Diversity of the GTS Groundwater
The total cell numbers measured were 4.9 ± 5.1 × 103 cells/ml for BOADUS96.001, 7.5 ± 3.5 × 103 cells/ml for US85.02, 2.9 ± 1.4 × 104 cells/ml for JGP09.002, and 3.0 ± 0.9 × 104 cells/ml for LCS08.002. It should be noted that microbial cells were more abundant in the two recently drilled boreholes than in the two old boreholes. The total cell numbers have been reported to range from 103 to 105 cells/ml in other deep granitic aquifers [13, 34, 36]; therefore, the GTS groundwater appears to have average cell concentrations.
16S rRNA gene sequence analysis of groundwater samples collected from the four boreholes was conducted. No amplification with the Archaea-specific primer set was obtained. The low abundance of Archaea that includes methanogens is expected for groundwater not depleted in sulfate. In contrast, a total of 113 bacterial 16S rRNA gene sequences that were assigned to 28 phylotypes (GTS_B1-28) based on 97 % similarity were obtained from the four boreholes (Table 3 and Supplementary Table 1). In samples from the recently drilled boreholes (LCS and JGP), β-proteobacterial phylotypes related to Methyloversatilis universalis (93.7 % similarity), Hydrogenophaga pseudoflava (98.5 % similarity) and Dechloromonas hortensis (99.0 % similarity) were abundant. As shown in Table 3, four phylotypes representing 26 clones were related to “Candidatus Magnetobacterium bavaricum” of the phylum Nitrospirae with a similarity ranging from 90.5 to 93.1 %. Two phylotypes representing seven clones were related to Thermodesulfovibrio spp. of the Phylum Nitrospirae with a similarity ranging from 91.1 to 92.0 %. Phylotypes without cultivable relatives with a similarity of <90 % were defined as unclassified.
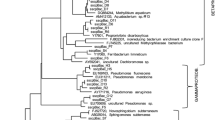
The phylogenetic affiliations of the unclassified GTS phylotypes were further analyzed by comparing their unambiguously aligned sequences against sequences from environmental clones and cultured species. A neighbor-joining tree was constructed to visualize the phylogenetic relationship among GTS phylotypes and related sequences (Fig. 2). The unclassified GTS phylotypes GTS_B2, 7, 9, and 14 were related to the Candidate Division OP5, while the unclassified phylotype GTS_B13 was related to the phylum Chlorobi (Fig. 2). The cell abundance of the groundwater of the four boreholes is compared in Fig. 3. The compositions of microbial communities based on the phylogenetic affiliations of the GTS phylotypes are proportionally integrated in the vertical bars representing the cell abundance. We tentatively assigned phylotypes GTS_B2, 7, 9 and 14 to the Candidate Division GTS. It is clear from Fig. 3 that although the phylotypes affiliated within Nitrospirae and the Candidate Division GTS were evenly distributed in the four boreholes, regardless of borehole age, the β-proteobacterial phylotypes accounted for the increased cell abundance observed in the recently drilled boreholes.
Neighbor-joining tree of the GTS phylotypes and related sequences. Blue, green, orange, and red GTS phylotypes are from JGP09.002, LCS08.002, US85.02 and BOADUS96.001, respectively. Scale bar = 0.1 expected change per nucleotide position. Bootstrap values of >50 % are shown at branching points. Taxonomic groups associated with and without GTS phylotypes are indicated by shaded and white trapezoids, respectively
To assess whether microbial cells detected by DNA staining with SYBR Green I were alive and to verify the compositions of microbial communities revealed by 16S rRNA gene sequence analysis, we conducted CARD-FISH analysis. Representative fluorescent microscopic photographs for samples from the old borehole BOADUS96.001 and the recently drilled borehole LCS08.002 are shown in Fig. 4. In the LCS08.002 sample, cells stained with EUB(I–III) had various fluorescent intensities (Fig. 4a). The clearly stained cells with Bet42a were observed (Fig. 4b), and their morphologies were short rods, which were also observed in EUB(I–III)-stained cells. The fluorescent intensities of stained cells with EUB(I–III) in the BOADUS96.001 sample (Fig. 4c) were dark as compared to those in the LCS08.002 sample. No β-proteobacterial cells were detected with Bet42a in the BOADUS96.001 sample (Fig. 4d). Those results suggest that bacterial cells contained cellular ribosomal RNA and were alive, while alive β-proteobacterial cells occurred only in the recently drilled borehole, which agrees well with the microbial compositions based on results from 16S rRNA gene sequence analysis.
Fluorescent microscopic photographs of CARD-FISH and DAPI stained cells. Left panels depict probe signals (green) and right panels depict DAPI staining (blue). a LCS 08.002 stained with EUB(I–III). b LCS 08.002 stained with Bet42a. c BOADUS 96.001 stained with EUB(I–III). d BOADUS 96.001 stained with Bet42a. Scale bars correspond to 10 μm
To quantify microbial cells stained with the probes EUB(I–III) and Beta42a by CARD-FISH, the numbers of microscopic views (85 × 65 μm) and cells observed were increased for groundwater from all the boreholes (Table 4). The ratio of cells hybridized with Bet42a to those with EUB(I–III), which were based on 123–131 microscopic views, were 0.06 (2.1 × 102 to 3.5 × 103 cells/ml) for US85.02, 0.05 (1.2 × 102 to 2.3 × 103 cells/ml) for BOADUS96.001, 0.76 (1.3 × 103 to 1.7 × 103 cells/ml) for LCS08.002 and 0.22 (2.1 × 103 to 9.3 × 103 cells/ml) for JGP09.002, respectively. The Bet42a to EUB(I–III) ratios for all the samples were largely consistent with those from 16S rRNA gene sequence analysis. Although the cell number hybridized with EUB(I–III) from LCS08.002 was one order of magnitude lower than the total cell number, the β-proteobacterial dominance in the recently drilled boreholes was supported by the two independent molecular methods.
Factors Controlling Microbial Ecology in the GTS Groundwater
The physiological properties of microorganisms were inferred from traits commonly associated with their close relatives (>97 % similarity). With regard to the β-proteobacterial phylotypes exclusively detected from the recently drilled boreholes, the phylotypes related to H. pseudoflava (98.5 % similarity) and D. hortensis (99.0 % similarity) are likely capable of aerobic hydrogen oxidation [42] and oxidation of chlorinated organic compounds coupled to oxygen and nitrate reduction [44], respectively. The most abundant phylotype, GTS_B1, was distantly related to M. universalis (93.7 % similarity). GTS_B1 belongs to the family Rhodocyclaceae, which contains many members with the ability to degrade halogenated compounds coupled to denitrification and reduction of (per)chlorates [18]. Therefore, it is speculated that microorganisms represented by the phylotype GTS_B1 mediate microaerobic metabolisms that involve halogenated compounds. These physiological inferences suggest that the intrusion of oxygen into the boreholes and surrounding rock fractures during drilling stimulated the growth of aerobic chemoautotrophs and aerobic to microaerobic heterotrophs that degrade refractory organic compounds as energy sources.
To constrain factors favorable for the occurrence of GTS phylotypes with low homologies to known relatives (<97 %), it is necessary to specify their closely related environmental clones and the characteristics of habitats in which these clones were detected. As shown in Fig. 2, the dominant Nitrospirae phylotypes GTS_B4 and 6, which were found in the GTS groundwater from boreholes JGP09.002, LCS08.002, and US85.02 were somewhat related to environmental clones from wet soils, freshwater sediments, and shallow aquifers. The most closely related environmental clone was detected from 3- to 5-cm-deep low salinity tidal sediment [10]. The Nitrospirae phylotypes GTS_B5 and 11 were only detected from borehole BOADUS96.001. These phylotypes were related to environmental clones detected from groundwater contaminated with coal tar or oil. The similarity between phylotypes GTS_B5 and 11 and their most closely related environmental clones was less than 95 %. In contrast, the remaining Nitrospirae phylotypes GTS_B10 and 12 were closely related to environmental clones found in deep groundwater from a South Africa gold mine (97.6 % similarity; [22]) and the Tono uranium mine (96.5 % similarity; [26]), respectively (Fig. 2). These findings suggest that the microorganisms represented by Nitrospirae phylotypes from the GTS groundwater are only found in the terrestrial subsurface. Four unclassified phylotypes, GTS_B2, 7, 9, and 14, which were tentatively assigned to the Candidate Division GTS and distantly related to the Candidate Division OP5, were closely related to environmental clones from deep groundwater from the Tono uranium mine (∼95.0 % similarity: [26]). The remaining unclassified phylotype, GTS_B13, which was phylogenetically clustered within the phylum Chlorobi (Fig. 2), was also distantly related to environmental clones from a South Africa gold mine (93.3 % similarity).
The decrease in β-Proteobacteria over time and ubiquitous occurrence of Nitrospirae and the Candidate Division GTS suggest that the latter represents the indigenous subsurface community stably colonized in the granitic aquifer. The occurrence of environmental clones related to the Candidate Division GTS with >90 % similarity was limited to groundwater from the Tono uranium mine. The Tono groundwater was collected from a lignite-bearing sedimentary formation at a depth of 160 m [16, 27]. As shown in Table 2, the Tono groundwater is similar to the GTS groundwater in terms of pH, salinity and temperature. Although the types of host rock appear to be different, DOC contents of groundwater from the GTS and the Tono mine are similar, with a narrow range of 24.7 to 82.7 μM (Table 2).
It is counterintuitive that granitic groundwater that was recharged at the bare surface harbors microorganisms that were closely related to those detected from the Tono uranium mine, which is naturally enriched with refractory organic matter (lignite). This correlation indicates that, despite the low supply of DOC, the primary energy could be derived from organic compounds rather than inorganic counterparts derived from rock-water interactions. Environmental clones related to the Nitrospirae phylotypes GTS_B10 and 12 were detected from a 1.4-km-deep borehole located in the Beatrix Gold mine, South Africa [22]. Although Beatrix groundwater is relatively similar to the GTS groundwater in terms of the pH and DOC (Table 2), high concentrations of dissolved ionic and gaseous species associate with the long residence time in Precambrian metamorphic and igneous rocks are distinctive.
It has been reported that the cell abundance in Scandinavian granitic groundwater associated with residual seawater was only correlated with the DOC concentration in the groundwater [32]. It was also clarified that the numbers of cultivable Fe(III)- and sulfate-reducing heterotrophs increase with that of heterotrophic acetogen, since many of Fe(III) and sulfate reducers are capable of metabolizing acetate as a carbon source and an electron donor [14]. These results strengthen our inference that organic matter fuels the activities of microorganisms in the GTS groundwater. However, recent studies on the Scandinavian granitic groundwater indicated that H2 concentrations are positively correlated with most probable numbers of heterotrophic and autotrophic anaerobes [13].
As for the four GTS boreholes that harbor geochemically similar groundwater, the abundance and diversity of microbial populations were correlated to the borehole age. The proteobacterial dominance is common in boreholes newly drilled from the land surface [39] as well as deep underground tunnels [28], which cautions that the population dynamics should be monitored after drilling to exclude non-indigenous microbes. As the GTS groundwater samples from the 14- and 25-years old boreholes were geochemically and microbiologically similar, statistical approaches to elucidate their correlation are not applicable. The study of geographically distant groundwater that is geochemically similar to that of the GTS might provide insight into the global distribution and potential metabolism of bacteria represented by the novel GTS phylotypes.
Conclusions
We investigated the geochemical and microbiological properties of deep granitic groundwater from boreholes that varied in age from 1 to 25 years at the GTS. Although the geochemical properties were almost uniform among the four investigated boreholes, the abundance and diversity of microbial populations varied according to the borehole ages. Whereas phylotypes closely related to β-Proteobacteria were only abundant in 1- to 2-year-old boreholes, novel phylotypes affiliated with the phylum Nitrospirae and tentatively assigned to the Candidate Division GTS were ubiquitous in boreholes, regardless of age. Our results from population dynamics in the study area suggest that boreholes with a wide range of drilling age are suitable to clarify microbial populations stably colonizing under borehole conditions, the novel lineages of which might be indigenous to and widely distributed in the deep terrestrial subsurface.
References
Alonso EE, Alcoverro J, Coste F, Malinsky L, Merrien-Soukatchoff V, Kadiri I, Nowak T, Shao H, Nguyen TS, Selvadurai APS, Armand G, Sobolik SR, Itamura M, Stone CM, Webb SW, Rejeb A, Tijani M, Maouche Z, Kobayashi A, Kurikami H, Ito A, Sugita Y, Chijimatsu M, Börgesson L, Hernelind J, Rutqvist J, Tsnag C-F, Jussila P (2005) The FEBEX benchmark test: case definition and comparison of modeling approaches. Int J Rock Mech Min Sci 42:611–638
Balkwill DL, Reeves RH, Drake GR, Reeves JY, Crocker FH, King MB, Boone DR (1997) Phylogenetic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol Rev 20:201–216
Brockman FJ, Kieft TL, Fredrickson JK, Bjornstad BN, Li SMW, Spangenburg W, Long PE (1992) Microbiology of vadose zone paleosols in south-central Washingtone-state. Microb Ecol 23:279–301
Canfield DE (2001) Isotope fractionation by natural populations of sulfate-reducing bacteria. Geochim Cosmochim Acta 65:115–125
Colwell FS, Stormberg GJ, Phelps TJ, Birnbaum SA, McKinley J, Rawson SA, Veverka C, Goodwin S, Long PE, Russell BF, Garland T, Thompson D, Skinner P, Grover S (1992) Innovative techniques for collection of saturated and unsaturated subsurface basalts and sediments for microbiological characterization. J Microbiol Methods 15:279–292
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M (1999) The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444
Degueldre C, Pfeiffer HR, Alexander W, Wernli B, Bruetsch R (1996) Colloid properties in granitic groundwater systems.1. Sampling and characterisation. Appl Geochem 11:677–695
Delong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89:5685–5689
Dyrssen D, Sillén LG (1967) Alkalinity and total carbonate in sea water. A plea for p-T independent data. Tellus 19:113–121
Edmonds JW, Weston NB, Joye SB, Mou X, Moran MA (2009) Microbial community response to seawater amendment in low-salinity tidal sediments. Microb Ecol 58:558–568
Fukuda A, Hagiwara H, Ishimura T, Kouduka M, Ioka S, Amano Y, Tsunogai U, Suzuki Y, Mizuno T (2010) Geomicrobiological properties of ultra-deep granitic groundwater from the Mizunami Underground Research Laboratory (MIU), Central Japan. Microb Ecol 60:214–225
Fredrickson JK, Balkwill DL (2006) Geomicrobial processes and biodiversity in the deep terrestrial subsurface. Geomicrobiol J 23:345–356
Hallbeck L, Pedersen K (2008) Characterization of microbial processes in deep aquifers of the Fennoscandian Shield. Appl Geochem 23:1796–1819
Haveman SA, Pedersen K (2002) Distribution of culturable microorganisms in Fennoscandian Shield groundwater. FEMS Microbiol Ecol 39:129–137
Ijiri A, Tsunogai U, Gamo T (2003) A simple method for oxygen-18 determination of milligram quantities of water using NaHCO3 reagent. Rapid Commun Mass Spectrom 17:1472–1478
Iwatsuki T, Xu S, Mizutani Y, Hama K, Saegusa H, Nakano K (2001) Carbon-14 study of groundwater in the sedimentary rocks at the Tono study site, central Japan. Appl Geochem 16:849–859
Jain DK, Stores-Gascoyne S, Providenti M, Tanner C, Cord I (1997) Characterization of microbial communities in deep groundwater form granitic rock. Can J Microbiol 43:272–283
Kalyuzhnaya MG, De Marco P, Bowerman S, Pacheco CC, Lara JC, Lidstrom ME, Chistoserdova L (2006) Methyloversatilis universalis gen. nov., sp. nov., a novel taxon within the Betaproteobacteria represented by three methylotrophic isolates. Int J Syst Evol Microbiol 56:2517–2522
Kaplan IR, Rittenberg SC (1964) Microbiological fractionation of sulfur isotopes. J Gen Microbiol 34:195–212
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lieb RW (1989) Presentation of the Grimsel Test Site. Nucl Eng Des 116:7–9
Lin LH, Hall J, Onstott TC, Gihring T, Sharwood Lollar B, Boice E, Pratt L, Lippman-Pipke J, Bellamy RES (2006) Planktonic microbial communities associated with fracture-derived groundwater in a deep gold mine of South Africa. Geomicrobiol J 23:475–497
Lin LH, Wang PL, Rumble D, Lippmann-Pipke J, Boice E, Pratt LM, Lollar BS, Brodie EL, Hazen TC, Andersen GL, DeSantis TZ, Moser DP, Kershaw D, Onstott TC (2006) Long-term sustainability of a high-energy, low-diversity crustal biome. Science 314:479–482
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH (1992) Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria—problems and solutions. Syst Appl Microbiol 15:593–600
Miyoshi T, Iwatsuki T, Naganuma T (2005) Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl Environ Microbiol 71:1084–1088
Murakami Y, Fujita Y, Iwatsuki T, Naganuma T (2002) Abundance and viability of subsurface microbial communities in sedimentary and igneous rock aquifers. Microbes Environ 17:63–74
Onstott TC, Moser DP, Pfiffner SM, Fredrickson JK, Brockman FJ, Phelps TJ, White DC, Peacock A, Balkwill D, Hoover R, Krumholz LR, Borscik M, Kieft TL, Wilson R (2003) Indigenous and contaminant microbes in ultradeep mines. Environ Microbiol 5:1168–1191
Pernthaler A, Pernthaler J, Amann R (2002) Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68:3094–3101
Pedersen K (1997) Microbial life in deep granitic rock. FEMS Microbiol Rev 20:399–414
Pedersen K, Arlinger J, Ekendahl S, Hallbeck L (1996) 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Aspo hard rock laboratory, Sweden. FEMS Microbiol Ecol 19:249–262
Pedersen K, Ekendahl S (1990) Distribution and activity of bacteria in deep granitic groundwaters of Southeastern Sweden. Microb Ecol 29:37–52
Pedersen K, Hallbeck L, Arlinger J, Erlandson AC, Jahromi N (1997) Investigation of the potential for microbial contamination of deep granitic aquifers during drilling using 16S rRNA gene sequencing and culturing methods. J Microbiol Methods 30:179–192
Pedersen K, Arlinger J, Eriksson S, Hallbeck A, Hallbeck L, Johansson J (2008) Numbers, biomass and cultivable diversity of microbial populations relate to depth and borehole-specific conditions in groundwater from depths of 4–450 m in Olkiluoto, Finland. ISME J 2:760–775
Richardson SM, McSween HY Jr (1989) Geochemistry: pathways and processes. Prentice Hall, New Jersey
Sahl JW, Schmidt RH, Swanner ED, Mandernack KW, Templeton AS, Kieft TL, Smith RL, Sanford WE, Callaghan RL, Mitton JB, Spear JR (2008) Subsurface microbial diversity in deep-granitic-fracture water in Colorado. Appl Environ Microbiol 74:143–152
Sclater JG, Jaupart C, Galson D (1980) The heat-flow through oceanic and continental-crust and the heat-loss of the Earth. Rev Geophys 18:269–311
Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R (2003) An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol 69:2928–2935
Stroes-Gascoyne S, Hamonet CJ, Audette-Stuart M, Beaton D, King-Sharp K, Festarini A, Serran M, McMullin D, Kramer- Tremblay S, Rose S, Bellan L (2011) Microbial characterization of groundwater from boreholes CR9 and CR18 at CRL(2007–2009)—Implications for a possible future repository for radioactive non-fuel waste. In: Proceedings of CNS Conference on Waste Management, Decommissioning and Restoration for Canada’s Nuclear Activities, September 11–14, Toronto, ON, Canada
Takai K, Nakamura K, Toki T, Tsunogai U, Miyazaki M, Miyazaki J, Hirayama H, Nakagawa S, Nunoura T, Horikoshi K (2008) Cell proliferation at 122 degrees C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc Natl Acad Sci U S A 105:10949–10954
Tsunogai U, Yoshida N, Ishibashi J, Gamo T (2000) Carbon isotopic distribution of methane in deep-sea hydrothermal plume, Myojin Knoll Caldera, Izu-Bonin arc: implications for microbial methane oxidation in the oceans and applications to heat flux estimation. Geochim Cosmochim Acta 64:2439–2452
Wen AM, Fegan M, Hayward C, Chakraborty S, Sly LI (1999) Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int J Syst Bacteriol 49:567–576
Whiticar MJ (1999) Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol 161:291–314
Wolterink A, Kim S, Muusse M, Kim IS, Roholl PJM, van Ginkel CG, Stams AJM, Kengen SWM (2005) Dechloromonas hortensis sp. nov. and strain ASK-1, two novel (per)chlorate-reducing bacteria, and taxonomic description of strain GR-1. Int J Syst Evol Microbiol 55:2063–2068
Acknowledgments
We thank Dr. Rueedi, Dr. Martin, Dr. Blechschmidt, and Mr. Brand (Nagra) for coordination of our sampling. We are grateful to Mr. Tonooka Mrs. Shuin, and Mrs. Nakamura for their technical assistance with geochemical and microbial analysis. This study was supported by grants from the Nuclear and Industrial Safety Agency (NISA).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 80 kb)
Rights and permissions
About this article
Cite this article
Konno, U., Kouduka, M., Komatsu, D.D. et al. Novel Microbial Populations in Deep Granitic Groundwater from Grimsel Test Site, Switzerland. Microb Ecol 65, 626–637 (2013). https://doi.org/10.1007/s00248-013-0184-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-013-0184-5