Abstract
Water and nutrient availability are the major limiting factors of biological activity in arid and semiarid ecosystems. Therefore, perennial plants have developed different ecophysiological adaptations to cope with harsh conditions. The chemical profile of the root exudates varies among plant species and this can induce variability in associated microbial populations. We examined the influence of two shrubs species, Artemisia sieberi and Noaea mucronata, on soil microbial diversity. Soil samples were collected monthly, from December 2006 to November 2007, near canopies of both shrubs (0–10-cm depth). Samples were used for abiotic tests and determination of soil bacterial diversity. No significant differences were found in the abiotic variables (soil moisture, total organic matter, and total soluble nitrogen (TSN)) between soil samples collected from under the two shrubs during the study period. No obvious differences in the Shannon–Weaver index, evenness values, or total phylogenetic distances were found for the soil microbial communities. However, detailed denaturing gradient gel electrophoresis (DGGE) clustering as well as taxonomic diversity analyses indicated clear shifts in the soil microbial community composition. These shifts were governed by seasonal variability in water availability and, significantly, by plant species type.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The Negev Desert is a xeric environment in which the timing of rainfall is predictable (October to late April), while rainfall amount, distribution, and intensity are unpredictable. One of the most predictable sources of moisture is dew (210 days/year, mainly in late summer and autumn), which enhances water availability and triggers an active phase in a wide range of biological populations.
Soil organic matter content is known to be relatively low in these ecosystems (~100 g m−2) due to low nutrient availability [1–3]. According to Larson and Pierce [4], Steinberger and Loboda [5], Steinberger [6], and Cornwell et al. [7], the presence of organic matter, its patchy distribution, and its variation in chemical composition play an important role in soil biotic composition and activity. The interaction between moisture and organic matter availability is one of the major features determining soil biotic functions in arid and semiarid ecosystems [8–11].
In desert ecosystems, water and organic matter availability are among the major factors limiting primary production and soil biotic ability to fulfill their biological functions [2, 12–14]. Therefore, plants and soil biota have developed an adaptive ability to perform their biological role and function to suitable conditions in time and space in order to overcome the extreme environment [11]. This feature is similar to that found in perennial plants, in which ecophysiological adaptations also evolved in order to cope with the same harsh environment. These ecophysiological adaptations, which include salt (Reaumuria negevensis) and chemical (Artemisia sieberi) compound excretion beneath the plant canopy, provide an allelopathic environment that affects organic matter and soil moisture levels.
Recent studies by Zhou et al. [15] and Bagayoko et al. [16] contributed to our understanding of the effect of soil parameters such as soil type, soil temperature, water content, organic matter, pH, and nutrient availability on the soil microorganism community and its diversity. Additional studies have described the effect of plants on soil bacterial community composition [17–20], emphasizing the impact of the plant roots as one of the most important factors affecting soil biota dynamics [21–24]. According to Haichar et al. [24], plants produce diverse chemical compounds and signaling molecules to defend themselves against pathogens and attract useful organisms. Moreover, according to Smalla et al. [18], Sørensen [25], Nunan et al. [23], Jaeger et al. [26], and Lynch and Whipps [27], root exudates can stimulate the growth of bacteria under the plant and can affect composition and functional diversity in the rhizosphere.
The objective of this study was to determine the effect of plant ecophysiological adaptation on soil microbial diversity in a natural desert environment. In order to achieve this, soil samples were collected under the canopy of two widespread shrub populations representing the Negev arid ecosystem, A. sieberi and Noaea mucronata. The A. sieberi shrub is known to excrete chemical compounds (allelopathic adaptation) that act as germination inhibitors for other plants in their surroundings [28], while the second shrub, N. mucronata, is not known to induce allelopathic effects. Both shrubs share the same biotope and are found to have a similar density.
Based on the above, we hypothesized that the significant differences in plant ecophysiological allelopathic adaptation induce a considerable effect on soil bacterial community structure. Similarity analyses would also reveal the role of water activity in the soil bacterial community.
Study Site
The study site was located in the northern Negev Desert at the M. Evenari Runoff Research Farm (34°46′E/30°47′N), Avdat, Israel. The area consists of loess plain rocky slopes with shallow, saline, gray, lithogenic, calcareous soils. The soil at the study site is an alkaline (pH 7.8), deep, fine-textured loessial sierozem, with small amounts of organic carbon (0.47 %) and large amounts of carbonate (40 %). The climate is Mediterranean, with mild, rainy winters (5–14 °C in January) and hot summers (18–32 °C in June). The multiannual evaporation rate is 2,615.3 mm. The rainy season usually begins in October and ends in late April, with most of the rainfall occurring as scattered showers between December and February. The multiannual mean rainfall is 90 mm and fluctuates between 34 mm in a drought year and 187 mm in an extremely rainy year. The multiannual mean amount of dew at this location contributes an amount of water equal to 35 mm of rainfall. Dewfall occurs during approximately 210 nights per year, mainly in late summer and autumn. The study site is dominated by desert–shrub associations in which the most common species are R. negevensis, Tamarix aphylla, A. sieberi, and N. mucronata [12].
Materials and Methods
The study was conducted over a period of 12 months, between December 2006 and November 2007. The data obtained were divided into four seasonal periods: winter (December 2006 and January and February 2007), spring (March, April, and May 2007), summer (June, July, and August 2007), and autumn (September, October, and November 2007).
Soil samples were randomly collected at the study site below individual plant canopies (n = 4) of N. mucronata and A. sieberi from the 0–10-cm soil layer with a soil auger (8-cm diameter). The distance between each shrub was a minimum of 3 m in order to prevent intershrub interactions. All shrubs had similar canopy size and there was no significant difference in the root system between the plant types.
At the field site, each soil sample was placed in an individual plastic bag and stored in a cool insulated box until arrival at the laboratory. After sieving through a 2-mm mesh in order to remove stones, roots, and other organic debris, the soil samples were stored at 4 °C until they were used for abiotic and biotic analyses. DNA was extracted from the soil samples within a maximum 7 days after collection.
Each soil sample was subjected to the following analyses:
Soil Moisture
Gravimetric determination of soil water content was carried out by drying the samples at 105 °C for 48 h (Fig. 1).
Total Organic Matter
Total organic matter was determined by oxidation with dichromate in the presence of H2SO4 without application of external heat, as described by Rowell [29] and Lahav and Steinberger [30].
Total Soluble Nitrogen
Total soluble nitrogen (TSN) was determined by chemical extraction and color reactions using a Skalar autoanalyzer [31] (Fig. 1).
DNA Extraction
DNA was extracted from the soil samples using the BioLab kit according to the manufacturer’s purification protocol. The DNA was then purified by electrophoresis on 1 % agarose gel and isolated with a NucleoTrap gel extraction kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s purification protocol. The quality and concentration of DNA were confirmed by gel electrophoresis on a 1 % agarose gel.
Polymerase Chain Reaction Amplification
For DNA sequence diversity analyses, 16S rRNA genes were amplified from extracted DNA samples. Each 50 μl reaction had 0.6–1 DNA μl template, 1.5 U Taq DNA polymerase (Red Taq; Sigma-Aldrich, St. Louis, MO, USA), 5 μl Taq buffer (Sigma-Aldrich), 6.25 μg bovine serum albumin (BSA) (Roche Diagnostics, Mannheim, Germany), 2 μM polymerase chain reaction (PCR) nucleotide mixture (dNTP) (Larova, Telpow, Germany), 2 mM MgCl2 (AMRESCO, USA), and 50 μM of the bacterial primer pair 341F (GCC TAC GGG AGG CAG CAG) and 907R (GTT TGA TCM TGG CTC AG). The program included initial denaturation at 95 °C for 5 min, followed by 33 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 30 s. The cycling was completed with a final elongation step at 72 °C for 5 min. The PCR reaction was performed in the TGradient Thermocycler (Whatman Biometra, Göttingen, Germany). The PCR products were visualized on 1.5 % agarose gel and purified with a Nucleospin plasmid purification kit (Macherey-Nagel) according to the manufacturer’s purification protocol. The purified products were then ligated using the Topo PCR cloning kit, which includes the pCRII-TOPO TA vector (Invitrogen/Life Technologies, Carlsbad, CA, USA). Amplicons for DGGE analysis were obtained using the same primer set but with a GC clamp attached to the 5′ end of the forward primer (Fig. 2).
Sequence Analysis
Nucleotide sequences of rDNA clones were obtained with an ABI 3730XL BigDye terminator v3.1 cycle sequencer at the Genome Center at Washington University. Plasmid DNA was extracted with the Perfectprep Plasmid 96 (×4) Vac Kit (5prime).
Each set of unique sequences associated with a soil sample was compared against the National Center for Biotechnology Information (NCBI) database via BLASTN [32] through the Community Cyberinfrastructure for Advanced Microbial Ecology Research & Analysis site (CAMERA, http://camera.calit2.net/; [33]. All sequences are available at CAMERA under the accession number CAM_P_0000948.
Microbial Diversity
We employed several levels of analysis, from simple to complex, in order to gain understanding of the microbial diversity within our system.
Denaturing Gradient Gel Electrophoresis Analysis
Denaturing gradient gel electrophoresis (DGGE) of the 566-bp 16S rRNA PCR amplicons was carried out on a DCode system (Bio-Rad, Hercules, CA, USA) using the following components: 1× TAE buffer (40 mM Tris–HCl, 20 mM acetic acid, 1 mM ethylene diamine tetraacetic acid (EDTA) [pH 8.3]), and 1-mm-thick polyacrylamide gels (6 %). The gels contained a 30–60 % denaturing gradient and were electrophoresed for 17 h at 100 V at 60 °C. The gels were stained with ethidium bromide and photographed on an ultraviolet (UV) transillumination table (302 nm) with a Kodak digital camera (Kodak Co., New Haven, CT, USA), with imaging software provided by the supplier. Image analysis was carried out with LabImage software, version 2.7.1 (Kapelan GmbH, Halle, Germany).
Ecological Diversity Index
Initially, sequences were manually assigned to taxonomic levels. Species level was considered for sequences indicating a 96–100 % similarity and query coverage. A 93–95 % similarity was attributed to genus and up to 90 % to families. If less than 90 % similarity was found, the sequence was named soil bacteria. This dataset was used to assess species diversity (α diversity) calculated by the Shannon–Weaver formula [34]:
where P i is the ratio of the population of a particular bacterial species and the sum of the bacterial species.
Species evenness was calculated using Pielou’s evenness index [35]. A value close to 1 means that the sample is diverse, while a value close to 0 means that the sample is not diverse:
The similarity between the samples (β diversity) was calculated by Sørensen’s similarity index [36].
Taxonomic Diversity
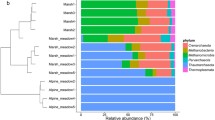
The taxonomic classification network trees for all samples were then directly compared in MEGAN 4 [37]. For this, the BLASTN files were imported into MEGAN 4, where each query sequence was assigned, according to its BLASTN similarity and bit score, to the appropriate node in the NCBI taxonomic network tree (a summary of phyla and class assignments is available in Table 1). Given the uniqueness of each sequence in a given sample set, we used a minimum taxonomic support of 1 for the least common ancestor algorithm [38]. Thus, each sequence was, in effect, a distinct operational taxonomic unit (OTU) assigned to an NCBI node according to its putative identity (Table 1). The normalized Euclidean distance matrix was exported into SplitsTree [39] and visualized as a UPGMA phylogram using the least square weights, the closest tree splits filter, and tree selector transformation (Fig. 3). Note that this approach eliminated about 22.8 % of the OTUs that could only be assigned to the kingdom Bacteria. Of the remaining OTUs, 476 could be assigned to a phylum and 433 could be assigned to a class.
Aggregate taxonomic distance network obtained from the comparison of the sum of NCBI-based taxonomic distances as assigned in MEGAN [37]. Note that 22.8 % of all sequences that were assigned to Kingdom Bacteria were not used. Fit = 0.94 (ratio between the total length of the fitted distances versus the total length of the taxonomic distances according to the NCBI network)
Total Taxonomic Diversity via Operational Taxonomic Unit Alignment
As the approach presented above did not include the OTUs assigned only at kingdom level, an alignment of all OTU sequences was performed to estimate the total diversity independent of putative identities. Duplicate sequences were clustered using the CD-HIT-454 program [40], as integrated in CAMERA. Sequences that were found to be similar over more than 96 % of their length were considered similar and thus clustered together. Only the longest representative sequence from each cluster was retained for alignment. The obtained FASTA file was used to produce an alignment using ClustalW [41], as integrated in Geneious Pro 5.4.6 (Biomatters Ltd., Auckland, New Zealand). A maximum likelihood tree was then prepared by employing the PhyML program in Geneious with a Jukes–Cantor substitution model and bootstrap (100 cycles) optimized for tree topology, branch length, and base substitution rate [42]. The sequence sample identities removed during the CD-HIT-454 processing were manually reassigned to the appropriate location in the tree. This tree, thus, integrated all sequences without inferring any taxonomic assignment (see Supplementary material). This allowed for the calculation of total phylogenetic diversity [43, 44] in SplitsTree (Table 2).
Relationship Between Taxonomic Diversity, Shrub Species, and Sampling Season
The assignments obtained via the MEGAN 4 pipeline, as described above, were used to assess the role of plant species and season in the taxonomic richness of specific taxonomic units. For this, assignments for each season and plant treatment were grouped at phyla or class levels and used in principal component analysis (PCA) based on factor covariance using Minitab® and Genstat™. The datasets represent counts of putatively unique taxa, OTUs. Thus, the PCA compares variability in the putative taxonomic diversity within each phylum or class as induced by the considered treatments. Phyla with the greatest variances among the samples are likely to be the ones that best explain variability. The PCA results are presented in Figs. 4 and 5.
Principal component analysis of the variability in taxonomic diversity within bacterial phyla as associated with shrub species and seasons. Acidobacteria, Cyanobacteria, Deinococcus-Thermus, Planctomycetales, and Verrucomicrobia had very low richness (each was represented by a total of less than 4 OTUs for all samples) and thus, not included in this analysis
Principal component analysis of the variability in taxonomic diversity within bacterial classes as associated with shrub species and seasons. Bacteroidia, Opitutae, Anaerolinea, Chloroflexi, Dehalococcoidetes, Deinococci, Bacilli, Negativicutes, and Planctomycetacia had very low richness (each represented by a total of less than 3 OTUs for all samples) and thus, not included in this analysis
Results
Rainfall
During the study period (December 2006 to November 2007), a total rainfall amount of 170.42 mm, which is almost twofold higher than the multiannual average of 90 mm year−1, was measured. The rainfall levels ranged from 3.9 mm in May to a maximum of 57.3 mm in April, with relatively high cumulative monthly rainfall.
Soil Moisture and Organic Matter
No significant differences (p > 0.05) in soil moisture were found between the soil samples collected in the vicinity of N. mucronata and A. sieberi during the study period (Table 3, Fig. 1). The highest values in the two different soil samples were obtained in winter and spring (ranging from 5.46 to 7.18 %) and the lowest values were obtained in summer and autumn (ranging from 1.64 to 2.07 %) (Fig. 1).
Similar to soil moisture, no significant differences in total organic matter were found (p > 0.05) between the soil samples collected under the A. sieberi and N. mucronata plants, with values ranging from 0.44 to 1.18 %. Furthermore, no significant differences in TSN were obtained between the two shrubs throughout the year (Table 3, Fig. 1). TSN values in samples collected under N. mucronata ranged from 3.46 to 7.55 μg TSN g−1 dry soil. While not significantly different, TSN in samples collected under A. sieberi exhibited smaller fluctuations, with values ranging from 5.13 to 8.07 μg TSN g−1 dry soil in comparison to N. mucronata.
Total Soil Bacterial Population Diversity
Seasonal differences in H′ values (p > 0.05) were found between the soil samples collected in the vicinity of N. mucronata and A. sieberi (Table 3, Fig. 6). Evenness values ranged between 0.89 and 0.98 E H in samples collected in the vicinity of the two shrubs. This sheds light on the existence of broad microbial diversity (Fig. 6). No significant seasonal effect on evenness values was obtained between N. mucronata and A. sieberi (Table 3).
Significant maximal Sørensen’s similarity values were found between the soil samples collected in the vicinity of N. mucronata and A. sieberi in winter, with 0.4 β diversity. In the summer, spring, and autumn, the values were almost twofold lower than in winter (0.19, 0.19, and 0.21 β diversity, respectively). This is very similar to the results obtained from evenness analysis (Fig. 6).
Cluster analysis (DGGE) indicated that the effects of the shrub-associated soil microbial diversity are seasonally dependent. This was shown by the close relationship between the fingerprints obtained during the spring, summer, and autumn while the winter fingerprints clustered independently (Fig. 2). Aggregate taxonomic distance assessment for the sequences assigned to taxonomic levels by MEGAN 4 confirmed the distinctiveness of the winter diversity profiles from the diversity profiles of the rest of the year (Fig. 3). Moreover, within both clusters, plant species played a clearly distinct role in the differentiation of the bacterial diversity profile. Reviewing the DGGE clustering in light of the information from Fig. 3 strengthens this conclusion. These results also show that DGGE, although a relatively lower sensitivity method, did largely delineate seasonal variability in bacterial diversity, similar to more detailed sequencing-based methods but was less sensitive to the role of plant species.
Total phylogenetic diversity, which did not account for taxonomic identities and thus included all OTUs, was calculated as a total distance between all OTUs aligned in the phylogenetic tree, as described (see Supplementary material). These values were only slightly different across the considered variables (Table 2), suggesting comparable total diversities. Overall, A. sieberi soil samples were associated with a slightly greater phylogenetic diversity index, but the functional or statistical significance of this is unclear.
Thus, substantially similar total phylogenetic diversities (Table 3) were associated with more significant differences, as can be induced from the results of tests that considered sequence diversity (Fig. 6) and taxonomic assignments (Fig. 3). This indicates that while total diversity parameters may not vary greatly across the considered experimental variables, differences in soil bacterial diversity across seasons and plant species are more likely induced by shifts in taxonomic identities.
Soil Bacterial Taxonomic Diversity Shifts
We employed PCA to verify the role of our assessed parameters, plant species and season, in any shift in taxonomic identities (Figs. 4 and 5). Across phyla breakdown, of the total OTUs, 35.7 % were assigned to Actinobacteria, 17.2 % to Proteobacteria, 12.3 % to Bacteroidetes, 4.9 % to Gemmatimonadetes, 3.4 % to Chloroflexi, and 2.1 % to Firmicutes (Table 1).
The phylum-level PCA analysis shows that Bacteroidetes and Actinobacteria dominate the first two principal components, which describe over 91 % of the taxonomic diversity among the samples (Fig. 4). A minor role, and thus associated with a smaller taxonomic diversity shift, can also be attributed to Proteobacteria and Gemmatimonadetes. A more detailed analysis at class level shows that under the observed natural conditions, most taxonomic diversity variability were associated with Actinobacteria (Actinobacteria), Flavobacteria and Cytophagia (both Bacteroidetes), and Gemmatimonadetes. The latter is a relatively novel taxonomic group, classified at phylum level with no defined subdivisions and represented mainly by environmental isolates distributed both in soil and water environments [45]. Given its overall significance at the phylum level, it was also used in the analysis at class level. Proteobacteria phylum exhibited increased taxonomic diversity during the warmer seasons, with little obvious difference between the two plant species. Among the Proteobacteria phyla, the variations in the taxonomic diversity of Alphaproteobacteria were the better predictors of seasonality. During this whole test period, only Deltaproteobacteria seemed to be more diverse under A. sieberi (Table 1) but overall, this still described a very small proportion of the total diversity shift. The PC4, which was most obviously associated with Deltaproteobacteria and also with Gammaproteobacteria and Betaproteobacteria, explained less than 4 % of the total taxonomic variability (Fig. 5).
For A. sieberi-associated soil samples, the winter season correlated with the largest increase in the taxonomic diversity within Bacteroidetes phylum (Figs. 4, 5, and Supplementary material), specifically Flavobacteria and Cytophagia classes, and with decreases in the taxonomic diversity of Actinobacteria. At the same time, the taxonomic diversity within the Proteobacteria declined. For N. mucronata, the pattern of changes in taxonomic diversity was more complex and less obvious; increases in the diversity of Gemmatimonadetes and Sphingobacteria as well as decreases in Actinobacteria diversity seemed to predict winter conditions for this shrub species. Thus, shifts in the taxonomic diversity associated with Actinobacteria phylum and some classes within Bacteroidetes (Flavobacteria and Cytophagia) were the ones most responsive to variations in the tested conditions, while the variability in the taxonomic diversity within Proteobacteria was less affected. These patterns indicate commonalities likely associated with the effect of seasonal variability on water activity and differences likely associated with the effect of the plant species.
Given that it is very likely that recovery of sequences from soil may preferentially be skewed towards the more dominant bacteria [46], these results do not necessarily indicate absolute absence of certain OTUs but clearly represent the OTUs that dominate the tested systems. The lack of significance in the amounts of TSN or organic matter content between treatments (Table 3), while simplifying the test system, precluded an estimation of their role for any taxonomic diversity shift.
Discussion
As mentioned by McGuire and Treseder [47], any given ecosystem can potentially colonize an area via vertical and horizontal dispersion and migration, where abiotic (climate and soil parent material) and plant cover divergence on a temporal scale will actively affect soil biotic function and composition (diversity). Hence, shrub cover plays a multifunctional role in xeric systems, first, being one of the main organic matter donors and second, providing a physical barrier (shelter) that allows prolonged soil biotic activity [11, 14, Berg and Steinberger, 2011—personal communication]. Moreover, the above factors are among the major causes inducing change in soil bacterial diversity [48–51]. It is known that plants contribute carbohydrate compounds originating from roots, dead cells, and foliage to the soil under their canopy. According to Steinberger et al. [52], carbohydrates constitute 10 % of soil organic matter. Therefore, plant rhizospheres are hotspots concentrating the largest amounts of C compounds in soil [53, 54]. The chemical profiles of the root exudates may vary with plant species even for plants exposed to similar environmental conditions, and this may affect the diversity of organisms associated with exudating roots [55, 56].
The work of Jacobson and Jacobson [57] conducted in the Namib Desert indicates that one of the limiting factors affecting decomposition processes in desert systems is water availability. Moreover, according to Bachelet et al. [58], Reynolds et al. [59, 60], and Whitford [14], it is not the size of the rainfall pulse or even the cumulative amounts that trigger biological activity, but rather a combination of timing and other events.
The standard ecological diversity indices and total phylogenetic diversities, as calculated here, do not seem to confirm the above findings, as no significant differences were found in the soil bacterial diversity based on the Shannon (H′) index and evenness analyses, or based on the total phylogenetic diversity between A. sieberi and N. mucronata shrubs or across seasons. On the other hand, analyses that consider the shifts in taxonomic identity, such as DGGE fingerprinting of 16S rDNA amplicons (Fig. 2), the aggregate taxonomic distance between the sequences’ assigned taxonomic identities (Fig. 3), and the PCA analyses (Figs. 4 and 5), confirm the role of seasonal variability in soil moisture and also the role of the plant species in inducing taxonomic diversity shifts for similar total phylogenetic diversities. The latter analysis also confirms the interactive role of season and plant species with A. sieberi, inducing obvious shifts in the diversity of Bacteroidetes during the winter. During the dry seasons, plant species also induced differential diversity in associated soils (Fig. 3) but this may have been obvious mostly for Actinobacteria. Any microbial diversity divergence induced by the specific composition of the exudates of the two plant species was mitigated by the advent of the wet season.
No significant differences were found in the abiotic variables (soil moisture, total organic matter, and TSN) between the two plant species. On the other hand, the similarity of the bacterial communities between the two different soil sample sources (Sørensen’s similarity) was low for most of the year (spring, summer, and autumn). While this varied for the other tests that we employed here, mostly as a consequence of the variable level of detail afforded by each of the specific analyses that we used, all results point to the differences in the soil bacteria population in the vicinity of the plant canopies to be induced by the plants themselves as a consequence of their different ecophysiological adaptations to the desert conditions. Along these lines, the chemical compounds excreted by A. sieberi below the canopy have greater influence on the bacterial genetic diversity while not necessarily on the bacterial functional diversity.
The high evenness values (0.89–0.98 E H) in the soil samples collected in the rhizosphere of the two shrubs and the relatively consistent phylogenetic diversity indices during all seasons indicate a relatively high abundance of species. Both DGGE analysis and the aggregate taxonomic distance network evaluation demonstrated that the soil bacterial diversities in spring, summer, and autumn were distinctly different from the winter samples. Based on the above, we assume that although the species richness and total phylogenetic diversities for the soil samples collected under the N. mucronata and A. sieberi canopies were similar during all seasons, the bacterial taxonomic diversities in the soil community were dissimilar.
A combination of lower exudate rates in the winter [60] and increased precipitation likely had a dilution effect on the root zone and thus, the differential impact induced by the two plant species declined.
In spite of the fact that no significant differences were found in the abiotic variables (soil moisture, total organic matter, and TSN) between the samples collected beneath the two shrub species, microbial diversity, including the major phyla, was different. This only strengthens our assumption that the ecophysiological adaptations of the shrubs influence the soil microbial community below the canopy.
According to Janssen [61], significant variability in microbial phyla may be associated with soil sampling location, although it is often difficult to state what the main factors governing such variability are, given the controlling role of microniche variability in determining diversity. In a study on canola soil samples from two different soil and climatic zones, Germida et al. [62] found that H′ and E H values were similarly related to bacterial diversity. However, lower H′ and E H values were obtained in soil samples collected from the wheat rhizosphere. They, therefore, suggested that bacterial diversity might be dependent on plant species and not on soil factors [62]. On the other hand, their research cannot confirm whether this observation is limited to canola or is valid for other plants as well.
According to Germida and Siciliano [63], Di Cello et al. [64], Latour et al. [65], and Westover et al. [66], bacterial diversity is influenced by root exudation and changes according to season. The different root exudates and the composition of the root cell components are linked to plant diversity [67]. The variety of organic compounds released by plants has been postulated to be one of the key factors affecting the diversity of microorganisms in the rhizosphere of different plant species [67–70]. However, little is known about the interaction between factors that influence soil microbial composition and the relationship between plant diversity and soil microbial diversity [71].
Conclusions
Our results support the assumption that there is a direct correlation between seasonal variability in abiotic conditions and microbial community diversity. Moreover, our study expounds the importance of rainfall as ‘one’ of the primary triggers leading to major changes in bacterial diversity. It was obvious that plant has also induced variability in the microbial community diversity, an effect mitigated by variability in water availability.
Additional and more targeted studies are required to elucidate the interactive role of nonmicrobial biota on soil microbial diversity, and especially its activity, in order to allow the development of tools and management strategies to alleviate the effect of shifts in plant community composition, trophic shifts, and variable nutrient availability induced by plant physiological tradeoffs in xeric ecosystems.
References
Noy-Meir I (1973) Desert ecosystems: environment and producers. Ann Rev Ecol Syst 4:25–51
Noy-Meir I (1974) Desert ecosystems: higher trophic levels. Ann Rev Ecol Syst 5:195–214
West NE, Skujins J (1978) Nitrogen in desert ecosystems, US/IBP no. 9. Dowden, Hutchinson and Ross, Stroudsburg
Larson WE, Pierce FJ (1994) The dynamics of soil quality as measure of sustainable management. In: Doran DW, Coleman DC, Bendicek DF, Stewart BA (eds) Defining soil quality for sustainable environment. Soil Science Society of America Inc. and American Society of Agronomy Inc., Madison, pp 37–51
Steinberger Y, Loboda I (1991) Nematode population dynamics and trophic structure in a soil profile under the canopy of the desert shrub Zygophyllum dumosum. Pedobiologia 35:191–197
Steinberger Y (1995) Soil fauna in arid ecosystems: their role and functions in organic matter cycling. Adv GeoEcol 28:29–36
Cornwell WK, Cornelissen JHC, Amatangelo K, Dorrepaal E, Eviner VT, Godoy O, Hobbie SE, Hoorens B, Kurokawa H, Perez-Harguindeguy N, Quested HM, Santiago LS, Wardle DA, Wright IJ, Aerts R, Allison SD, van Bodegom P, Brovkin V, Chatain A, Callaghan TV, Diaz S, Garnier E, Gurvich DE, Kazakou E, Klein JA, Read J, Reich PB, Soudzilovskaia NA, Vaieretti MV, Westoby M (2008) Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol Lett 11:1065–1071
Buyanovsky G, Dicke M, Berwick P (1982) Soil environment and activity of soil microflora in the Negev Desert. J Arid Environ 5:13–28
Coleman DC (1985) Through a ped darkly: an ecological assessment of root-soil-microbial-faunal interactions. In: Fitter AH, Atkinson D, Read DJ, Usher MB (eds) Ecological interactions in soil. Blackwell, Cambridge, pp 1–21
Beare MH, Parmelee RW, Hendrix PF, Cheng W (1992) Microbial and fungal interactions and effects on litter nitrogen and decomposition in agroecosystems. Ecol Monogr 62:569–591
Sarig S, Steinberger Y (1993) Immediate effect of wetting event on microbial biomass and carbohydrate production mediated aggregation in desert soil. Geoderma 56:599–607
Evenari ME, Shanan L, Tadmor W (1982) The Negev: the challenge of a desert. Harvard University Press, Cambridge
Shmida A, Evenari M, Noy-Meir I (1986) Hot desert ecosystems. An integrated view. In: Evenari M, Noy-Meir I, Goodall DW (eds) Ecosystems of the world: hot deserts and arid shrublands. Elsevier, Amsterdam, pp 379–387
Whitford WG (2002) Ecology of desert systems. Academic, New York
Zhou JZ, Davey ME, Figueras JB, Rivkina E, Gilichinsky D, Tiedje JM (1997) Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913–3919
Bagayoko M, Alvey S, Neumann G, Buerkert A (2000) Root-induced increases in soil pH and nutrient availability to field-grown cereals and legumes on acid sandy soils of Sudano-Sahelian West Africa. Plant Soil 225:117–127
Stephan A, Meyer AH, Schmid B (2000) Plant diversity affects culturable soil bacteria in experimental grassland communities. J Ecol 88:988–998
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751
Costa R, Gotz M, Mrotzek N, Lottmann J, Berg G, Smalla K (2006) Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol Ecol 56:236–249
Loranger-Merciris G, Barthes L, Gastine A, Leadley P (2006) Rapid effects of plant species diversity and identity on soil microbial communities in experimental grassland ecosystems. Soil Biol Biochem 38:2336–2343
McCaig AE, Glover LA, Prosser JI (1999) Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol 65:1721–1730
Kowalchuk GA, Buma DS, de Boer W, Klinkhamer PGL, van Veen JA (2002) Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Anton Leeuw Int J G 81:509–520
Nunan N, Daniell TJ, Singh BK, Papert A, McNicol JW, Prosser JI (2005) Links between plant and rhizoplane bacterial communities in grassland soils, characterized using molecular techniques. Appl Environ Microbiol 71:6784–6792
Haichar FE, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230
Sorensen J (1997) The rhizosphere as a habitat for soil microorganisms. In: van Elsas JD, Trevors JT, Wellington EMH (eds) Modern soil microbiology. Marcel Dekker, New York, pp 21–45
Jaeger CH, Lindow SE, Miller S, Clark E, Firestone MK (1999) Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and Tryptophan. Appl Environ Microbiol 65:2685–2690
Lynch JM, Whipps JM (1990) Substrate flow in the rhizosphere. Plant Soil 129:1–10
Friedman J (1995) Allelopathy, autotoxicity, and germination. In: Kigel J, Galili G (eds) Seed development and germination. Marcel Dekker, NY, pp 599–628
Rowell DL (1994) Soil science: methods and applications. Longman, London
Lahav I, Steinberger Y (2001) Soil bacterial functional diversity in a potato field. Eur J Soil Biol 37:59–67
S.F.A.S. (1995) Manual—SAN Plus analyzer. Skalar Analytical, The Netherlands
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Sun S, Chen J, Li W, Altinatas I, Lin A, Peltier S, Stocks K, Allen EE, Ellisman M, Grethe J, Wooley J (2011) Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res 39(Suppl 1):D546–D551
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Pielou EC (1966) The measurement of diversity in different types of biological collections. Statistical Research Service, Canada Department of Agriculture, Ottawa
Sorensen T (1948) A method of establishing groups of equal amplitude in plant society based on similarity of species content. K Danske Vidensk Selsk Biol SKR 5:1–34
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Niu B, Fu L, Sun S, Li W (2010) Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinform 11:187
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10
Faith DP (2006) The role of the phylogenetic diversity measure, PD, in bio-informatics: getting the definition right. Evol Bioinform 2:277–283
Zhang H, Sekiguchi Y, Hanada S, Hugenholtz P, Kim H, Kamagata Y, Nakamura K (2003) Gemmatimonas aurantiaca gen. nov., sp nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int J Syst Evol Microbiol 53:1155–1163
Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, Catroux G (2001) DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol 67:2354–2359
McGuire KL, Treseder KK (2010) Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol Biochem 42:529–535
Maloney PE, van Bruggen AHC, Hu S (1997) Bacterial community structure in relation to the carbon environment in lettuce and tomato rhizosphere and in bulk soil. Microb Ecol 34:109–117
Hertenberger G, Zampach P, Bachmann G (2002) Plant species affect the concentration of free sugars and free amino acids in different types of soil. J Plant Nutr Soil Sci 165:557–565
Buckley DH, Schmidt TM (2003) Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ Microbiol 5:441–452
Johnson MJ, Lee KY, Scow KM (2003) DNA fingerprinting reveals links among agricultural crops, soil properties, and the composition of soil microbial communities. Geoderma 114:279–303
Steinberger Y, Vishnevetsky S, Barness G, Lavee H (1998) Effects of topoclimatic gradient on soil dehydrogenase activity in a Judean Desert ecosystem. Arid Soil Res Rehab 12:387–393
Dilly O, Buscot F, Varma A (2005) Microbial energetics in soils. Soil biology, volume 3, Microorganisms in soils: roles in genesis and functions. Springer, Berlin, pp 123–138
Jones DL, Murphy DV (2007) Microbial response time to sugar and amino acid additions to soil. Soil Biol Biochem 39:2178–2182
Broeckling CD, Broz AK, Bergelson J, Manter DK, Vivanco JM (2008) Root exudates regulate soil fungal community composition and diversty. Appl Environ Microbiol 74:738–744
Brimecombe MJ, De Leij FA, Lynch JA (2001) The effect of root exudates on rhizosphere microbial populations. In: Pinton R, Varanini Z, Nannipieri P (eds) The rhizosphere: biochemistry and organic substances at the soil–plant interface. Marcel Dekker, Inc., NY, pp 95–104
Jacobson KM, Jacobson PJ (1998) Rainfall regulates decomposition of buried cellulose in the Namib Desert. J Arid Environ 38:571–583
Bachelet D, Brugnach M, Neilson RP (1998) Sensitivity of a biogeography model to soil properties. Ecol Model 109:77–98
Reynolds JF, Virginia RA, Kemp PR, de Soyza AG, Tremmel DC (1999) Impact of drought on desert shrubs: effects of seasonality and degree of resource island development. Ecol Monogr 69:69–106
Reynolds JF, Kemp PR, Ogle K, Fernandez RJ (2004) Modifying the ‘pulse-reserve’ paradigm for deserts of North America: precipitation pulses, soil water, and plant responses. Oecologia 141:194–210
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Germida JJ, Siciliano SD, de Freitas JR, Seib AM (1998) Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.). FEMS Microbiol Ecol 26:43–50
Germida JJ, Siciliano SD (2001) Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol Fertil Soils 33:410–415
Di Cello F, Bevivino A, Chiarini L, Fani R, Paffetti D, Tabacchioni S, Dalmastri C (1997) Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl Environ Microbiol 63:4485–4493
Latour X, Corberand TS, Laguerre G, Allard F, Lemanceau P (1996) The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl Environ Microbiol 62:2449–2456
Westover KM, Kennedy AC, Kelley SC (1997) Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J Ecol 85:863–873
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Bowen GD, Rovira AD (1991) The rhizosphere—the hidden half of the hidden half. In: Waisel Y, Eshel A, Kafkafi U (eds) Plant roots—the hidden half. Marcel Dekker, New York, pp 641–649
Bolton H Jr, Fredrickson JK, Elliott LF (1994) Microbial ecology of the rhizosphere. In: Metting FB Jr (ed) Soil microbial ecology. Marcel Dekker, New York, pp 27–63
Grayston SJ, Wang S, Campbell CD, Edwards AC (1998) Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30:369–378
Carney KM, Matson PA (2005) Plant communities, soil microorganisms, and soil carbon cycling: does altering the world belowground matter to ecosystem functioning? Ecosystems 8:928–940
Acknowledgments
The authors thank Lilach Iasur, Hila Elifantz, and Patricia Bucki (students of Dr. Dror Minz), and Dr. Einav Mayzlish-Gati for consultation and guidance. Many thanks go to Dr. Orit Shaul for providing the necessary equipment. Special thanks to Ms. Sharon Victor for language editing.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper is dedicated in loving memory to Saul Saul (father of V.S.-T.).
Subject category: Microbial population and community ecology: environmental factors (biotic and abiotic) defining the distribution and abundance of microbial populations.
Rights and permissions
About this article
Cite this article
Saul-Tcherkas, V., Unc, A. & Steinberger, Y. Soil Microbial Diversity in the Vicinity of Desert Shrubs. Microb Ecol 65, 689–699 (2013). https://doi.org/10.1007/s00248-012-0141-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0141-8