Abstract
In this report, the diversity of oil-degrading bacteria and alkB gene was surveyed in the seawater around Xiamen Island. Forty-four isolates unique in 16S rRNA sequence were obtained after enrichment with crude oil. Most of the obtained isolates exhibited growth with diesel oil and crude oil. alkB genes were positively detected in 16 isolates by degenerate polymerase chain reaction (PCR). And for the first time, alkB genes were found in bacteria of Gallaecimonas, Castellaniella, Paracoccus, and Leucobacter. Additional 29 alkB sequences were retrieved from genomic DNA of the oil-degrading communities. Phylogenetic analysis showed that the obtained alkB genes formed five groups, most of which exhibited 60–80% similarity at the amino acid level with sequences retrieved from the GenBank database. Furthermore, the abundance of alkB genes in seawater was examined by real-time PCR. The results showed that alkB genes of each group in situ ranged from about 3 × 103 to 3 × 105 copies L−1, with the homologs of Alcanivorax and Pseudomonas being the most predominant. Bacteria of Alcanivorax, Acinetobacter, and Pseudomonas are important oil degraders in this area; while those frequently reported in other area, like Oleiphilus spp., Oleispira spp., and Thalassolituus spp. were not found in our report. These results indicate that bacteria and genes involved in oil degradation are quite diverse, and may have restriction in geographic distribution in some species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Alkane-degrading bacteria are ubiquitous in environments and play a crucial role in oil removal. In marine environments, they are frequently documented as, but not restricted to, Alcanivorax spp., Oleiphilus spp., Oleispira spp., and Thalassolituus spp. [3]. They can use alkanes, of various chain lengths, branched and/or straight, as sole carbon and energy sources [28, 29]. Recently, bacteria of Brachybacterium, Idiomarina, Leifsonia, Martelella, Kordiimonas, Parvibaculum, and Tistrella have been detected as new oil degraders from pelagic surface water of Atlantic Ocean [31].
While the mechanism of alkane mineralization has been not fully understood, it is known that alkane hydroxylase (AlkB) is one of the key enzymes in the process [15]. It has been detected in α-, β-, and γ-proteobacteria as well as the Actinobacteria [21, 27, 30]. Although several hydroxylases have been confirmed recently to be involved in alkane degradation, such as P450 [10], AlmA [23, 34], and Lad A [1], AlkB is most important and prevalent in aerobic oil-degrading bacteria. Actually, little is known about the diversity of alkB genes in marine environments. Recently, from the sediments of the Timor Sea, novel AlkB genes were retrieved, with identities averaged only 73% with those previously identified from marine α-proteobacteria [32]. Quite diverse alkB genes were also found in the bacteria isolated from oil-degrading communities across Atlantic Ocean; the alkBs from Salinisphaera isolates formed a separate cluster and shared only 54–69% amino acid identity with that from Nocardia farcinica IFM10152 as a closest relative in GenBank [31].
AlkB gene can be used as molecular marker to detect the ecological role of oil-degrading bacteria in environments. All AlkB proteins are conserved in six hydrophobic stretches that are likely to span the cytoplasmic membrane [18], and eight to nine histidines that are essential for the alkane-hydroxylizing activity [26]. Based on these conserved moieties, degenerate primers for polymerase chain reaction (PCR) detection of alkB genes have been designed [21]. With these primers, the alkB gene has been detected in a variety of environments, including Mississippi shallow aquifers [20], California soil [19], Arctic and Antarctic soil [35], Brazilian soils [9], German barley fields and grassland soil [7], Mediterranean beaches [14], Alaskan and Antarctic marine sediments [6], and recently in the Timor Sea [32]. Moreover, the alkB gene can be used to predict the potential of bioattenuation of oil-polluted environments. A relationship between alkB gene abundance and n-alkane degradation has been confirmed [4, 17]. Quantification of alkB can be developed to monitor microbial community change during bioremediation of hydrocarbon-contaminated Antarctic soil [13]. The knowledge about the diversity of alkB gene as well as their hosting bacteria in certain area helps to evaluate the potential of this area to recover from an oil spill accident.
Oil spill occurs along the coastline here and there and unpredictably. Xiamen Island is a unique niche to examine marine oil-degrading bacterium, as it locates the subtropical area in the Taiwan Straits which links East China Sea and Southern China sea. Moreover, it is also a large international port, and the estuarine of Jiu-Long River. Pollutions of various sources are unavoidable and rouse public concerns [5]. The ranges for petroleum hydrocarbons in the sediments of Xiamen Harbour was 133–943 μg g−1 (dw) [12], and 10 ∼ 14 μg L−1 in surface water (2005, unpublished). However, little is known about the oil-degrading bacteria in this area. In 2005, we collected the surface water around the island to examine the diversity of oil-degrading bacteria, as well as their abundance reflected by the copy numbers of alkB genes in situ.
Materials and Methods
Sampling
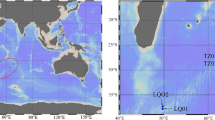
Surface seawater was collected with sterilized bottles in June 2005 around Xiamen Island, which is located at the mainland side of the Taiwan Straits (118.06–118.10°E, 24.40–24.50°N) (Fig. 1). Water samples were collected at 11 sites, one of which (arrowhead in Fig. 1) was used for quantification of the copy numbers of alkB genes. Other sites were used to enrich oil-degrading bacteria. For quantification of alkB genes, three bottles of seawater samples were collected from the same site and treated separately throughout the experiments. The seawater quality parameters were measured according to universal methods [4, 33]: the chemical oxygen demand (COD) was 0.5 ∼ 0.9 mg/L, the concentration of hydrocarbons was 10 ∼ 14 μg/L, N in the form of nitrate was 2 ∼ 4 mg/L, and chlorophyll a was 3 ∼ 6 μg/L. Representatively, at the site labeled an arrow head: the pH was 7.8, the temperature was 27°C, the COD was 0.7 mg/L, the salinity was 2.9%, the concentration of hydrocarbons was 12 μg/L, N in the form of nitrate was 3 mg/L, and chlorophyll a was 4 μg/L (2005, unpublished).
Enrichment of Oil-Degrading Bacteria
About 1 ml of sterilized crude oil was added to a 100 ml sample in a 250-ml flask to start the enrichment of oil-degrading bacteria with a sterilized seawater treatment as a control. After 5 days, all samples exhibited oil degradation indicated by bacteria growth and oil emulsification, and 2% of the inoculum was transferred into 100 ml fresh artificial sea water medium (ASM) [8] with 1% crude oil as the carbon source and shaking (150 rpm) for 10 days at 28°C. Meanwhile, a control without inoculum was paralleled. Repeated transfers were done twice more. The bacteria were isolated on high-salt Luria–Bertani (HLB) agar plates [8] and characterized by the 16S rRNA gene.
Assays for Oil Biodegradation
Isolates were tested for their ability to utilize both 1% (v/v) sterilized crude and diesel oils in ASM, medium (no carbon sources) by measuring the cell density at 600 nm; growth was compared with a control containing no carbon source and a non-inoculated control. The isolates were first activated by growing in HLB liquid medium. Then, approximately 6 × 108 cells were collected, washed twice with sterilized ASM, and used to inoculate ASM medium. The assay for biodegradation was conducted in 100-ml Erlenmeyer flasks containing 30 ml sterile ASM medium, incubated at 25°C with shaking (150 rpm) for about a week.
Genomic DNA Extraction
Bacterial genomic DNA of isolates was extracted with the TIANamp Bacteria DNA kit (TIANGEN, China) according to the manufacturer’s instruction. To extract the DNA from the enrichment communities, a 3-ml culture was centrifuged, and the cell pellet was resuspended in 120 μl 1 × Tris-ethylenediaminetetraacetic acid (EDTA) buffer (10 mM Tris, pH 7.5, 1 mM EDTA) before the addition of lysozyme at 37°C for 1 h. Then, 80 μl 10% sodium dodecyl sulfate was added, and the sample was incubated at 65°C for 1.5 h and then extracted with the same kit.
To quantify the copy number of the alkB gene in seawater, 3 l of surface seawater was collected and sieved through a phytoplankton net (10-μm mesh size, 23-cm mouth diameter, and 40-cm long) to remove large granules and through a nitrocellulose membrane (0.22 μm) to collect the remaining cells. The cell pellet was stored at −70°C and thawed, incubated with lysozyme at 37°C for 30 min. DNA was then extracted with TIANamp Bacteria DNA Kit according to the manufacturer’s protocol (TianGen, China).
Bacterial 16S rRNA Gene Sequencing
The 16S rRNA gene was amplified by PCR using the universal primer set 16SF (positions 8 ∼ 27 of the Escherichia coli numbering; 5′-AGAGTTTGATCCTGGCTCAG-3′) and 16SR (positions 1,512 ∼ 1,493; 5′-ACGGCTACCTTGTTACGACT-3′) [8]. Approximately 1.5-kb product was obtained, and sequenced by Invitrogen (Shanghai, China) using an ABI model 3730 DNA sequencer. The following thermal cycling parameters were used: a 5-min hot start at 95°C; followed by 32 cycles of denaturation for 1 min at 94°C, annealing at 55°C for 1 min, and extension at 72°C for 1.5 min; and a final extension of 20 min at 72°C.
Polymerase Chain Reaction Detection of alkB Genes
To survey the diversity of alkB genes, PCR was carried out with a pair of degenerate primers, alkBwf (5′-AAYACNGCNCAYGARCTNGGVCAYAA-3′) and alkBwr(5′-GCRTGRTGRTCHGARTGNCGYTG-3′), which were designed based on well-conserved motifs of AlkB in the N- (NTXHELGHK) and C-terminal (LQRHSDHHA) domains [7]. In addition, another pair of degenerate primers, monf (5′-TCAAYACMGSNCAYGARCT-3′) and monr (5′-CCGTARTGYTCNAYRTARTT-3′), was also used, designed based on the same N-terminal motif (NTXHELGHK) as above and a different C-terminal motif (INYIEHYGLL) [8]. These primer pairs were expected to generate a PCR product of about 550 and 420 bp, respectively.
The PCR mix of 50 μl contained the following: 5 μl 10× buffer (provided with Taq polymerase), 1.5 μl MgCl2 (50 mM), 4–5 μl dNTP mix (2.5 mM each), 5–6 μl of each primer (10 μM), 10–15 ng of purified DNA from cultured strains or 15–20 ng from total community DNA, and 2.5 U Taq DNA polymerase (Invitrogen, Karlsruhe, Germany). Cycling was performed with an initial denaturation for 5 min at 94°C followed by 32 cycles of 30 s at 94°C, 30 s at 50–55°C, and 45 s at 72°C and a final elongation step for 5 min at 72°C. PCR products were separated in a 1.0% agarose gel.
Polymerase Chain Reaction Product Cloning and Library Analysis
The PCR product of the alkB gene from each isolate was purified prior to cloning. The PCR product was run in an agarose gel, and the band of the expected size was cut out and extracted with the TIANGEN Mini Purification kit (TIANGEN, China). DNA was eluted in a final volume of 20 μl. The purified PCR products were cloned into a PMD-19 T vector (TaKaRa Bio, China) and transformed into E. coli DH5α cells. A fast screening of the transformants was performed by colony PCR [7]. Plasmid DNA was extracted with the Plasmid Mini kit (Qiagen, Hilden, Germany) and used for sequencing.
With the genomic DNA of each oil-enrichment community as template, alkB gene was PCR cloned to detect the more homologues other than those form isolates. The alkB PCR products from all the communites were blended, and cloned into a PMD-19 T vector and transformed into E. coli DH5α cells. The libraries were screened via restriction analysis by digestion with HaeIII and MspI (TaKaRa Bio, China), and clones with unique restriction patterns were sequenced. The sequencing processes were conducted with an ABI model 3730 DNA sequencer (Invitrogen, Shanghai, China).
Primer Design for PCR Quantification of alkB Genes
To quantify alkB in the surface seawater, five pairs of specific non-degenerate primers targeting different groups of alkB genes were designed for real-time PCR based on the results of the phylogenetic analyses in this report (Table 1). The specificity of these primers was examined by BLAST searches in public databases (NCBI) and further tested by standard real-time PCR using SYBR green with reference strains of known geno- or phenotype (Table 3). Additionally, the specificity of the above five pairs of primers for each alkB group was reconfirmed using the DNA extracted from surface seawater samples as a template by cloning, sequencing, and phylogenetic analysis of the PCR products. Briefly, purified PCR products (QIAquick PCR Purification Kit, Qiagen) were cloned into the pCR2.1 vector using the TA cloning kit (Invitrogen). A total of 100 clones were randomly chosen from the libraries of each Groups I, II, III, IV, and V for sequencing (MWG Biotech). In addition, melt curve analysis was used to check for the production of PCR products or secondary products, such as primer dimers, in these assays.
The amplification efficiency for the real-time PCR primer sets was estimated with a gradient of annealing temperatures (from 52 to 62°C) against DNA from reference strains of pure bacterial cultures (Table 4). Standards for quantative PCR (Q-PCR) calibration were constructed according to universal methods with reference to Powell and colleagues [13]. The standard curves for all assays were linear (r 2 > 0.95) over four orders of magnitude for the alkB assay. The r 2 values were consistently greater than 0.95 and usually greater than 0.99 for both assays. The primer concentration was optimized; the efficiency of the reactions was between 0.9 and 1.1 (or 90–110%). Runs that fell outside of these parameters (r 2 < 0.95; 0.9 < efficiency < 1.1) were repeated. There was no signal in the negative control (template absent) in the alkB assay.
Quantitative PCR
Real-time quantitative PCR was performed on an IQ™ 5 Multicolor Real-Time PCR Detection system (Bio-Rad, California, USA) with the following program: 2 min at 50°C (uracil-N-glycosylase activation), 10 min at 95°C (activation of the Taq polymerase) and 45 cycles of denaturation (10 s, 95°C), annealing and elongation (30 s, 56–61°C). Fluorescence data were acquired at the end of the elongation step. The specificities of accumulated products were verified by melting curve analysis. The following reagents were added: 12.5 μl of 2× SYBR Green Supermix (Bio-Rad, California, USA), 1.0 μl of each forward and reverse primer (10 mM), 0.5 μl of template DNA, and PCR-grade water to a final volume of 25 μl. A negative treatment control (template absent) was included during each real-time PCR experiment to check the quality of the reagents used. For each gene, three biological replicates and three technical replicates were performed. Data shown represent the mean value from nine runs of real-time PCR and have a variation coefficient \( \left( {{\hbox{CV}} = {\hbox{standard deviationvalue}} \times {1}00/{\hbox{mean value}}} \right) \) ranging from 6.2–24.5%, which corresponds to less than 0.2 Log errors for the quantification values. The statistical determination of significance (a = 0.05) was determined with Microsoft Office Excel 2003 using a one sample Student’s t test on the biological repeats of each experimental condition.
Sequencing, Phylogenetic and Diversity Analysis
Sequencing of the inserted fragments was carried out on a model 377 automated DNA sequencer using a BigDye Terminator Cycle Sequencing kit (Applied Biosystems). The obtained sequences were aligned using DNAMAN (version 5.1, Lynnon Biosoft) with alkB sequences retrieved from GenBank. A phylogenetic tree of the derived protein sequences was constructed by the neighbor-joining method using the PHYLIP package [16] and DNAMAN programs. Bootstrap analysis was used to evaluate the tree topology of the neighbor-joining data with 1,000 re-samplings. Jukes–Cantor evolutionary distances were calculated using DNADIST of the PHYLIP 3.68 package.
Sequences
Sequences of all 16S rRNA genes can be retrieved from the GenBank database under accession numbers GU593614 to GU593656. All alkB sequences were deposited in the GenBank database with accession numbers of GU593657 and from EU853379 to EU853422.
Results and Discussion
AlkB Genes from the Isolates and Enrichments
From the oil-enrichment communities, 44 strains were obtained that differed from each other in 16S rRNA gene sequence. Most of the obtained isolates exhibited oil degradation. They were affiliated to 26 genera, including Alcanivorax, Acinetobacter, Pseudomonas, Flavobacterium, Corynebacterium, Gallaecimonas, Castellaniella, Paracoccus, and Leucobacter, etc. Table 2 shows the isolates that were positively detected of alkB genes, including six isolates of Alcanivorax.Interestingly, alkB gene was detected for the first time according to our knowledge in the bacteria of following genera, Gallaecimonas, Castellaniella, Paracoccus, and Leucobacter; moreover, these isolates exhibited vigorous growth with crude oil (Table 2). However, other isolates which exhibited growth with crude oil were negatively detected of alkB genes, maybe harbored other alkane monooxygenases.
Additionally, from the alkB libraries derived from the community DNA, 45 different alkB sequences were obtained (Fig. 2), 16 of which were same to those obtained from the isolates (Table 2), others were most probably from bacteria in the community but not successfully isolated. All the sequences from both community and isolates contained eight to nine histidines that are essential for the alkane-hydroxylizing activity and well conserved. Phylogenetical analysis showed that they are clustered to alkB genes previously reported from Pseudomonas putida, Pseudomonas fluorescens, Alcanivorax, Rhodococcus, Acinetobacter, Mycobacterium, and Nocardia.
Phylogenetic tree based on amino acid sequences of AlkB homologs obtained from the Xiamen coastal area and references. AlkB reference sequences from the database are labeled with strain names and the GenBank accession numbers in parentheses (protein). AlkB proteins that have been confirmed for the function of alkane hydroxylase are indicated by (+). Sequences named alkB1 to 27 were derived from bacterial community DNA used in this report; others were from isolates and labeled with the isolate No. The numbers in parentheses are the clone number of each sequence in 100 clones derived with group-specific Q-PCR primers from surface seawater samples (see “Materials and Methods”). Scale bar, 0.05 substitutions per amino acid site
Most of the obtained alkB genes in this report exhibited 60–80% similarity at the amino acid level with sequences retrieved from the GenBank database. Compared to the surface water across the Atlantic Ocean [31], the alkB sequences in Xiamen coastal area were even more diverse (for more details, see “Phylogenetic Analysis” below). They are also different from those detected in soils; such as in land soil of Germany, alkB was detected mainly from Rhodococcus, Acinetobacter, Mycobacterium, and Nocardia [7]; while in Arctic and Antarctic soils, alkB sequences were closely related to those from P. putida GPo1, Rhodococcus sp. strain Q15, and Acinetobacter sp. strain ADP-1 [35].
Novel Oil-Degrading Bacteria in Xiamen Area
Several isolates as follows showed 16Sr RNA similarity below 98% with type strains. Isolate 1-C-1 showed only 94.5% similarity in 16S rRNA gene with Flavobacterium indicum GPTSA100-9T. As a possible novel species of this genus, Isolate 1-C-1 exhibited vigorous growth with diesel oil and crude oil (Table 2); moreover, an alkB gene was detected, which is most closely related to that of P. putida strain GPo1 at the amino acid level, but only of 70% identity.
Isolate II-C-7 was also proved as oil degrader, and had 97% similarity in 16S rRNA gene with Castellaniella ginsengisoli DCY36T. And for the first time of this genus, alkB gene was detected; moreover, it harbored two alkB genes that had 97% and 68% similarities at the amino acid level with those of Alcanivorax dieselolei B-5T and P. putida strain GPo1, respectively.
Isolate 7-C-7 is possibly a novel species of Alcanivorax. It is only of 94.9% similarity in 16S rRNA gene with Alcanivorax venustensis ISO4T, while harbored an alkB homolog that had 80% similarity at the amino acid level with that of P. fluorescens CHA0.
Isolate 1-D-2 and 2-D-2 displayed 97.8% and 97.4% similarities in 16S rRNA gene with Acinetobacter junii LMG998T and Pseudomonas pseudoalcaligenes subsp. pseudoalcaligenes DSM 50188T. Both isolates exhibited vigorous growth with diesel oil and crude oil. Isolate 1-D-2 harbored an alkB gene of a high similarity with that of Acinetobacter sp. M-1. Isolate 2-D-2 harbored a quite novel alkB gene that had only 46% similarity at the amino acid level with that of P. putida (CAB54050) as the closest relative.
Phylogenetic Analysis
Our alkB homologs formed five major groups named groups I to V in the phylogenetic tree, base on their deduced amino acid sequences (Fig. 2). In addition, some sequences distantly related with the big clusters and formed several separate branches, named A to G in the tree. As mentioned above, these sequences are quite diverse and novel. Group I included 6 homologs derived from 6 isolates of Alcanivorax, Castellaniella, and Flavobacterium; they shared 73–75% similarity with the AlkB of Marinobacter sp. ELB17 (EAZ98470). Group II was composed of five AlkBs sharing 80–100% similarity with those of P. putida (CAB54050) and Alcanivorax borkumensis SK2 (BAC98365). For short, groups III, IV, and V were centered with previously reported AlkBs of P. fluorescens (CAB51045), Rhodococcus sp. Q15 (AF388182) and Acinetobacter spp., respectively (Fig. 2). The AlkB from Acinetobacter is also called AlkM, which has been proved to act on long-chain alkanes, such as in encoded by A. venetianus strain M-1 and ADP1 [35].
Interestingly, distantly related bacteria can possess similar or even identical alkB genes, as observed in “group I”, which contained nearly identical alkB genes. This is probably owing to horizontal gene transfer, as proposed previously [25].
Groups II and IV were mainly composed of those genes directly retrieved from the community libraries; other groups and branches also included some alkB genes directly retrieved from the community libraries. In other words, their host bacteria have not been isolated. Compared with alkB genes of isolates, those alkB genes were quite divergent. Moreover, they were divergent from the published sequences (Fig. 2).
Group Specificity of alkB Primers
The group specificity of alkB primers designed for quantification of alkB genes by Q-PCR in seawater was first examined with genomic DNA of both bacterial strains and seawater. Group-wise alignment of all alkB sequences obtained in this report allowed for the design of several specific primer pairs (Table 1; Electronic Supplementary Material, Fig. S1). Then, each primer pair was tested on corresponding reference strains including our isolates (Table 3). All PCR products derived from alkane-degrading bacteria showed a single band of the expected size, while there was no amplification from non-target bacteria.
Furthermore, the group specificity was also tested on surface seawater samples. Cloning, sequencing and phylogenetic analysis of the PCR products were conducted for each group (“Materials and Methods”). In total, with Q-PCR, 96, 91, 93, 87, and 83 target alkB clones were obtained from the 5 libraries, respectively (Table 4). In general, the absolute majority of the sequences are identical to those detected by degenerate PCR described above in Fig. 2. For more details, with the alkBIf/r primers, 30, 22, and 20 copies in Group I were identical or nearly identical to the alkBs of strain 3-C-3, 6-D-6 and 1-C-1, respectively. With alkBIIf/r primers of group II, alkB genes mainly composed of “alkb21” and “alkb1” with 44 and 35 copies, respectively, both were from uncultivated bacteria in Fig. 2. Similarly, in other three libraries, the group specificity was also confirmed. Overall, sequences close to Pseudomonas, Alcanivorax, Rhodococcus, and Acinetobacter occupied larger proportions in the five libraries, implies they are relatively abundant in the coastal area. However, “alkb3”, “alkb9”, “alkb13”, and “alkb26” were not retrieved, indicating their low copy number in situ.
Worth to note, only two clones amplified with group I primers actually fell in group II; likely, in the other libraries, five to nine clones fell in non-targeted groups as well. However, only two, one, and two non-alkB sequences were found in groups I, IV, and V libraries, respectively (Table 4). In general, these results proved that the feasibility of these primer sets to quantify alkB genes with group specificity.
Valid Copy Number Range of alkB for Q-PCR
A preliminary Q-PCR experiment was conducted prior to in situ quantification. The results showed that for each group, alkB genes were amplified with the same efficiency (see “Materials and Methods”), and standard curves were linear for about six orders of magnitude (data not shown). This permits quantifying alkB in a range of four to six orders of magnitude. In details, the valid range for Group I, Group IV and Group V was 102 to 106 copies per tube (25 μl real-time PCR reagents), and 102 to 108 copies for groups II and III. These ranges were referred below in examination of alkB genes of in situ seawater.
Quantification of alkB Genes in the Coastal Surface Water
The validated Q-PCR assay was applied to quantification of alkB genes of seawater in situ. The real-time PCR quantification results are shown in Fig. 3. For each alkB group, the gene copy number (mean threshold values ± S.D.) was determined ranging from about 3 × 103 to 3 × 105 L−1 in seawater. Specifically, alkB homologs in groups I to V were determined to be 4.19 (±0.26) × 104, 3.27 (±0.57) × 105, 4.70 (±0.54) × 105, 4.75 (±0.93) × 103, and 3.47 (±0.85) × 103, respectively. Apparently, the copy numbers of groups II and III were significantly higher than the other groups.
As group I, II, and III contained alkB genes from Alcanivorax spp. and Pseudomonas spp., bacteria of two genera are deduced abundant correspondingly in seawater. Consistently, A. dieselolei was found to be one of the most predominant members in these oil-degrading communities (unpublished). In this report, four isolates of A. dieselolei of 16S rRNA gene similarity above 98% were obtained in the plate cultivation. These results indicate that A. dieselolei bacteria might play an important role in this area. However, A. borkumensis, which has been recognized to play an important role in oil removal from marine systems [2], was not isolated successfully. Interestingly, the clone numbers of Group I, III and V were significantly higher than the other groups in community libraries (data not shown). Group V contained alkB genes were obtained from Acinetobacter spp., which was also found to be one of the most predominant members in the oil-degrading communities (unpublished). Thus, Acinetobacter bacteria might play an important role in this area in case of oil spill occurrence.
Although the abundance of alkB genes partially reflected their role in alkane mineralization in situ, whether they actually function or dictate the synthesis of alkane monooxygenase requires further investigation, for example, by quantification of the mRNA or AlkB directly.
In addition to the pathway initiated by AlkB monooxygenase, other alkane monooxygenase systems should exist, such as cytochrome P450 [10], which also contribute to alkane degradation and oil removal in situ. In addition, AlmA has been confirmed as an alkane monooxygenase of the flavin-binding family and is involved in long-chain n-alkane metabolism [23, 34]. To gain an overview of the genes responsible for oil pollutant bioattenuation in the coastal area, P450 and almA genes, which have been confirmed as alkane monooxygenases, should also be taken into consideration.
In summary, the alkB genes were quite diverse in the coastal surface seawater around Xiamen Island. They exist in each milliliter of the surface water with about 1,000 copies, hosted in bacteria including Alcanivorax, Acinetobacter, Pseudomonas, Gallaecimonas, Castellaniella, Paracoccus, and Leucobacter, etc. These bacteria are thought active in removal of oil in situ, especially Alcanivorax, Pseudomonas, and Acinetobacter in this area. However, bacteria of Oleiphilus spp., Oleispira spp., and Thalassolituus spp., which frequently reported in other areas, were not found in our report. This indicates that oil-degrading bacteria may have restriction in geographic distribution in some species.
References
Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, Tang Y, Liu X, Han W, Peng X, Liu R, Wang L (2007) Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci USA 104:5602–5607
Golyshin PN, Martins Dos Santos VA, Kaiser O, Ferrer M, Sabirova YS, Lunsdorf H, Chernikova TN, Golyshina OV, Yakimov MM, Puhler A, Timmis KN (2003) Genome sequence completed of Alcanivorax borkumensis, a hydrocarbon-degrading bacterium that plays a global role in oil removal from marine systems. J Biotechnol 106:215–220
Head IM, Jones DM, Roling WF (2006) Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182
Heiss-Blanquet S, Benoit Y, Marechaux C, Monot F (2005) Assessing the role of alkane hydroxylase genotypes in environmental samples by competitive PCR. J Appl Microbiol 99:1392–1403
Islam KS, Rahman MM, Eda LEH (2009) Pollution status and sustainable management of Xiamen Bay in China: a brief review. Int J Ocean Syst Manage 1:155–168
Kuhn E, Bellicanta GS, Pellizari VH (2009) New alk genes detected in Antarctic marine sediments. Environ Microbiol 11:669–673
Kloos K, Munch JC, Schloter M (2006) A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J Microbiol Methods 66:486–496
Liu C, Shao Z (2005) Alcanivorax dieselolei sp. nov., a novel alkane-degrading bacterium isolated from sea water and deep-sea sediment. Int J Syst Evol Microbiol 55:1181–1186
Luz AP, Pellizari VH, Whyte LG, Greer CW (2004) A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can J Microbiol 50:323–333
Maier T, Forster HH, Asperger O, Hahn U (2001) Molecular characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem Biophys Res Commun 286:652–658
Marin MM, Smits TH, van Beilen JB, Rojo F (2001) The alkane hydroxylase gene of Burkholderia cepacia RR10 is under catabolite repression control. J Bacteriol 183:4202–4209
Ou S, Zheng J, Richardson BJ, Lam PK (2004) Petroleum hydrocarbons and polycyclic aromatic hydrocarbons in the surficial sediments of Xiamen Harbour and Yuan Dan Lake, China. Chemosphere 56:107–112
Powell SM, Ferguson SH, Bowman JP, Snape I (2006) Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb Ecol 52:523–532
Quatrini P, Scaglione G, De Pasquale C, Riela S, Puglia AM (2008) Isolation of Gram-positive n-alkane degraders from a hydrocarbon-contaminated Mediterranean shoreline. J Appl Microbiol 104:251–259
Rojo F (2009) Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sei K, Sugimoto Y, Mori K, Maki H, Kohno T (2003) Monitoring of alkane-degrading bacteria in a sea-water microcosm during crude oil degradation by polymerase chain reaction based on alkane-catabolic genes. Environ Microbiol 5:517–522
Shanklin J, Whittle E, Fox BG (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochem 33:12787–12794
Siciliano SD, Fortin N, Mihoc A, Wisse G, Labelle S, Beaumier D, Ouellette D, Roy R, Whyte LG, Banks MK, Schwab P, Lee K, Greer CW (2001) Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl Environ Microbiol 67:2469–2475
Stapleton RD, Ayler GS (1998) Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microbial Ecol: 36:49–361
Smits TH, Röthlisberger M, Witholt B, van Beilen JB (1999) Molecular screening for alkane hydroxylase genes in gram-negative and gram-positive strains. Environ Microbiol 1:307–317
Tani A, Ishige T, Sakai Y, Kato N (2001) Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J Bacteriol 183:1819–1823
Throne-Holst M, Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB (2007) Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl Environ Microbiol 73:3327–3332
van Beilen JB, Wubbolts MG, Witholt B (1994) Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161–174
van Beilen JB, Smits THM, Whyte LG, Schorcht S, Röthlisberger M, Plaggemeier T, Engesser KH, Witholt B (2002) Alkane hydroxylases in gram-positive strains. Environ Microbiol 4:676–682
van Beilen JB, Duetz WA, Schmid A, Witholt B (2003) Practical issues in the application of oxygenases. Trends Biotechnol 21:170–177
van Beilen JB, Li Z, Duetz WA, Smits THM, Witholt B (2003) Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci Technol 58:427–440
van Beilen JB, Funhoff EG (2005) Expanding the alkane oxygenase toolbox: new enzymes and applications. Curr Opin Biotechnol 16:308–314
van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21
Vomberg A, Klinner U (2000) Distribution of alkB genes within n alkane-degrading bacteria. J Appl Microbiol 89:339–348
Wang L, Wang W, Lai Q, Shao Z (2010) Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ Microbiol 12(5):1230–1242
Wasmund K, Burns KA, Kurtboke DI, Bourne DG (2009) Novel alkane hydroxylase gene (alkB) diversity in sediments associated with hydrocarbon seeps in the Timor Sea, Australia. Appl Environ Microbiol 75:7391–7398
WEF AA (1998) Standard Methods for the Examination of Water and Wastewater (20th ed.), APHA-AWWA-WEF, Washington, DC
Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB, Throne-Holst M (2007) Bacterial metabolism of long-chain n-alkanes. Appl Microbiol Biotechnol 76:1209–1221
Whyte LG, Schultz A, Van Beilen JB, Luz AP, Pellizari D, Labbé D, Greer CW (2002) Prevalence of alkane monooxygenase genes in arctic and antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol Ecol 41:141–150
Whyte LG, Smits TH, Labbe D, Witholt B, Greer CW, van Beilen JB (2002) Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl Environ Microbiol 68:5933–5942
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (30670051), the Science and Technology Program of Fujian Province of China (2009H0029), and the China Scholarship Council Program (2008631036).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
Multiple alignment of each alkB group to show the conserved regions used for Q-PCR primer design. Q-PCR primers of each group were designed based on the two conserved regions labeled in grey pane. a. Group I-like alkB gene; b. Group II-like alkB gene; c. Group III-like alkB gene; d. Group IV-like alkB gene; e. Group V-like alkB gene. (GIF 750 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Wang, L. & Shao, Z. Diversity and Abundance of Oil-Degrading Bacteria and Alkane Hydroxylase (alkB) Genes in the Subtropical Seawater of Xiamen Island. Microb Ecol 60, 429–439 (2010). https://doi.org/10.1007/s00248-010-9724-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9724-4