Abstract
Treacher Collins syndrome (TCS) is a genetic disorder affecting facial development, primarily caused by mutations in the TCOF1 gene. TCOF1, along with NOLC1, play important roles in ribosomal RNA transcription and processing. Previously, a zebrafish model of TCS successfully recapitulated the main characteristics of the syndrome by knocking down the expression of a gene on chromosome 13 (coding for Uniprot ID B8JIY2), which was identified as the TCOF1 orthologue. However, database updates renamed this gene as nolc1 and the zebrafish database (ZFIN) identified a different gene on chromosome 14 as the TCOF1 orthologue (coding for Uniprot ID E7F9D9). NOLC1 and TCOF1 are large proteins with unstructured regions and repetitive sequences that complicate alignments and comparisons. Also, the additional whole genome duplication of teleosts sets further difficulty. In this study, we present evidence that endorses that NOLC1 and TCOF1 are paralogs, and that the zebrafish gene on chromosome 14 is a low-complexity LisH domain-containing factor that displays homology to NOLC1 but lacks essential sequence features to accomplish TCOF1 nucleolar functions. Our analysis also supports the idea that zebrafish, as has been suggested for other non-tetrapod vertebrates, lack the TCOF1 gene that is associated with tripartite nucleolus. Using BLAST searches in a group of teleost genomes, we identified fish-specific sequences similar to E7F9D9 zebrafish protein. We propose naming them “LisH-containing Low Complexity Proteins” (LLCP). Interestingly, the gene on chromosome 13 (nolc1) displays the sequence features, developmental expression patterns, and phenotypic impact of depletion that are characteristic of TCOF1 functions. These findings suggest that in teleost fish, the nucleolar functions described for both NOLC1 and TCOF1 mediated by their repeated motifs, are carried out by a single gene, nolc1. Our study, which is mainly based on computational tools available as free web-based algorithms, could help to solve similar conflicts regarding gene orthology in zebrafish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biomedical researchers have used the favorable characteristics of the zebrafish, Danio rerio, to study a wide range of topics including its development, natural biology, and behavior (Parichy 2015). Additionally, the zebrafish has been an invaluable model for understanding gene function in humans, particularly in regards to disease-related genes. The ability to perform gene ablation, to study mutant and morphant phenotypes, and to efficiently characterize gene expression and function in zebrafish has greatly enhanced our understanding of human gene function (Santoriello and Zon 2012; Bradford et al. 2017; Collin and Martin 2017). To evaluate gene functions across species, distinguishing between orthologs (genes derived from a common ancestor) and paralogs (genes arising from duplication events) is crucial. Various methods have been proposed to determine orthologies, such as the reciprocal best hits (RBH), phylogenetic analysis, and conserved synteny analysis (Postlethwait 2006; Nichio et al. 2017). While these approaches have proven valuable, they can pose challenges in situations with uneven or rapid rates of evolution, concerted evolution, partial sequences, insertions, deletions, translocations, and duplications, including whole genome duplications (WGD) as observed in teleosts (Locascio et al. 2002; Wall et al. 2003). Furthermore, proteins without well-defined structures, such as intrinsically disordered proteins (IDPs), raise difficulties in sequence similarity and phylogenetic studies due to their higher evolutionary rates and compositional biases (Midic et al. 2009; Lange et al. 2016).
The biogenesis of ribosomes is one of the most important processes in the cell. It is responsible for as much as 95% of total transcription and consumes more than 60% of cellular energy (Grzanka and Piekiełko-Witkowska 2021). NOLC1 (Nucleolar and Coiled-body phosphoprotein 1, also known as Nopp140, nucleolar protein, 130-KD, p-130) and TCOF1 (Treacle ribosome biogenesis factor, also known as treacle) are believed to function as molecular chaperones, delivering small nucleolar ribonucleoprotein complexes (snoRNPs) to the nucleolus where pre-ribosomal RNA is synthesized, cleaved, chemically modified, and assembled into large and small ribosomal subunits (He and DiMario 2011). These proteins have been related as paralogs (He and DiMario 2011; Werner et al. 2015; Sochacka et al. 2022) although no systematic study has been published yet. Orthologues of NOLC1 have been identified in an evolutionarily wide range of eukaryotes from yeast to human, but TCOF1 appears to be more restricted (He and DiMario 2011).
Proteins containing stretches of amino acids that lack a stable tertiary structure and provide relative motional freedom are known as IDPs or natively unstructured protein (Van Der Lee et al. 2014). These regions are considered to be an inherent characteristic of the protein, and are found in many different types of proteins. Two examples of IDPs are NOLC1 and TCOF1 (Na et al. 2018), which contain extensive unstructured regions (IDRs, intrinsically disordered regions) spanning more than 80% of their sequences. Both proteins share an amino-terminal Lissencephaly Type-1 homology (LisH) motif in their amino termini, and a large unstructured central repeat domain consisting of alternating acidic and basic motifs (He and DiMario 2011). NOLC1 and TCOF1 may be multifunctional: the large central domains of these proteins serve as scaffolds for delivering and positioning snoRNPs within nucleoli. NOLC1 and TCOF1 interact with Pol I transcription machinery (He and DiMario 2011; Werner et al. 2015). Nolc1 is required for normal development in Drosophila, while TCOF1 is critical for mammalian neural crest cell development (He and DiMario 2011). Loss of TCOF1 function causes a nucleolar stress response that initiates p53-mediated apoptosis in embryonic neural epithelial and neural crest cells leading to the Treacher Collins–Franceschetti Syndrome (TCS) in humans, a collection of craniofacial malformations (Dixon et al. 1997). Beyond the roles described above for TCOF1 and NOLC1, it is likely that, taking into account their size and modular nature, their biological functions have not yet been fully described.
Recently, we modeled TCS in zebrafish inducing ZDB-GENE-030131-6349 knock-down by translation-blocking Morpholino injection (De Peralta et al. 2016; Rosas et al. 2019). This zebrafish TCS model recapitulated all the pathological hallmarks described for the human syndrome and the murine model. However, lately, in genomic databases including The Zebrafish Information Network (ZFIN), ZDB-GENE-030131-6349 was annotated as a NOLC1 orthologue and not as a TCOF1 orthologue as stated by Weiner et al. (Weiner et al. 2012). Moreover, in ZFIN and other databases (e.g., NCBI), there is a gene annotated as TCOF1 orthologue, ZDB-GENE-141219-12 (coding for Uniprot ID E7F9D9). In this study, utilizing freely available computational tools as web-based algorithms, we present evidence that confirms the paralogy between NOLC1 and TCOF1 and challenges the classification of ZDB-GENE-141219-12 (ENSDARG00000087555) as a ortholog of TCOF1, at least at the functional level. Additionally, we demonstrate that Uniprot ID B8JIY2 coded by ZDB-GENE-030131-6349 (ENSDARG00000024561) possesses the appropriate structural characteristics to fulfill both NOLC1 and TCOF1 nucleolar functions, and exhibits a compatible expression profile during embryonic development in zebrafish. Considering the significance of accurate gene annotation in biomedical research, along with the challenges posed by IDPs and duplicated genomes in zebrafish, we consider that our analysis can provide valuable insights to the scientific community.
Materials and Methods
Sequences and Alignments
Protein sequences were mostly obtained from the Uniprot database (https://www.uniprot.org/) [Last accessed: 2/15/2024]. The IDs of the TCOF1 and NOLC1 proteins selected for this work were, respectively: Q13428 and Q14978 (human), O08784 and E9Q5C9 (mouse), A0A6I8SC14 and F6XPN6 (Xenopus tropicalis), Q5G8Z4 and Q91803 (Xenopus laevis). IDs selected for Danio rerio proteins were B8JIY2 (product of ENSDARG00000024561 gene) and E7F9D9 (product of ENSDARG00000087555 gene). Additional sequences included in the phylogenetic analysis are detailed below [Last accessed: 6/15/2024].
RBH analysis were performed manually using the TBLASTN and BLASTP algorithms in Ensembl (https://www.ensembl.org/Tools/Blast, human genome assembly GRCh38.p14 (GCA_000001405.29) and zebrafish assembly GRCz11 (GCA_000002035.4)) and in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi, human GRCh38.p14 reference assembly RS_2023_10 and zebrafish GRCz11 reference Annotation Release 106). Uniprot ID E7F9D9 (current zebrafish Tcof1 according ZFIN) and human TCOF1 (Q13428) were the queries.
Protein Uniprot sequences were aligned using the Align tool (https://www.uniprot.org/align) under default settings (program: clustalo; version: 1.2.4; guidetreeout: true, addformats: true; iterations: 0, Outfmt: clustal_num, order: input) and the results were downloaded as the percent identity matrix. Similarity values among these proteins were obtained using the Pairwise alignment tool from the Protein Information Resource (PIR) webpage (https://proteininformationresource.org/pirwww/search/pairwise.shtml) and the UniprotKB identifiers taken in pairs (Smith and Waterman 1981).
Multiple sequence alignments (MSA) for the molecular phylogenetic tree construction were obtained using the COBALT (COnstraint Based ALignment Tool) algorithm (Papadopoulos and Agarwala 2007) available at the NCBI site. Highly variable sites were removed using BMGE software (Criscuolo and Gribaldo 2010) with the following parameters: Estimated BLOSUM matrix = 62, sliding windows size = 3; Maximum entropy threshold = 0.5; Gap rate cut-off = 0.5; Minimum block size = 3 (see Suppl. Fig 1 for de MSAs). Amino Acid substitution model prediction was conducted using Aminosan (Tanabe 2011), and subsequent Bayesian phylogenetic inference analysis was performed with the software MrBayes 5D, a modified variant of MrBayes (Ronquist and Huelsenbeck 2003) which allows a wider range of amino acid substitution models (available at https://www.fifthdimension.jp/products/mrbayes5d/). The specified model was VTT + G + F (Yang 1994; Müller and Vingron 2001; Wang et al. 2008) for amino acid substitution. Two independent runs with four Markov Chain Monte Carlo simulations were performed for 5 million iterations, and trees were sampled every 1000th generation. The first 25% of trees were discarded as burn-in. Mixing and convergence were visually assessed by examining likelihood scores and estimated sample sizes of MCMC runs using the software Tracer (Rambaut et al. 2018). Finally, phylogenetic tree editing was conducted using FigTree-version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). This analysis involved 50 protein sequences. Additional protein sequences were downloaded from Ensembl and NCBI databases. See Suppl. Table 1 for more details. Tree rooting was performed using NOTUNG 2.9 (Chen et al. 2000) using a species tree generated by PhyloT (https://phylot.biobyte.de/). The tree graphic was created using the open-source vector graphics editor Inkscape (https://inkscape.org/).
Identification of Homologous Sequences
TBLASTX, which is a bioinformatic tool on Ensembl, was used for identifying homologous proteins in different species, even if the genes for those proteins have diverged significantly at the DNA sequence level. TBLASTX compares a genomic DNA sequence query translated to protein in all six possible reading frames to a translated nucleotide database using the BLAST algorithm (finds similar sequences by looking for short regions of high sequence identity). The query sequences, used to analyze NOLC1 and TCOF1 homologs in tetrapods, were nolc1.L 10.1 (XB-GENE-6252171) and tcof1.S 10.1 (XB-GENE-6253275) from Xenopus laevis. Configurations: Maximum E-value for reported alignments 1e-1. Options: with filters for low-complexity regions. In our quest to identify zebrafish E7F9D9 homologs (see below), we conducted additional BLASTP searches in both the NCBI and Ensembl (https://www.ensembl.org/Tools/Blast) databases, using E7F9D9 as a reference, against the available fish genomes. The proteins codified by the hits obtained in this way were further analyzed and specifically selected in terms of (i) presence of a LisH motif in the amino terminal, (ii) presence of intrinsically disordered low-complexity regions (identified by MOBI-DB) and (iii) the position of each hit's coding gene located near the “arsi” gene (mirroring the zebrafish E7F9D9 coding gene's location in the reference genome).
Selection Analyses
The ratio of non-synonymous (β) to synonymous (α) substitutions (dN/dS) was calculated using MrBayes (Ronquist and Huelsenbeck 2003) and several algorithms provided by the Datamonkey web server (Weaver et al. 2018) within the HyPhy software package (http://www.datamonkey.org) (Kosakovsky Pond et al. 2020). For MrBayes selection studies, we constructed three distinct protein alignments for each family under investigation by means of the COBALT algorithm. These alignments were then used to perform three codon-aware MSAs, integrating the corresponding coding sequences. This step was carried out using a custom-built program developed with Biopython (https://biopython.org/). The same codon-aware alignments were employed for the Fixed Effects Likelihood (FEL) (Kosakovsky Pond and Frost 2005), Branch-site Unrestricted Statistical Test for Episodic Diversification (BUSTED) (Murrell et al. 2015) and Fast Unconstrained Bayesian Approximation (FUBAR) (Murrell et al. 2013) algorithms of the Datamonkey server. Additionally, Contrast-FEL (Kosakovsky Pond et al. 2021) was applied using a full codon-wise MSA that included all three genes of interest. Significance level or posterior probability thresholds were set as default by each algorithm.
LisH Domain Detection
To evaluate the presence of LisH domains within the protein structures studied in this work we used the ScanProsite bioinformatic tool (https://prosite.expasy.org/scanprosite/) (de Castro et al. 2006). Protein sequences (in FASTA format) and the LisH motif (as the PROSITE accession PS50896) were provided and scanned them against each other. All the other options were left as default [Last accessed: 12/23/2022]. A match with a normalized score of 9.0 or higher is expected to occur about once in a database of one billion residues. A normalized score of 8.5 is typically defined as the default cut-off value in PROSITE profiles.
Syntenic Analysis
Visualization and analysis of the genomic context and evolutionary conservation of TCOF1 and NOLC1 genes were first done on Genomicus (Muffato et al. 2010; Thi et al. 2021), a freely available web-based tool at Genomicus v110.01-Gene Search. The genomic data used in Genomicus is obtained from the Ensembl database and the search for this data was conducted using gene names. The reference genes human NOLC1 (ENSG00000166197) and TCOF1 (ENSG00000070814) were compared to the root species “Vertebrata” that includes rhesus, mouse, chicken, frog, and the zebrafish genomic context. The AlignView tool was utilized to display an alignment between the genes contained within the genomic region of the reference gene and their respective orthologues in other species. The online tool displays a graphical representation of the reference gene in the center with 15 neighboring genes on both sides, as well as orthologs and paralogs of the query genes in their own respective genomic regions, also with 15 neighboring genes. To improve the readability of the figure, the image was cropped to show 3 of the syntenic blocks surrounding each gene of interest. The synteny results obtained from Genomicus, particularly the genes surrounding the LLCPs (that includes zebrafish E7F9D9, see below), were manually validated and cross-checked using genomic browsers (NCBI and Ensembl) for a more accurate examination of the chromosomal region housing the genes of interest. This meticulous process was undertaken to confirm the presence of genes in each contiguous chromosomal region that could not result properly rendered by Genomicus. Therefore, it was important not to depend exclusively on this computational resource.
Intrinsic Protein Disorder Analysis
MobiDB 5.0 database (Piovesan et al. 2023) (https://mobidb.bio.unipd.it/) was searched using the Uniprot IDs of D. rerio and human proteins. MobiDB disorder predictions are generated with MobiDB-lite. MobiDB-lite executes up to 9 different disorder predictors, collects the outputs and calculates a consensus. At least 62.5% of predictors must agree to assign disorder state to a residue. MobiDB provides extra information from various sources and prediction tools (https://mobidb.bio.unipd.it/about/vocabulary#publicationslist).
Repetitive Domain Analysis
Detection and alignment of internal repeats in TCOF1 and NOLC1 orthologues was performed using the web interface for RADAR (Rapid Automatic Detection and Alignment of Repeats) available at https://www.ebi.ac.uk/jdispatcher/pfa/radar (Heger and Holm 2000) and following the two steps indicated on the server.
Expression Analysis During D. rerio Developmental Stages
Baseline expression of ENSDARG00000024561 (currently annotated as nolc1) and ENSDARG00000087555 (si:ch73-308l14.2) from transcriptional profiling of zebrafish developmental stages (RNA-Seq mRNA baseline expression in TPM, transcripts per kilobase million) (White et al. 2017) was downloaded from Expression Atlas [Last accessed: 12/20/2022] (https://www.ebi.ac.uk/gxa/home) and presented as a line plot versus embryological stage. For demonstration purposes, we also include in this analysis two other ribosomal biogenesis factors: utp4 (ENSDARG00000017675) and rbm28 (ENSDARG00000025332).
Single-cell RNA sequencing expression data from zebrafish embryos during the first day of development was also obtained from Wagner et al. (2018) for nolc1, LOC100329277 (previous gene symbol of E7F9D9 coding locus) and sox10 (as a neural crest marker). The single cell expression data was explored using the SPRING tool (Dr. Allon Klein lab, Harvard University, (Weinreb et al. 2018)). SPRING plots of single-cell expression data were generated for different genes of interest at https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?cgi-bin/client_datasets/fish_embryo_timecourse/full. For comparison, a similar analysis was performed for frog embryos spanning zygotic genome activation through early organogenesis using the single cell expression data from Briggs et al. (Briggs et al. 2018) also available in SPRING-based interface (https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?cgi-bin/client_datasets/xenopus_embryo_timecourse_v2/full). Also, spatial and temporal transcriptomic data from microdissections taken from Xenopus laevis embryos in the course of neurulation was obtained using the EctoMap expression datasets available from Monsoro-Burq lab resources (https://monsoro-lab-ectomap.shinyapps.io/EctoMAP/, (Plouhinec et al. 2017). The expression counts data (in RPKM units) obtained for nolc1, tcof1, snai2 and pax3 genes were extracted from the original datasets and plotted against tissue origin at stage 12 and 14.
Results and Discussion
TCS is an autosomal dominant disorder of craniofacial development, and mutations in the TCOF1 gene are responsible for about 90% of TCS cases (Vincent et al. 2016). Although it is acknowledged that TCS is a ribosomopathy because TCOF1 mutations affect ribosome production (Robson et al. 2016; Falcon et al. 2022), the molecular mechanisms underlying the TCS are not well understood (Calo et al. 2018; Fitriasari and Trainor 2021; Falcon et al. 2022). To gain insight into the disease, and having in mind the advantages of zebrafish models in biomedical research, efforts were performed to identify and study Danio rerio putative TCOF1 ortholog by Weiner et al. (2012). After several in silico and in vivo studies the authors arrived at the conclusion that Uniprot B8JIY2 encoded by ZDB-GENE-030131-6349 in chromosome 13 might be the TCOF1 ortholog in zebrafish. However, later, this gene was re-named as nolc1 and a different gene, on chromosome 14 (ZDB-GENE-141219-12 encoding E7F9D9), was annotated as the TCOF1 orthologue in D. rerio.
To decipher these annotations, we conducted RBH analysis on the NCBI and Ensembl databases (Suppl. Fig. 2) using TBLASTN and BLASTP algorithms. Our queries were the zebrafish E7F9D9 and human TCOF1 proteins. While TBLASTN algorithm did not generate hits in any database, BLASTP results provided some reciprocity but the alignments were between 47 and 52 amino acid residues long (with 34–40% of identity). Taking into account that these proteins are large (628 to 1488 residues) and the alignments were only 40 to 50 residues long, that not all the algorithms and databases agreed, and that the NCBI database provided both E7D9F9 and NOLC1 (B8JIY2) proteins as significant alignments for human TCOF1, we decided to deepen our analysis on this ortholog/paralog conundrum. So, we conducted MSAs of the zebrafish proteins designated as orthologues of TCOF1 and NOLC1 (Uniprot identifiers E7F9D9 and B8JIY2, respectively) together with the NOLC1 and TCOF1 proteins from H. sapiens, M. musculus, X. laevis, and X. tropicalis (see Table 1). Zebrafish B8JIY2 shows more sequence identity to NOLC1 orthologues (40 to 50%) than to TCOF1 orthologues (26 to 30 %). E7F9F9 shows about 23 to 28% of identity to NOLC1 proteins and even less to TCOF1 proteins (19 to 23%). It is worth noting both B8JIY2 and E7E9F9 proteins show higher identity percentages to human NOLC1 than to TCOF1. Finally, it is worth mentioning that the original findings from Weiner et al. (2012) based on similarity (not identity) among these proteins indicated higher similarity of B8JIY2 with TCOF1 than with NOLC1 proteins. Via pairwise alignments we analyzed similarity among these proteins (Table 1). Using this algorithm, B8JIY2 exhibited more similarity to NOLC1 (63–65%) than to TCOF1 (52–54.5%) orthologues while E7F9D9 showed equivalent similarity values to both types of proteins (approx. 52%). Sequence homology is assigned based on the definition of a similarity matrix that allocates scores and over the years these matrices evolved and employed different approximations to compute similarity and to calculate scores. Also, as previously mentioned, NOLC1 and TCOF1 are IDPs, meaning they lack well-defined structural domains and a stable tertiary structure. Algorithms underlying existing MSA software are directly or indirectly based on knowledge obtained from studying 3D protein structures (Lange et al. 2016), and so, these procedures may not be fully reliable when performed on IDPs. Altogether, these results indicated that deeper analysis were necessary to disentangle these protein orthologies in zebrafish.
The unique structured motif that has been consistently identified in both NOLC1 and TCOF1 proteins is the LisH domain, predicted to be two α-helices (Emes 2001). The 33-residue LisH motif (see Logo in Suppl. Fig. 3) is found in eukaryotic intracellular proteins and is involved in microtubule dynamics, cell migration, nucleokinesis, and chromosome segregation (Gerlitz et al. 2005). Therefore, to investigate this motif in the zebrafish orthologues of these proteins, we searched for the LisH domain using the ScanPROSITE engine in the versions of TCOF1 and NOLC1 proteins from zebrafish (D. rerio), human (H. sapiens), mouse (M. musculus), X. tropicalis and X. laevis. The results (shown in Suppl. Fig. 3) indicate that all of the proteins have LisH motifs with similar good scores (all above 8).
In view of the previous results and beyond the limitations of MSAs with IDPs, we constructed a molecular phylogenetic tree based on COBALT alignment (MSA available in Suppl Fig. 1) and using Bayesian inference, incorporating statistical support such as posterior probabilities of branches. Before this, we verified the presence of orthologues in several vertebrates using the TBLASTX tool as described in Material and Methods. During this analysis, we noticed a significant finding concerning the tcof1 gene: the presence of a TCOF1 orthologue in coelacanths (Latimeria chalumnae) whose genome has been sequenced (Nikaido et al. 2013). The results confirmed that NOLC1 and TCOF1 have orthologues in a variety of vertebrates (He and DiMario 2011). In particular, results for TBLASTX on the Danio rerio genomic sequences endorsed that B8JIY2 has a good sequence identity to the Xenopus laevis query sequence (nolc1.L) as shown by Clustal alignment (Table 1). However, the percentage of identity for X. laevis tcof1.S hits in the zebrafish genome was low, raising again doubts regarding the phylogenetic relationships between these proteins. In order to gather more homologous sequences to calculate the phylogenetic tree, we conducted additional BLAST searches in both the NCBI and Ensembl databases, using zebrafish E7F9D9 as a reference, against the available fish genomes. Our search strategy identified proteins that mainly aligned with the LisH motif region. Since these proteins lack the S/E-rich repeats characteristic of NOLC1 and TCOF1 (See below and Suppl. Fig. 4), we suggest naming them LisH-containing Low-Complexity Proteins (LLCPs). We analyzed NOLC1, TCOF1 and LLCP protein sequences from a range of vertebrates, including mammals (human and mouse), reptiles (lizards and turtles), amphibians (X. laevis, X. tropicalis and spiny toad), a lobe-finned fish (coelacanth), cartilaginous fishes (elephant shark and great white shark), teleost fishes (zebrafish, cod, fugu, medaka, carp, barbel, tilapia, etc.) and non-teleost fishes (spotted gar and sea lamprey). We also included an invertebrate, Caenorhabditis elegans, in our analysis (refer to Suppl. Table 1 for gene names and IDs).
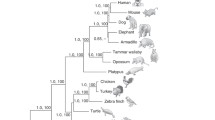
The calculated tree is displayed in Fig. 1. While the phylogeny demonstrates clear separation between these protein families, resolution is hampered within the NOLC1 cluster, particularly in fish species. Polytomies observed in this region might be due to difficulties in aligning the protein sequences, errors in the sequence data, or the absence of information from certain fish species. Coelacanth and channel catfish NOLC1 are closely related but also contribute to the complex branching pattern in this part of the tree. On the other side, TCOF1 and LLCP proteins form distinct clusters but share a common ancestral lineage that branches from a node shared by a subset of NOLC1 proteins (fish NOLC1). LLCP appears to have undergone more rapid evolutionary changes compared to TCOF1. In this context, a dN/dS analysis using several algorithms (Suppl. Fig. 5) suggests that NOLC1 genes are under stronger purifying selection, while LLCP genes are experiencing more diversifying selection, with TCOF1 genes in an intermediate position. Since divergence under positive selection is one of the key models by which duplicate genes contribute to new functions (Rosello and Kondrashov 2014), these dN/dS studies suggest that LLCP genes may be undergoing such a process.
Bayesian phylogenetic tree inferred from an alignment of 49 vertebrate protein sequences plus C. elegans (invertebrate) NOLC1 (See Suppl. Table 1 for full details). The tree displays species names and protein names are indicated to the right TCOF1 (red), NOLC1 (purple) and LLCP (blue). C. elegans NOLC1 was used as an outgroup for rooting the phylogenetic tree. Arrows highlight specific zebrafish proteins: LLCP (E7F9D9) and NOLC1 (B8JIY2). The scale bar represents the number of expected substitutions per site. Bayesian posterior probability values are indicated above the branches. The black arrows indicate zebrafish proteins (Color figure online)
While it has been previously suggested in the literature (He and DiMario 2011; Calo et al. 2018; Jaberi-lashkari et al. 2022) that TCOF1 may have originated from a duplication event of NOLC1, this is the first phylogenetic study confirming this paralogy. Even though phylogenetic relationships among coelacanths, lungfishes and tetrapods are still debated (Irisarri and Meyer 2016), previous reports have placed both sarcopterygians in a close evolutionary association with tetrapods. Lungfish is, at the date, the closest living relative to tetrapods (Meyer et al. 2021), but coelacanths were considered transitional species between fish and tetrapods and its genome has provided insights into tetrapod evolution (Amemiya et al. 2013; Nikaido et al. 2013). Unfortunately, our attempts to identify NOLC1 and TCOF1 orthologs in lungfish were unsuccessful. Lungfishes are sarcopterygian fish with notably large genomes (Meyer et al. 2021) so the absence of a BLAST hit could be attributed to a problem with the current assembly of its genome or the depth of sequencing coverage. Nonetheless, the presence of a TCOF1 ortholog in coelacanths suggests that it could have played a role in the evolution of unique morphological and physiological characteristics in tetrapods, potentially contributing to their transition from water to land. Referring to previous research (Jaberi-lashkari et al. 2022), the presence of TCOF1 orthologs correlates with the presence of a nucleolar fibrillar center (FC) in species that span the transition to amniotes. According to this study, the authors noted that Xenopus, which is not an amniote, has a TCOF1 ortholog and the ability to form a FC. Our findings suggest that the emergence of TCOF1 may have played a role in evolution of tetrapods rather than specifically in amniotes. Given the unique features observed in the tree, stemming from potentially unreliable protein alignments, we explored alternative approaches in our study in order to refine the results.
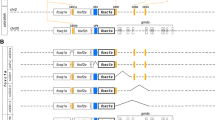
Conserved blocks of genomic sequences among species play a crucial role in identifying orthologous genes and support gene function annotation as well as understanding evolutionary relationships (Barbazuk et al. 2000). In line with this, we first conducted a synteny analysis using the Genomicus database (Thi et al. 2021). Genomicus is a conserved synteny browser that includes all genomes available at Ensembl in a phylogenetic context and it has the useful feature that it can reconstruct ancestral gene organizations (Thi et al. 2021). According to the results obtained from Genomicus, Rhesus nolc1 on chromosome (chr) 9 and mouse nolc1 on chr 19 show conserved synteny regarding human NOLC1 on chr 10. Similarly, chicken nolc1 exhibits conserved syntenic blocks on chr 6. Despite ongoing updates to the Genomicus database reflecting improvements in reference genomes and serving as a valuable resource for the molecular evolution community, challenges arose due to unplaced scaffolds in several species. Furthermore, upon conducting manual local synteny searches in data browsers for Danio rerio, frogs (Xenopus and Bufo bufo), and Coelacanth, we observed discrepancies between the genome browsers synteny and the synteny displayed in Genomicus (Genomicus). Upon manually examining the genomes of various bonny fish species (including zebrafish, common carp, indian medaka, zebra mbuna, spotted gar, and channel catfish), we identified a consistent pattern across these species. Specifically, we noted that two genes (mrps6 and psap) were consistently located to the left of nolc1 (Fig. 2), while two other genes (micu1 and mcu) were consistently located to the right, indicating a conservation of local synteny among them but showing a weakly conserved synteny with sharks, coelacanth, and tetrapods. According to databases, human gene TCOF1 is located on chr 5 (GRCh38 5: 150,357,629-150,400,308 [+]). The AlignView option of Genomicus revealed a strong conservation of TCOF1-associated syntenic blocks among humans, primates and rodents. In Genomicus, the chicken genome showed a high degree of synteny with the human genome in the regions associated with chr 5 (Hsa5) and chr 13 (not shown). The presence of unplaced scaffolds imposed the use of manual searches in the NCBI and Ensembl databases to identify the genes flanking Tcof1 in frogs (X. tropicalis chr 3, X. laevis chr 3.S) and Coelacanth (chr unknown) in order to establish synteny. Overall, we observed that the Tcof1 gene is located between arsi and cd74 genes and shares conserved local syntenies to Hsa5 in tetrapods. We also conducted manual searches on zebrafish chr 14, particularly the chromosome region of the D. rerio genome containing the gene coding for E7F9D9 (or zebrafish LLCP, as mentioned above). LLCP coding gene is adjacent to “acsl1b” (acyl-CoA synthetase long chain family member 1b) and “arsia” (arylsulfatase family, member Ia) to the left, followed by “f8” (coagulation factor VIII, procoagulant component) and “slc16a2” (solute carrier family 16 member 2) to the right. This region of zebrafish chr 14 does not share conserved synteny with Hsa5. Similarly, upon searching for NCBI orthologs of zebrafish LLCP (E7F9D9) coding genes (some of them are named “tcof1”) 89 fish genes are shown. Just by looking at a few of them, the LLCP-coding genes in fish species like common carp, indian medaka, zebra mbuna, channel catfish and the non-teleost fish spotted gar are located next to an orthologue of “arsia” but does not share a conserved synteny with Hsa5 nor with D. rerio chr 14.
Local synteny analysis of NOLC1, TCOF1 and LLCP-coding gene (fish homologs of E7F9D9). The alignment and visualization of the genomic sequences are shown in a schematic scale (image adapted from Genomicus Database). The reference genes are human NOLC1 (orange) and TCOF1 (green), with the zebrafish E7F9D9 (blue) as the reference gene in fishes. All their putative orthologs are centered. Chromosome segments show gene order, gene orientation, and gene loss or undefined gene (uncharacterized gene within conserved regions in the diverse vertebrates. Arsi (cyan), camk2a (purple) and ndst2 (blue) genes are highlighted to emphasize their positions. Arrows show zebrafish chromosomes. abcg2c, ATP-binding cassette sub-family G, member 2c; acsl1b, acyl-CoA synthetase long chain family member 1b; adad1, adenosine deaminase domain containing 1 (testis-specific); arhgap12, Rho GTPase activating protein 12; ARSI, arylsulfatase family member I; CAMK2A, calcium/calmodulin dependent protein kinase II alpha; CAMK2G, calcium/calmodulin dependent protein kinase II gamma; CD74, CD74 molecule; CDX1, caudal type homeobox 1; cetn4, centrin 4; cntnap3, contactin-associated protein-like 4; CSL2, L-rhamnose-binding lectin; ELOVL3, Very long chain fatty acid elongase 3; epc1b, enhancer of polycomb homolog 1; etf1, eukaryotic translation termination factor 1; f8, coagulation factor VIII, procoagulant component; GBF1, golgi brefeldin A resistant guanine nucleotide exchange factor 1; HPS6, biogenesis of lysosomal organelles complex 2 subunit 3; ik, IK cytokine; ik17B, interleukin-17B; il21, interleukin 21; itgb1b.1, integrin beta 1b.1; LDB1, LIM domain binding 1; LOC103181598, annexin A11 (elephant shark); lrrn4cl LRRN4 C-terminal like; mcu, mitochondrial calcium uniporter; micu1, mitochondrial calcium uptake 1; mrps6, mitochondrial ribosomal protein S6; ms4a4d, membrane-spanning 4-domains subfamily A member 4D; MYOZ1, myozenin 1; myoz3a, myozenin 3a; NDST1, N-deacetylase and N-sulfotransferase 1; NDST2A. N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a; NDST2, N-deacetylase and N-sulfotransferase 2; ndufa2, NADH:ubiquinone oxidoreductase subunit A2; NST1, stress response protein NST1; oit3, oncoprotein induced transcript 3; PITX3, paired like homeodomain 3; polr3a, polymerase (RNA) III (DNA directed) polypeptide A; PPRC1, PPARG related coactivator 1; ZBED8-like—LOC121005645 Bufo bufo; psap, prosaposin; rapgef6, Rap guanine nucleotide exchange factor 6; rnf20, ring finger protein 20, E3 ubiquitin protein ligase; rpl26, ribosomal protein L26; RPS14, ribosomal protein S14; SEC24 homolog C, COPII coat complex component; SLC16A2, solute carrier family 16 member 2; slc29a3,solute carrier family 29 member 3; SLC6A7, solute carrier family 6 member 7; SYNPO2L, synaptopodin 2 like; tmco6, transmembrane and coiled-coil domains 6; ugt8 2-hydroxyacyl sphingosine 1-beta-galactosyltransferase; Unch: uncharacterized; vsir, V-set immunoregulatory receptor (Color figure online)
Intriguingly, we were unable to detect a shark sequence coding for either TCOF1 or LLCP. Moreover, the elephant shark (also the white shark, not shown) displays local synteny around nolc1 (ndst2a/camk2g2a/arsi/nolc1) that is reminiscent of the local synteny detected for coelacanth´s nolc1 and tetrapod´s genes coding for TCOF1, as well as partially similar to the spotted gar´s gene coding for LLCP (see highlighted genes in Fig. 2 and Suppl. Fig 6 for MSAs of the proteins coded by camk2g and ndst2). Shark nolc1 local synteny contrasts with that presented by the NOLC1 genes of tetrapods (except B. bufo) and teleost fish. These findings suggest a possible evolutionary divergence in the region surrounding the NOLC1 gene in bony vertebrates.
Collectively, this data suggests that the current naming system for genes in zebrafish and other fish species may not accurately represent their relationships with genes in other species, particularly in terms of their genomic organization. In order to maintain caution in the annotation of these genes (which often impacts multiple databases and is difficult to reverse) we suggest the name LLCP for E7F9D9-like proteins until more information is available to support their homology relationships.
Given that synteny analysis alone is not conclusive, and taking into account that low sequencing coverage of genomes might impact the accuracy of the analysis (Liu et al. 2018), to further investigate the orthology of the zebrafish LLCP, we explored other tools available.
The functional specificity of proteins is expected to be conserved among orthologs and to be different among paralogs (Mirny and Gelfand 2002). So, amino acid sequences, particularly those that are related to the protein's function (e.g., the active site, regulatory/interacting regions) can provide valuable information in distinguishing orthologs from paralogs, even in IDPs. Given the assumption of specificity conservation among orthologs and differing specificity among paralogs, we directed our focus towards the disordered regions of these proteins. To explore the presence of IDRs in the zebrafish orthologues of these proteins, we used the MobiDB database (He and DiMario 2011; Piovesan et al. 2023). MobiDB elucidates the structural properties and amino acid composition of disordered interacting interfaces, revealing alternative binding modes distinct from the conventional lock-and-key mechanism observed in well-structured partners.
We studied the disorder and mobility predictions for these proteins from D. rerio and used the information of the human counterparts for comparison. As shown in Fig. 3, all four proteins exhibit a long, almost complete (more than 95% of the sequences) disordered region (tracts shaded in pink in the figure). MobiDB also provides an additional characterization of the predicted regions related to their functional role. A feature displayed by MobiDB-lite are LIPs (Lineal Interacting Peptides). IDRs that are involved in binding interactions are called LIPs and are classified according to different levels of evidence and features related to their binding modes (Piovesan et al. 2023). Numerous LIPs were identified throughout the sequences of both D. rerio and H. sapiens TCOF1 and NOLC1 proteins, indicating extensive available sequences for interaction with other proteins. This observation aligns with previous findings for proteins functionally linked to the nucleolus (Kastano et al. 2020). Another feature displayed by MobiDB highlights amino acid residue composition. Different types of disordered regions are classified according to the fraction of charged residues and net charge (Comp. biased, in Fig. 3). Q13428 (human TCOF1), Q14978 (human NOLC1) and B8JIY2 (zebrafish Nolc1) exhibit predominant low-complexity tracts with few negative or positive polyelectrolyte areas. In contrast, E7F9D9 (D. rerio LLCP) displays more polar tracts and positive regions distributed along the center of its sequence (see arrows in Fig. 3D). Based on these MobiDB results, and the known feature of these proteins to possess repeated alternating motifs that act as scaffolds for delivering and positioning snoRNPs (He and DiMario 2011), we further investigated whether the putative zebrafish orthologs of TCOF1 and NOLC1 contain internal repeats.
Structural disorder and composition analysis of NOLC1 and TCOF1 proteins. MobiDB results for A human TCOF1 (Uniprot Q13428), B human NOLC1 (Uniprot Q14978), C zebrafish Nolc1 (Uniprot B8JIY2) and D zebrafish Uniprot E7F9D9. The arrows indicate the unusual composition of E7F9D9 compared with the other proteins. The line below each graph indicates the amino acid sequence coordinates of each protein. On the left of each graph, the analyzed feature is indicated: disorder (pink) and structured (light blue) regions, obtained by consensus, Composition biased (MobiDB-lite, green: polar residues; light blue: positive polyelectrolyte, pink: basic residues; pale orange: basic and acidic residues, polyampholyte) and linear interacting peptides (LIP, purple). P, Predicted annotation by a sequence-based method (Color figure online)
The RADAR algorithm (Heger and Holm 2000) was used to detect and align repeats within the sequences of four proteins: Q13428 (human TCOF1), Q14978 (human NOLC1), B8JIY2 (D. rerio nolc1) and E7F9D9 (D. rerio LLCP in this work) (Fig. 4 and Supp. Fig. 4). Q13428, human TCOF1, presented 14 repeats of a sequence of 62 amino acid residues. These repeats exhibit a lysine/proline-rich basic area followed by a serine/glutamic-rich acidic region (S/E-rich). The RADAR tool also detected two other kinds of repeats of lower level and fewer repetitions (Fig. 4B). On the other hand, Q14978, human NOLC1, shows 10 repeats of a length of 47 amino acid residues, once again characterized with S/E-rich and proline-rich stretches. An additional type of repeat was identified, although it repeated only 3 times and had a low score. Similarly, to human TCOF1 and NOLC1, the RADAR algorithm detected 17 S/E-rich and proline-rich repeats in zebrafish Nolc1, along with two additional low score repeat types (Fig. 4A and B, and Suppl. Fig. 4). Lastly, zebrafish LLCP presented 3 types of repeats, each repeated 3–5 times in the sequence. Interestingly, none of these repeats exhibited the characteristic S/E-rich and proline-rich motifs. We applied the RADAR algorithm to all proteins depicted in the phylogenetic tree in Fig. 1. All of them, except for LLCPs proteins, presented the S/E-rich motifs repeated several times, including Tcof1 from coelacanth (Suppl. Fig. 4). Altogether, these results indicate that all the fish LLCPs included in the tree have a disordered low-complexity central domain but lacks the abundance of S/E-rich repeats, a characteristic feature of nucleolar proteins like NOLC1 and TCOF1, linked to their nucleolar function (He and DiMario 2011). In a recent work, Lee et al., showed the prevalence of glutamic (E)-rich LCR (low-complexity region) sequences among human nucleolar proteins and use LCR copy number analyses and LCR maps to identify E-rich LCR-containing proteins which act as scaffolds or clients in the nucleolus (Lee et al. 2022). Notably, TCOF1 and NOLC1 are among the most representative proteins of this group. In particular, Jaberi-lashkari et al. (2023) showed that TCOF1 function seems to depend on the number of repetitions of these LCRs, being 5 or less detrimental. Also, Velichko et al. (2024) demonstrated that TCOF1 forms the core of the nucleolus's fibrillar center through liquid-liquid phase separation mediated by homotypic associations. Both studies emphasize the critical role of TCOF1's central domain (which contains the repeated stretches of alternatingly charged amino-acid residues) in maintaining nucleolar structure and function. Given the bipartite nucleolus in fish and LLCP's lack of sufficient LCR repetitions, it likely cannot function as a nucleolar scaffold like TCOF1 or NOLC1
LisH motif and S/E-rich repeats in NOLC1 and TCOF1 proteins. A Schematic representation of the LisH motif and S/E rich repeats in both human and zebrafish NOLC1 and TCOF1 proteins. Each protein is represented by a sequence coordinate where each motif or repeat is represented according to its position in the sequence. B Table showing the different repeats detected along with the number of times that each repeat occurs. The repeat marked with a pentagon represents the best-scored S/E-rich repeat with the highest number of repetitions
Orthologous genes committed to the same function (e.g. ribosome biogenesis) should display similar temporal expression behavior during development. This sort of approach has already been used for the identification of functionally equivalent, orthologous genes in Arabidopsis thaliana (Das et al. 2016) where the authors demonstrated that even a small set of gene expression data in addition to sequence homologies are instrumental in the assignment of functional orthologs in the presence of multiple orthologs. To understand ribosomopathies, researchers have studied the transcriptional expression of ribosome biogenesis factors (including tcof1) in Xenopus laevis embryos using ultra-high temporal resolution RNASeq (Robson et al. 2016). Interestingly, biogenesis factors (including tcof1) generally exhibited three, variably pronounced, peaks of expression during development suggesting increased biological demand at these stages (Robson et al. 2016) emphasizing the critical role that these proteins play in the development and survival of the embryo. RNASeq mRNA baseline expression from transcriptional profiling of zebrafish developmental stages (White et al. 2017) is available through Expression Atlas (Papatheodorou et al. 2019). Figure 5 shows the expression level of the transcripts corresponding to zebrafish LLCP (E7F9D9) and Nolc1 (B8JIY2) proteins. As shown in Fig. 5, the B8JIY2-associated transcript shows three peaks in the expression corresponding to blastula (128 cells), gastrula and 20–25 somite stages. The inset of Fig. 5 shows the transcriptional expression data (from the same profile in Expression Atlas) of two well-known ribosome biogenesis factors (utp4 and rbm28). These transcripts show two clear peaks (at blastula and gastrula) and one less sharp peak, similar to the Nolc1 coding transcripts. On the contrary, the zebrafish LLCP-associated transcript shows low and decreasing expression along early zebrafish development without any clear peaking expression. Orthologues and paralogs that have diverged recently are expected to share similar functions, which requires them to have similar expression patterns. Our results suggest that the Nolc1 (B8JIY2) coding gene has an expression profile compatible with a ribosome biogenesis factor like TCOF1, but the expression data for zebrafish LLCP (E7F9D9) during early embryogenesis is flattened and inconclusive.
Expression of zebrafish Nolc1 (B8JIY2) and LLCP (E7F9D9) transcripts during zebrafish early development. mRNA expression data (in TPM, Transcripts Per kilobase Million) for transcripts corresponding to the proteins of interest was obtained from the Expression Atlas experiment named “Baseline expression from transcriptional profiling of zebrafish developmental stages”. Data is presented according to the developmental stages (shown in the list to the right). The arrows indicate three expression peaks and the corresponding stages are underlined in the list (see text for further information)
Following the same idea, but from the spatial perspective, we delved into single-cell transcriptomics data spanning zebrafish (Wagner et al. 2018) and Xenopus (as reference) embryos development (Briggs et al. 2018; Weinreb et al. 2018), employing the computer-based analytical tool SPRING. Supplementary Fig. 7 illustrates the expression profiles of zebrafish coding genes for Nolc1 (B8JIY2) and LLCP (E7F9D9) alongside Xenopus genes nolc1 and tcof1, juxtaposed with the neural crest marker sox10. Notably, these biogenesis factors exhibit ubiquitous expression patterns. A similar result can be observed (Suppl. Fig. 7) for transcriptomic analysis performed in microdissections of X. laevis embryos (Plouhinec et al. 2017). Despite the mutant’s specific phenotypes, reminiscent of many ribosomopathies, they do not result from localized expression within affected tissues (Yelick and Trainor 2015; Farley-Barnes et al. 2019). Consequently, discerning between tcof1 and nolc1 based on definitive expression features remains elusive. Nonetheless, it's noteworthy that in Xenopus, both genes exhibit robust expression in neural crest territories, whereas in zebrafish, only the Nolc1 coding gene displays pronounced expression in these cells.
Conclusions
In silico tools, like algorithms that produce sequence alignments and synteny analysis, are useful to assign orthologies and have been widely and successfully used (Mirny and Gelfand 2002; Postlethwait 2006; Nichio et al. 2017). However, in cases dealing with the presence of paralogs with IDRs and LCRs, these tools may not be sufficient to univocally assign and annotate orthologues and further examination is required. This is especially true for D. rerio and possibly for all teleost fish, which experienced an extra WGD. Recently, several computational tools and databases (e.g., MobiDB (Piovesan et al. 2023)) were developed and made public to assist with IDP characterization and analysis. Also, databases like Expression Atlas (Papatheodorou et al. 2019), a public repository that provides basal gene expression pattern data under different biological conditions, allow the in silico analysis of expression. In particular, Expression Atlas offers, at the moment, 17 different experiments of gene expression in zebrafish under different conditions, including a high-resolution mRNA expression time course of embryonic development (White et al. 2017). These new tools can be incorporated in the analysis to discriminate paralogs and annotate genes with more confidence. Curation of annotated genes in order to include these new data is essential to organisms like D. rerio, adopted by the scientific community to study and model human diseases. Figure 6 outlines our proposed approach for integrating novel computational tools and data from databases into the conventional analysis of alignments and tree construction. This strategy aims to tackle homology challenges encountered when studying proteins with intrinsically disordered regions and sequences of low-complexity composition.
Schematic diagram showing the suggested workflow for analyzing homology relations among Intrinsically Disordered Proteins (IDPs). Faced with a controversy regarding the homology among sequences of interest, the suggested steps are (i) to obtain the sequences and available information (identifiable domains, etc.) accessible in databases such as Protein information Resource (PIR); (ii) confirm and score identifiable domains (if present, they can also help in MSA (Multiple Sequence Alignments)); (iii) execute classic evaluation strategies such as Reciprocal Best Hit and MSAs with evaluation of identity (and similarity) percentages; (iv) an analysis of local synteny and (v) calculate a phylogenetic tree. For these purposes, it may be necessary to add further sequences from other species (obtainable through BLAST searches in databases). In the case of IDPs, (vi) analysis of the percentage of disordered sequences, the composition of amino acid residues and the presence of Linear Interacting Peptides (LIPs) can help distinguish functionally relevant sequences that can suggest a difference among them. The presence of LIPs and the low sequence complexity are suggestive of the presence of internal repeats (identifiable by RADAR). This information can also be used to revise MSAs. Finally, (vii) selection (dN/dS) can be assessed to detect differential diversifying or purifying pressures. Additionally, the expression of the genes of interest can be evaluated both spatially and temporally and compared with existing data. Bibliography revision along all these stages is essential to interpret the results and decide next steps. Eventually, the information generated may be the input for other analysis
In this study, we gathered results from different databases and employed web-based algorithms to disentangle the annotation of TCOF1 and NOLC1 orthologues in zebrafish. Overall, our findings suggest that the currently annotated TCOF1 orthologue in zebrafish, Uniprot ID E7F9D9 (or now LLCP), does not possess the requisite sequence characteristics or expression profile to fulfill the functions of the nucleolar protein TCOF1.
Based on our findings, we recommend that the gene ZDB-GENE-030131-6349 (ENSDARG00000024561), which encodes the protein with the Uniprot ID B8JIY2, be more accurately referred to as “nolc1” (for nucleolar and coiled-body phosphoprotein 1). Finally, and more importantly, we strongly discourage the annotation of ZDB-GENE-141219-12 (ENSDARG00000087555, coding for E7F9D9) as tcof1 until more functional information becomes available.
Although the primary aim of this article was to assess the Tcof1 nomenclature/annotation in zebrafish, the molecular phylogenetic analysis allowed us to perform a comparative study of the TCOF1 and NOLC1 genes in different species and to infer their evolutionary relationships. Our findings agree with existing literature proposing that TCOF1 and NOLC1 are paralogs and diverged from a common ancestral gene through duplication during the course of evolution. Rapidly evolving proteins present challenges in accurately identifying homologous sequences and aligning them, particularly for sequences in highly repetitive, low-complexity regions. Paralogs tend to diverge more rapidly due to relaxed selective pressure and may retain or lose function, leading to differential evolutionary rates among sites within a protein (Nunez-Castilla and Siltberg-Liberles 2020). To gain further insights, we included in the molecular phylogenetic tree sequences from the elephant shark (Callorhinchus milii), possessing the most slowly evolving vertebrate genome identified to date (Holland and Ocampo Daza 2018; Sacerdot et al. 2018) and the sea lamprey (Petromyzon marinus), which underwent the two WGDs at the base of vertebrates. These species only present the NOLC1 protein, lacking the presence of TCOF1 or LLCP. The spotted gar (Lepisosteus oculatus), which evaded the Teleost genome duplication event (TGD), intriguingly presents NOLC1 and LLCPs. And the coelacanth, an accepted ancestor of tetrapods (also a species that did not experience an extra WGD), presents NOLC1 and TCOF1. According to (Holland and Ocampo Daza 2018; Sacerdot et al. 2018), when studying the evolution of HOX genes, they found that during the evolution of bony vertebrates (euteleostomi) and amniotes, their lineages experienced additional chromosomal rearrangements. These events included chromosome fusions, segmental duplications, and independent gene gains and losses. In this connection, the observed synteny pattern surrounding shark and coelacanth nolc1 genes, more closely resembling the genomic context of TCOF1 and LLCP coding sequences than the other nolc1 genes, could be evidence of similar genomic rearrangements. These observations suggest a potential paralogous relationship among NOLC1, TCOF1 and LLCP, being a NOLC1 coding sequence the ancestor. However, due to gene duplications and losses, occurring both in Tetrapoda and fishes, and until further information becomes available, the homology relationships among these genes are currently difficult to establish definitively. Moreover, we cannot disregard the possibility of additional hidden paralogs influencing the phylogenetic landscape (Kuraku 2011; Holland and Ocampo Daza 2018). In this connection, it is intriguing that only one copy of the nolc1 gene is present in the teleost species analyzed, as they underwent the TGD. Finally, another striking aspect in the phylogenetic tree is that the NOLC1 family forms two clusters, only one of which has a common ancestor with the TCOF1 and LLCP families. Further molecular evolutionary deep analysis would be necessary to address these issues, but it would surely provide valuable insights into the function and evolution of the NOLC1 and TCOF1 genes in vertebrates.
As a final comment, we propose that in order to understand the phylogenetic relationships among these IDPs (and others), it is necessary to put together the information from multiple sources and look for details that clarify the evolutionary path undergone. Currently, to our knowledge there is no accessible tool alike BLAST that allows biologists to effortlessly perform this type of search that integrates not only sequence similarity but also synteny data or expression data or other data (e.g. interactor network, metabolic pathways) and then to correlate this information with phenotypes or a particular evolutionary history. This information has to be manually compiled and analyzed, which takes time and effort. Finally, in cases where evidence from different sources does not converge to a univocal solution, we recommend caution in the annotation, since once assigned it is usually replicated by other databases propagating the (mis)annotation and hindering further any correction.
Data Availability
All data generated or analyzed during this study are included in the manuscript and supporting files.
References
Amemiya CT, Alfoldi J, Lee AP et al (2013) The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311–316. https://doi.org/10.1038/NATURE12027
Barbazuk WB, Korf I, Kadavi C et al (2000) The syntenic relationship of the zebrafish and human genomes [1]. Genome Res 10:1351–1358
Bradford YM, Toro S, Ramachandran S et al (2017) Zebrafish models of human disease: gaining insight into human disease at ZFIN. ILAR J 58:4–16. https://doi.org/10.1093/ilar/ilw040
Briggs JA, Weinreb C, Wagner DE et al (2018) The dynamics of gene expression in vertebrate embryogenesis. Science 5780:1–17. https://doi.org/10.1126/science.aar5780.The
Calo E, Gu B, Bowen ME et al (2018) Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature 554:112–117. https://doi.org/10.1038/nature25449
Chen K, Durand D, Farach-Colton M (2000) NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol 7:429–447. https://doi.org/10.1089/106652700750050871
Collin J, Martin P (2017) Zebrafish as a research organism: danio rerio in biomedical research. Elsevier, Amsterdam
Criscuolo A, Gribaldo S (2010) BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. https://doi.org/10.1186/1471-2148-10-210
Das M, Haberer G, Panda A et al (2016) Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol 171:2343–2357. https://doi.org/10.1104/pp.15.01207
de Castro E, Sigrist CJA, Gattiker A et al (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362. https://doi.org/10.1093/NAR/GKL124
De Peralta MSP, Mouguelar VS, Sdrigotti MA et al (2016) Cnbp ameliorates Treacher Collins syndrome craniofacial anomalies through a pathway that involves redox-responsive genes. Cell Death Dis 7:e2397. https://doi.org/10.1038/cddis.2016.299
Dixon J, Edwards SJ, Anderson I et al (1997) Identification of the complete coding sequence and genomic organization of the Treacher Collins syndrome gene. Genome Res 7:223–234. https://doi.org/10.1101/gr.7.3.223
Emes RD (2001) A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet 10:2813–2820. https://doi.org/10.1093/hmg/10.24.2813
Falcon KT, Watt KEN, Dash S et al (2022) Dynamic regulation and requirement for ribosomal RNA transcription during mammalian development. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.2116974119
Farley-Barnes KI, Ogawa LM, Baserga SJ (2019) Ribosomopathies: old concepts, new controversies. Trends Genet 35:754–767. https://doi.org/10.1016/j.tig.2019.07.004
Fitriasari S, Trainor PA (2021) Diabetes, oxidative stress, and DNA damage modulate cranial neural crest cell development and the phenotype variability of craniofacial disorders. Front Cell Dev Biol 9:644410. https://doi.org/10.3389/fcell.2021.644410
Gerlitz G, Darhin E, Giorgio G et al (2005) Novel functional features of the Lis-H domain: role in protein dimerization, half-life and cellular localization. Cell Cycle 4:1632–1640. https://doi.org/10.4161/cc.4.11.2151
Grzanka M, Piekiełko-Witkowska A (2021) The role of TCOF1 gene in health and disease: beyond Treacher Collins syndrome. Int J Mol Sci 22:1–19. https://doi.org/10.3390/IJMS22052482
He F, DiMario P (2011) Structure and function of Nopp140 and treacle. Protein Rev 15:253–278. https://doi.org/10.1007/978-1-4614-0514-6_11
Heger A, Holm L (2000) Rapid automatic detection and alignment of repeats in protein sequences. Proteins Struct Funct Genet 41:224–237. https://doi.org/10.1002/1097-0134(20001101)41:2%3c224::AID-PROT70%3e3.0.CO;2-Z
Holland LZ, Ocampo Daza D (2018) A new look at an old question: when did the second whole genome duplication occur in vertebrate evolution? Genome Biol 19:2–5. https://doi.org/10.1186/s13059-018-1592-0
Irisarri I, Meyer A (2016) The identification of the closest living relative(s) of tetrapods: phylogenomic lessons for resolving short ancient internodes. Syst Biol 65:1057–1075. https://doi.org/10.1093/sysbio/syw057
Jaberi-lashkari N, Lee B, Aryan F, Calo E (2022) TCOF1 is a single-component scaffold of the nucleolar fibrillar center. bioRxiv. https://doi.org/10.1101/2022.10.16.512422
Jaberi-Lashkari N, Lee B, Aryan F, Calo E (2023) An evolutionarily nascent architecture underlying the formation and emergence of biomolecular condensates. Cell Rep 42:112955. https://doi.org/10.1016/j.celrep.2023.112955
Kastano K, Erdős G, Mier P et al (2020) Evolutionary study of disorder in protein sequences. Biomolecules 10:1–17. https://doi.org/10.3390/biom10101413
Kosakovsky Pond SL, Frost SDW (2005) Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. https://doi.org/10.1093/molbev/msi105
Kosakovsky Pond SL, Poon AFY, Velazquez R et al (2020) HyPhy 2.5—a customizable platform for evolutionary hypothesis testing using phylogenies. Mol Biol Evol 37:295–299. https://doi.org/10.1093/molbev/msz197
Kosakovsky Pond SL, Wisotsky SR, Escalante A et al (2021) Contrast-FEL—a test for differences in selective pressures at individual sites among clades and sets of branches. Mol Biol Evol 38:1184–1198. https://doi.org/10.1093/molbev/msaa263
Lange J, Wyrwicz LS, Vriend G (2016) KMAD: knowledge-based multiple sequence alignment for intrinsically disordered proteins. Bioinformatics 32:932–936. https://doi.org/10.1093/bioinformatics/btv663
Lee B, Jaberi-Lashkari N, Calo E (2022) A unified view of low complexity region(LCRs) across species. Elife 11:1–32. https://doi.org/10.7554/eLife.77058
Liu D, Hunt M, Tsai IJ (2018) Inferring synteny between genome assemblies: a systematic evaluation. BMC Bioinform 19:1–13. https://doi.org/10.1186/s12859-018-2026-4
Locascio A, Manzanares M, Blanco MJ, Nieto MA (2002) Modularity and reshuffling of snail and slug expression during vertebrate evolution. Proc Natl Acad Sci USA 99:16841–16846. https://doi.org/10.1073/pnas.262525399
Meyer A, Schloissnig S, Franchini P et al (2021) Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590:284–289. https://doi.org/10.1038/s41586-021-03198-8
Midic U, Dunker AK, Obradovic Z (2009) Protein sequence alignment and structural disorder: a substitution matrix for an extended alphabet. In: Proceedings of the KDD-09 workshop on statistical and relational learning in bioinformatics, StReBio ’09. pp 27–31
Mirny LA, Gelfand MS (2002) Using orthologous and paralogous proteins to identify specificity determining residues. Genome Biol. https://doi.org/10.1186/gb-2002-3-3-preprint0002
Muffato M, Louis A, Poisnel CE, Crollius HR (2010) Genomicus: a database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics 26:1119–1121. https://doi.org/10.1093/bioinformatics/btq079
Müller T, Vingron M (2001) Modeling amino acid replacement. J Comput Biol 7:761–776. https://doi.org/10.1089/10665270050514918
Murrell B, Moola S, Mabona A et al (2013) FUBAR: a fast, unconstrained bayesian AppRoximation for inferring selection. Mol Biol Evol 30:1196–1205. https://doi.org/10.1093/molbev/mst030
Murrell B, Weaver S, Smith MD et al (2015) Gene-wide identification of episodic selection. Mol Biol Evol 32:1365–1371. https://doi.org/10.1093/molbev/msv035
Na J-HH, Lee W-KK, Gyu YuY, Yu YG (2018) How do we study the dynamic structure of unstructured proteins: a case study on nopp140 as an example of a large, intrinsically disordered protein. Int J Mol Sci. https://doi.org/10.3390/ijms19020381
Nichio BTL, Marchaukoski JN, Raittz RT (2017) New tools in orthology analysis: a brief review of promising perspectives. Front Genet 8:1–12. https://doi.org/10.3389/fgene.2017.00165
Nikaido M, Noguchi H, Nishihara H et al (2013) Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res 23:1740–1748. https://doi.org/10.1101/gr.158105.113
Papadopoulos JS, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073–1079. https://doi.org/10.1093/bioinformatics/btm076
Papatheodorou I, Moreno P, Manning J et al (2019) Expression atlas update: from tissues to single cells. Nucleic Acids Res 48:77–83. https://doi.org/10.1093/nar/gkz947
Parichy DM (2015) Advancing biology through a deeper understanding of zebrafish ecology and evolution. Elife 4:1–11. https://doi.org/10.7554/eLife.05635
Piovesan D, Del Conte A, Clementel D et al (2023) MobiDB: 10 years of intrinsically disordered proteins. Nucleic Acids Res 51:D438–D444. https://doi.org/10.1093/NAR/GKAC1065
Plouhinec JL, Medina-Ruiz S, Borday C et al (2017) A molecular atlas of the developing ectoderm defines neural, neural crest, placode, and nonneural progenitor identity in vertebrates. PLOS Biol. https://doi.org/10.1371/journal.pbio.2004045
Postlethwait JH (2006) The zebrafish genome: a review and msx gene case study. Genome Dyn 2:183–197. https://doi.org/10.1159/000095104
Rambaut A, Drummond AJ, Xie D et al (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/sysbio/syy032
Robson A, Owens NDL, Baserga SJ et al (2016) Expression of ribosomopathy genes during Xenopus tropicalis embryogenesis. BMC Dev Biol 16:1–13. https://doi.org/10.1186/s12861-016-0138-5
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Rosas MG, Lorenzatti A, Porcel de Peralta MS et al (2019) Proteasomal inhibition attenuates craniofacial malformations in a zebrafish model of Treacher Collins syndrome. Biochem Pharmacol 163:362–370. https://doi.org/10.1016/j.bcp.2019.03.005
Rosello OPI, Kondrashov FA (2014) Long-term asymmetrical acceleration of protein evolution after gene duplication. Genome Biol Evol 6:1949–1955. https://doi.org/10.1093/gbe/evu159
Sacerdot C, Louis A, Bon C et al (2018) Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol 19:1–15. https://doi.org/10.1186/s13059-018-1559-1
Santoriello C, Zon LI (2012) Hooked! modeling human disease in zebrafish. J Clin Invest 122:2337–2343. https://doi.org/10.1172/JCI60434
Sochacka M, Karelus R, Opalinski L et al (2022) FGF12 is a novel component of the nucleolar NOLC1/TCOF1 ribosome biogenesis complex. Cell Commun Signal 20:1–14. https://doi.org/10.1186/s12964-022-01000-4
Tanabe AS (2011) Kakusan4 and aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour 11:914–921. https://doi.org/10.1111/j.1755-0998.2011.03021.x
Thi N, Nguyen T, Vincens P et al (2021) Genomicus in 2022: comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab1091
Van Der Lee R, Buljan M, Lang B et al (2014) Classification of intrinsically disordered regions and proteins. Chem Rev 114:6589–6631
Velichko AK, Kovina AP, Luzhin AV et al (2024) Treacle’s ability to form liquid phase condensates is essential for nucleolar fibrillar center assembly, efficient rRNA transcription and processing, and rRNA gene repair. Elife. https://doi.org/10.7554/eLife.96722.1
Vincent M, Geneviève D, Ostertag A et al (2016) Treacher Collins syndrome: a clinical and molecular study based on a large series of patients. Genet Med 18:49–56. https://doi.org/10.1038/gim.2015.29
Wagner DE, Weinreb C, Collins ZM et al (2018) Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360:981–987. https://doi.org/10.1126/science.aar4362
Wall DP, Fraser HB, Hirsh AE (2003) Detecting putative orthologs. Bioinforma Appl 19:1710–1711. https://doi.org/10.1093/bioinformatics/btg213
Wang HC, Li K, Susko E, Roger AJ (2008) A class frequency mixture model that adjusts for site-specific amino acid frequencies and improves inference of protein phylogeny. BMC Evol Biol 8:1–13. https://doi.org/10.1186/1471-2148-8-331
Weaver S, Shank SD, Spielman SJ et al (2018) Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol 35:773–777. https://doi.org/10.1093/molbev/msx335
Weiner AMJ, Scampoli NL, Calcaterra NB (2012) Fishing the molecular bases of Treacher Collins syndrome. PLoS ONE 7:e29574. https://doi.org/10.1371/journal.pone.0029574
Weinreb C, Wolock S, Klein AM (2018) SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34:1246–1248. https://doi.org/10.1093/bioinformatics/btx792
Werner A, Iwasaki S, McGourty CA et al (2015) Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525:523–527. https://doi.org/10.1038/nature14978
White RJ, Collins JE, Sealy IM et al (2017) A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 6:e30860. https://doi.org/10.7554/ELIFE.30860
Yang Z (1994) Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol 39:306–314. https://doi.org/10.1007/BF00160154
Yelick PC, Trainor PA (2015) Ribosomopathies: global process, tissue specific defects. Rare Dis 3:e1025185. https://doi.org/10.1080/21675511.2015.1025185
Funding
This research was funded by CONICET (Grant number PIP 2020-2023-0505), Universidad Nac. de Rosario (80020180300039UR), Universidad Nac. de Tucumán (PIUNT D712), and Foncyt-ANPCYT (Grant numbers PICT 2019-00307, PICT2018-1370, PICT2019-0187). G.H.T. is a CONICET research fellow.
Author information
Authors and Affiliations
Contributions
G.C.; Conceptualization (lead); writing—original draft (lead); formal analysis (lead); writing—review and editing (equal). G.H.T.; formal analysis and figure construction; writing—review and editing (equal). M.J.A.; writing—review and editing (equal). J.P. and M.L.F.-tree construction - review and editing. All the authors read and approved the submitted article.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Handling editor: Cara Weisman.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hill-Terán, G., Petrich, J., Falcone Ferreyra, M.L. et al. Untangling Zebrafish Genetic Annotation: Addressing Complexities and Nomenclature Issues in Orthologous Evaluation of TCOF1 and NOLC1. J Mol Evol (2024). https://doi.org/10.1007/s00239-024-10200-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00239-024-10200-0