Abstract
Many reproductive proteins show signatures of rapid evolution through sequence divergence and duplication. These features of reproductive genes may complicate the detection of orthologs across taxa, making it difficult to connect studies in model systems to human biology. In mice, ZP3r/sp56 is a binding partner to the egg coat protein ZP3 and may mediate induction of the acrosome reaction, a crucial step in fertilization. In rodents, ZP3r, as a member of the Regulators of Complement Activation cluster, is surrounded by paralogs, some of which have been shown to be evolving under positive selection. Although primate egg coats also contain ZP3, sequence divergence paired with paralogous relationships with neighboring genes has complicated the accurate identification of the human ZP3r ortholog. Here, we phylogenetically and syntenically resolve that the human ortholog of ZP3r is the pseudogene C4BPAP1. We investigate the evolution of this gene within primates. We observe independent pseudogenization events of ZP3r in all Apes with the exception of Orangutans, and independent pseudogenization events in many monkey species. ZP3r in both primates that retain ZP3r and in rodents contains positively selected sites. We hypothesize that redundant mechanisms mediate ZP3 recognition in mammals and ZP3r’s relative importance to ZP recognition varies across species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Complex molecular interactions between the sperm and egg mediate fertilization (Swanson and Vacquier 2002; Carlisle and Swanson 2020). Although recent discoveries have described many important molecules mediating mammalian sperm-egg plasma membrane fusion, the molecular mediators of sperm–egg coat interactions remain ambiguous (Carlisle and Swanson 2020). The glycoproteinaceous egg molecules ZP2 and ZP3 have been shown to bind sperm in a species-specific manner, indicating that these molecules may be involved in sperm–egg interactions (Bleil and Wassarman 1980; Litscher, et al. 2009; Avella, et al. 2014; Carlisle and Swanson 2020). While there is no known sperm protein binding partner of ZP2, ZP3r (formally known as sp56) is described as the receptor of ZP3 in mice (Buffone, et al. 2008; Wassarman 2009). ZP3r is a sperm acrosomal protein that becomes transiently exposed on the sperm head post-capacitation in mice (Muro, et al. 2012). Isolated ZP3r inhibits sperm binding by binding mouse eggs in vitro and specifically binds ZP3 as shown by photoaffinity cross-linking (Bleil and Wassarman 1990; Buffone, et al. 2008). Despite these compelling results, mouse knockouts of ZP3r do not result in observable reductions in fertility; however, this may be due to alternative assays being needed to observe ZP3r’s function (Adham 1998; Muro, et al. 2012; Okabe 2018). For example, the sperm protein PKDREJ, while not causing infertility in male mice knockouts, does lead to a delay in the induction of the acrosome reaction by ZP recognition and a reduction in male fertility compared to wild-type animals in sequential mating trials (Sutton, et al. 2006, 2008; Miyata, et al. 2016). Multiple proteins, including ZP3r, may contribute to sperm recognition of the egg coat, and have redundant functions.
Identification of the human ortholog of ZP3r has been controversial. Previous studies have misidentified human ZP3r as either SELENBP1 or C4BPA, due to nomenclature confusion or difficulties in establishing orthology, respectively (Morgan, et al. 2010, 2017; Morgan and Hart 2019). In rodents, ZP3r is found in chromosome 1 among paralogous protein-coding genes that make up the Regulators of Complement Activation (RCA) cluster (Hourcade, et al. 1989; Krushkal, et al. 2000). The RCA cluster contains a tandem array of immunity genes which function in the complement system (Hourcade, et al. 1989; Krushkal, et al. 2000). Many of the proteins within this structure are paralogous complement control proteins (CCPs) that are defined by containing tandem arrays of CCP domains (also called Sushi domains) (Ojha, et al. 2019). CCP domains are small (~ 60 amino acid) beta sandwich domains with 2 conserved disulfide bonds and are found in adhesion proteins as well as complement system proteins (Ojha, et al. 2019). Many of the genes within the RCA cluster are diverging rapidly in sequence between species (Hart, et al. 2018). This sequence divergence of paralogs can complicate accurate ortholog identification. In this study, we demonstrate using syntenic and phylogenetic analysis that the pseudogene C4BPAP1 is the primate ortholog of ZP3r. We examine the evolution of ZP3r in primates and uncover a pattern of recurrent independent pseudogenizations of ZP3r in great apes and monkeys. Finally, we discuss how redundant mechanisms of gamete recognition may lead to species-specific loss events of ancestral fertilization genes.
Results and Discussion
C4BPAP1 is the Ortholog of Mouse ZP3r
Although well characterized in mice, the identification of primate ZP3r has been contentious. In mice, ZP3r is located within the RCA cluster (Fig. 1A) between its paralog C4BP (human ortholog C4BPA) and CD55. C4BPA/C4BP is a large glycoprotein that acts as an inhibitor within the complement system (Okroj 2018). An examination of the syntenic genomic region in humans reveals the gene C4BPAP1, a known pseudogenized paralog of C4BPA. No mRNA transcripts of C4BPAP1 are present within the NCBI blast database. Human C4BPAP1 shares a domain structure and sequence similarity to human C4BPA but contains a premature stop codon in Exon 2 that is fixed in humans. Human C4BPA and C4BPAP1 are composed of 11 exons which contain 8 CCP/sushi domains and a C-terminal transmembrane domain (Hofmeyer, et al. 2013). Both C4BPAP1 and C4BPA contain an additional CCP domain to mouse ZP3r which is missing CCP domain 7. A recent investigation identified C4BPA as the human ortholog of ZP3r, proposing that ZP3r arose from a duplication of C4BP that is not ancestral to primates (Morgan and Hart 2019). However, this study overlooks C4BPAP1 in its analysis, likely due to C4BPAP1 being a pseudogene in humans. Human C4BPA is known to function in immunity and its highest tissue expression is in the liver, inconsistent with a function as the sperm fertilization gene ZP3r (Carithers and Moore 2015; Okroj 2018). Meanwhile, although pseudogenized, C4BPAP1 shows its highest RNA expression in the testis, consistent with an ancestral function as a sperm fertilization gene (Carithers and Moore 2015). Genes that have been recently pseudogenized are often still expressed until completely knocked out (Bekpen, et al. 2009).
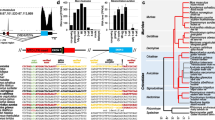
Syntenic and phylogenetic analysis indicates that C4BPAP1 is the human ortholog of mouse ZP3r. A Syntenic comparison of the genomic region between Mus musculus and Homo sapiens reveals that C4BPAP1 is syntenic to mouse ZP3r. B A protein alignment of C4BPAP1 and C4BPA from Homo sapiens and Macaca mulatta and ZP3r and C4BP from Mus musculus and Cavia porcellus was used to create a maximum likelihood phylogeny with bootstrapping. In the phylogeny, primate C4BPAP1 and rodent ZP3r cluster separately from primate C4BPA and rodent C4BP, indicating that in humans C4BPAP1, not C4BPA, is the ortholog of rodent ZP3r
Using phylogenetic analysis and syntenic mapping, we showed that C4BPAP1 is the human ortholog of mouse ZP3r (Fig. 1). A protein alignment of Homo sapiens and Macaca mulatta C4BPAP1 and C4BPA and Mus musculus and Cavia porcellus ZP3r and C4BP sequences was used to construct a maximum likelihood phylogeny. Primate C4BPAP1 and Rodent ZP3r clustered separately from Primate C4BPA and Rodent C4BP, indicating that Primate ZP3r (C4BPAP1), not C4BPA, is the ortholog of rodent ZP3r (Fig. 1B). We further supported the orthologous relationship between ZP3r and C4BPAP1 by constructing a phylogeny using additional protein sequences of primate C4BPA and C4BPAP1 and rodent C4BP and ZP3r (Supplementary Fig. 1). This phylogeny again showed clustering of rodent ZP3r with primate C4BPAP1 and rodent C4BP and primate C4BPA. Further, we used the best reciprocal tblastn hits of ZP3r/C4BPAP1 (stop codon removed), C4BPA, and neighboring RCA cluster gene transcripts between the mouse and human genome to establish orthology and syntenic relationships. Syntenic comparison between the region of the human and mouse RCA clusters containing C4BPAP1 and ZP3r, respectively, support C4BPAP1 as the human ZP3r ortholog. In humans, C4BPAP1 is located between C4BPA and CD55, as is ZP3r in rodents (Fig. 1A) (Kent, et al. 2002).
Previous research identified elevated linkage disequilibrium between the region of the human genome containing ZP3r/C4BPAP1 and the genomic region containing ZP3 (Rohlfs, et al. 2010). This linkage disequilibrium was hypothesized to suggest coevolution between ZP3 and its potential receptor located in humans syntenically to where ZP3r is in mice (Rohlfs, et al. 2010). Since ZP3r is pseudogenized in humans, this was possibly a false positive result or a complex association. An alternative hypothesis would be that the human sperm receptor for ZP3 is located nearby the pseudogenized human ZP3r. However, none of the annotated genes within the region shown to be in LD with ZP3 show testes-specific expression (Carithers and Moore 2015). Therefore, it seems likely that neither ZP3r or its close CCP domain-containing paralogs function in fertilization in humans.
ZP3r has Been Repeatedly and Rapidly Pseudogenized in Apes
C4BP/C4BPA and ZP3r/C4BPAP1 are members of the RCA cluster, the genes in this locus are largely conserved across even distantly related species, with sequence variation between species being driven by positive selection, indels, and intragenic domain duplications and losses (Sanchez-Corral, et al. 1993; Heinen, et al. 2006; Wu, et al. 2012; Garcia-Fernandez et al. 2021). However, some variation in RCA cluster gene content driven by clade-specific duplication or loss events has also been observed (Sanchez-Corral, et al. 1993; Pardo-Manuel de Villena 1995; Wu, et al. 2012). Notably, C4BPB is pseudogenized in mice, and there is evidence of two additional pseudogenized duplications of C4BPA found in humans (C4BPAP2 and C4BPAP3) (Pardo-Manuel de Villena 1995; Kent, et al. 2002). However, primate ZP3r/C4BPAP1 is unique in independently acquiring pseudogenization events in most apes and several monkey species (Fig. 2). Although there are examples in the literature of repeated pseudogenization events of genes across species, it is rare for independent events to occur within a closely related clade (Bainova, et al. 2014; Velova, et al. 2018). Remarkably, since the common ancestor of all apes (~16–20 mya), at least four unique pseudogenization events of ZP3r have occurred (Fig. 2) (Chatterjee, et al. 2009).
Recurrent and independent pseudogenization events of ZP3r in apes and monkeys. In primates, we extracted the coding DNA sequences of ZP3r/C4BPAP1 from available genomes and determined that many primates’ sequence contain pseudogenizing mutations. On the right side is a not-to-scale cartoon of the CCP domains (Green ovals) and C-terminal transmembrane domain (Blue ovals) found in ZP3r in each species. The loss of a CCP domain caused by a missing structurally important cysteine is shown as Yellow ovals. Red crosses indicate an insertion/deletion mutation causing a frameshift mutation; Black crosses indicate a premature stop codon causing pseudogenization. Darker colored CCP domains indicate regions of ZP3r that would not be translated due to a pseudogenization event. Within some CCP domains, multiple mutations have occurred. The cladogram on the left has pseudogenization events marked. For branches with multiple mutations, the order in which these mutations occurred is unknown (Color figure online)
Parsimony analysis of the pseudogenized ZP3r sequences indicates 9 independent pseudogenization events have occurred in primates. Remarkably, many of these pseudogenizing mutations occurred independently in closely related species and are located in distinct codons (Supplementary Fig. 2). With the exception of orangutan (Pongo abelli), C4BPAP1/ZP3r has been pseudogenized in all apes (Human, Chimpanzee, Bonobo, Gorilla, Gibbons) (Fig. 2). This rapid, repeated pseudogenization appears to be an extreme example of gene loss in apes. Gorillas, humans, and gibbons all have premature stop codons within CCP domain 2, all in different codons (Supplementary Fig. 2). Orangutan’s ZP3r does not have any pseudogenizing mutations, but its second CCP domain lacks a conserved and potentially structurally important cysteine that may disrupt the overall structure of the protein. In 10 New World monkey (NWM), 13 Old World monkey (OWM), and one tarsier genome assemblies, we identified the full ZP3r locus. Out of the 10 NWM genomes examined, 4 contained pseudogenizing mutations unique to that NWM species (Fig. 2). In OWMs, only one species, Colobus angolensis, had a pseudogenizing mutation within ZP3r (Fig. 2).
Recurrent, lineage-specific gene loss events between closely related species are suggestive of strong selection for gene loss. There is no obvious correlation between ZP3 sequence and glycosylation state and ZP3r loss in primates, and therefore, it is still unclear what is driving the loss of ZP3r in primates. Phylogenetic analysis of all individual CCP domains found in human C4BPA and C4BPAP1 and mouse C4BP and ZP3r indicates no evidence of concerted evolution between or within genes that could explain the repeated pseudogenization events (Supplementary Fig. 3). Further, a tblastn search of the human genome of rodent ZP3r and human C4BPAP1 (stop codon removed) reveals no new duplications of ZP3r that could be fulfilling its receptor function. However, a more distantly related paralog with low sequence similarity could be performing ZP3r’s function. Protein structure changes more slowly than protein sequence, and therefore, a paralog with low sequence identity may still retain similar function.
ZP3r Evolves Under Positive Selection
A recurrent feature of gamete recognition proteins is signatures of positive selection, potentially created through sexual selection or sexual conflict between the sperm and the egg (Carlisle and Swanson 2020). Genes mediating immune system functions are also frequently undergoing positive selection due to host–pathogen interactions driving arms race dynamics (Lazzaro 2012). So, it is unsurprising that previous studies have determined that both ZP3 and C4BPA are both undergoing positive selection in rodents and primates (Swanson, et al. 2001; Swann, et al. 2007, 2017; Rohlfs, et al. 2010; Hart, et al. 2018; Morgan and Hart 2019). In this study, we estimated values of dN/dS for ZP3r, C4BPA, and ZP3 in rodents and primates using the codeml program of PAML 4.8 (Yang 1997, 2007). For primate ZP3r, we only analyzed full coding sequences, no pseudogenized primate sequences were included. We compared models of selection using a likelihood ratio test (LRT) between neutral models and models with positive selection. Specifically, we compared M1 v. M2, M7 v. M8, and M8a v. M8 (Yang, et al. 2000; Swanson, et al. 2003) (Table 1, Supplementary Table 1).
We detected positively selected sites in ZP3r, C4BP, and ZP3 in rodents and ZP3r and C4BPA in primates, using the M8a v M8 comparison (Table 1). Signatures of positive selection in rodent and primate ZP3r are suggestive of functionally important genetic innovation being selected for within both clades. Because interacting reproductive proteins must co-evolve to maintain reproductive compatibility, ZP3r’s rapid evolution could be driven by the evolution of its putative binding partner ZP3. Although positively selected sites were not detected in primate ZP3 (M8a v M8) in this study, previous population genetic analysis has detected positive selection of ZP3 in humans (Rohlfs, et al. 2010; Hart, et al. 2018). Since ZP3r is undergoing positive selection in primates, ZP3r’s repeated and independent pseudogenization in primates is likely not driven by relaxed selection.
Conclusion
Despite their functional importance, the molecular mediators of fertilization have been poorly described in mammals, particularly for identifying sperm proteins mediating egg coat recognition (Carlisle and Swanson 2020). Difficulty in finding sperm receptors to egg coat proteins may be driven by functional redundancy causing many fertilization genes to be nonessential contributors to gamete recognition. Typically, protein functional redundancy refers to paralogous proteins that are structurally similar, that maintain the same interaction partners, and whose loss can be compensated for by their paralog. However, proteins can also be functionally redundant without being paralogous or structurally similar. For example, the acrosomal sperm proteins Zona Pellucida Binding Protein (ZPBP/sp38) and acrosin are structurally unrelated proteins that competitively interact with the ZP in boars (Mori, et al. 1993; Lin, et al. 2007). Functional redundancy of genes mediating fertilization could lead to clade-specific gene loss events or changes in relative functional importance between species. Again, reflecting on acrosin, knockouts of acrosin in mice (Mus musculus) result in infertility; yet, in hamsters (Mesocricetus auratus) acrosin is essential for zona penetration (Baba, et al. 1994; Hirose, et al. 2020). Together, these results indicate that functional redundancy between ZPBP, acrosin, and potentially other unknown sperm proteins allows the relative importance of acrosin to sperm bypassing the ZP to vary between species.
In this study, we demonstrate that the testes-expressed pseudogene C4BPAP1 is the human ortholog of rodent ZP3r using phylogenetic and syntenic analysis. While ZP3r is associated with ZP binding in mice, ZP3r shows repeated pseudogenization in primates (at least 9 times), most notably in apes. Recurrent independent pseudogenizations of a rapidly evolving protein are rarely discussed in the literature, and their existence is surprising. While rapid divergence is usually focused on sequence diversification, changes in gene content caused by gene gains and loss events could also be a significant contributor to molecular diversity and tolerated due to functional redundancy (Carlisle 2021). ZP3r is a nonessential fertilization gene in mice, and it may be one of many proteins interacting with ZP3 (Muro, et al. 2012; Miyata, et al. 2016; Okabe 2018). ZP3r’s repeated loss in many primates, particularly apes, despite being subject to positive selection in other primate species, indicates that the relevant importance of ZP3r to fertilization differs across primates. This difference could be due to the emergence or increase in relative importance of a different fertilization gene mediating ZP3 binding in primates. Differences in relative functional importance between clades may also partially explain why reproductive proteins are rapidly evolving in some clades and not others (Carlisle and Swanson 2020). This study highlights the potential variability of molecular mechanisms of fertilization even within mammals and emphasizes the value of using diverse model systems for investigating mechanisms of fertilization.
Methods
Identification of Sequences and Annotation of Domains
When possible, we used publicly available coding DNA sequences from Genbank for our analysis. If not available, we predicted the coding DNA sequences of ZP3, C4BPA/C4BP, and ZP3r/C4BPAP1 from rodent and primate genomes using the Protein2Genome command of the program Exonerate version 2.2.0 (Slater & Birney 2005). The top scoring prediction from Exonerate was used to define the paralogs’ exons. Supplementary File A lists the Genbank IDs of sequences used or the publicly available species’ genomes from which sequences were extracted. Our query sequences for primates were the amino acid sequences of human ZP3 (NP_001103824.1), C4BPA (NP_000706.1), and C4BPAP1 with the stop codon in CCP2 removed. For rodents, our query protein sequences were Mus musculus ZP3 (NP_035906.1), ZP3r (NP_001407586.1), and C4BP (NP_001406911.1). To get the human C4BPAP1 protein sequence, we used the Exonerate Protein2Genome command using human C4BPA as the protein query against the region of the human genome containing C4BPAP1 (Gene ID: 727,859). The highest scoring coding DNA sequence identified was translated into an incomplete amino acid sequence and used as a query in NCBI tblastn program against all great apes’ sequences (taxid:9604) (Johnson, et al. 2008). From this search the full-length Pongo abelii mRNA sequence was identified (XM_054521592.2) and used as a query to identify the predicted C4BPAP1 sequence in humans, which includes a premature stop codon in CCP2. We identified CCP domains using InterProScan (v5.68–100) (Jones et al. 2014). Alignments between protein sequences were used to identify CCP domains that had lost conserved cysteines belonging to structurally crucial disulfide bonds. For proteins that were pseudogenized, premature stop codons were removed for CCP domain annotation to determine the domain location of these codons.
Construction of Phylogenetic Trees
A protein alignment of C4BPAP1 and C4BPA from Homo sapiens and Macaca mulatta and ZP3r and C4BP from Mus musculus and Cavia porcellus was constructed using Clustal Omega (Siever and Higgins. 2014). The ends of the alignment were trimmed to remove regions where > 25% of sequences had gaps. All supplementary phylogenies were similarly constructed using RAxML-NG. For Supplementary Fig. 1 a Clustal Omega alignment was made of all collected rodent and primate C4BPAP1/ZP3r and C4BPA/C4BP amino acid sequences, all sites where > 50% of sequences had a gap were removed. For Supplementary Fig. 3, a Clustal Omega protein alignment of the isolated CCP domains. All protein alignments were used to construct a maximum likelihood phylogeny with bootstrapping. The phylogenetic inference tool RAxML-NG was used to construct the phylogenetic tree with the LG substitution matrix (Kozlov, et al. 2019). RaxML-NG conducts maximum likelihood based phylogenetic inference and provides branch support using non-parametric bootstrapping. The best scoring topology of 20 starting trees (10 random and 10 parsimony-based) was chosen. RAxML-NG was used to perform non-parametric bootstrapping with 1000 re-samplings that were used to re-infer a tree for each bootstrap replicate MSA. All alignments, tree files, and sources of sequences are available in the supplementary materials.
Detection of Positive Selection
We estimated dN/dS values for primate and rodent ZP3r, C4BPA/C4BP, and ZP3 using the codeml program of PAML 4.8 (Yang 2007). We compared models of selection using a likelihood ratio test (LRT) between neutral models and models with positive selection. The model comparisons were made between M1 v. M2, M7 v. M8, and M8a v. M8. The likelihood ratio (LRT) statistic was calculated as twice the negative difference in likelihoods between nested models. For M1a v M2a or M7 v Model 8, the LRT was compared to the χ2 distribution with 2 degrees of freedom (Yang 2007). Twice the negative difference in likelihoods between the models, M8a v. M8 comparison was compared. The LRT statistic was approximated by the 50–50 mixture distribution of 0 and χ2 with one degree of freedom (Swanson, et al. 2003). All input, output, and control files for codeml analysis can be found in the supplement.
Syntenic Comparison and Gene Expression Patterns
Using the annotated Mus musculus and Homo sapiens genomes available on the UCSC genome browser, syntenic comparison was performed for the genomic regions containing ZP3r/C4BPAP1 and C4BP/C4BPA. Genes shown in Fig. 1A passed reciprocal best blast (RBB). For RBB analysis, coding DNA sequences were queried against human and mouse genomes. Annotated genes and pseudogenes in the human genome were examined for their tissue-specific expression patterns using GTEx Analysis Release V8 (Carithers and Moore 2015).
References
Adham IM, Nayernia K, Engel W (1998) Spermatozoa lacking acrosin protein show delayed fertilization. Mol Reprod Dev 46:370–376
Avella MA, Baibakov B, Dean J (2014) A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J Cell Biol 205:801–809
Baba T, Azuma S, Kashiwabara S, Toyoda Y (1994) Sperm from mice carrying a targeted mutation of the acrosin gene can penetrate the oocyte zona pellucida and effect fertilization. J Biol Chem 269:31845–31849
Bainova H, Kralova T, Bryjova A, Albrecht T, Bryja J, Vinkler M (2014) First evidence of independent pseudogenization of toll-like receptor 5 in passerine birds. Dev Comp Immunol 45:151–155
Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, Ventura M, Kidd JM, Siswara P, Howard JC, Eichler EE (2009) Death and resurrection of the human IRGM gene. PLoS Genet 5:e1000403
Bleil JD, Wassarman PM (1980) Mammalian sperm-egg interaction: identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell 20:873–882
Bleil JD, Wassarman PM (1990) Identification of a ZP3-binding protein on acrosome-intact mouse sperm by photoaffinity crosslinking. Proc Natl Acad Sci USA 87:5563–5567
Buffone MG, Zhuang T, Ord TS, Hui L, Moss SB, Gerton GL (2008) Recombinant mouse sperm ZP3-binding protein (ZP3R/sp56) forms a high order oligomer that binds eggs and inhibits mouse fertilization in vitro. J Biol Chem 283:12438–12445
Carithers LJ, Moore HM (2015) The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank 13:307–308
Carlisle JA, Glenski, M.A., Swanson, W.J. 2021. Recurrent Duplication and Diversification of Acrosomal Fertilization Proteins in Abalone. BioRxiv.
Carlisle JA, Swanson WJ (2020) Molecular mechanisms and evolution of fertilization proteins. J Exp Zool B Mol Dev Evol. https://doi.org/10.1002/jez.b.23004
Chatterjee HJ, Ho SY, Barnes I, Groves C (2009) Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol 9:259
Garcia-Fernandez J, Vilches-Arroyo S, Olavarrieta L, Perez-Perez J, Rodriguez de Cordoba S (2021) Detection of genetic rearrangements in the regulators of complement activation RCA cluster by high-throughput sequencing and MLPA. Methods Mol Biol 2227:159–178
Hart MW, Stover DA, Guerra V, Mozaffari SV, Ober C, Mugal CF, Kaj I (2018) Positive selection on human gamete-recognition genes. PeerJ 6:e4259
Heinen S, Sanchez-Corral P, Jackson MS, Strain L, Goodship JA, Kemp EJ, Skerka C, Jokiranta TS, Meyers K, Wagner E et al (2006) De novo gene conversion in the RCA gene cluster (1q32) causes mutations in complement factor H associated with atypical hemolytic uremic syndrome. Hum Mutat 27:292–293
Hirose M, Honda A, Fulka H, Tamura-Nakano M, Matoba S, Tomishima T, Mochida K, Hasegawa A, Nagashima K, Inoue K et al (2020) Acrosin is essential for sperm penetration through the zona pellucida in hamsters. Proc Natl Acad Sci U S A 117:2513–2518
Hofmeyer T, Schmelz S, Degiacomi MT, Dal Peraro M, Daneschdar M, Scrima A, van den Heuvel J, Heinz DW, Kolmar H (2013) Arranged sevenfold: structural insights into the C-terminal oligomerization domain of human C4b-binding protein. J Mol Biol 425:1302–1317
Hourcade D, Holers VM, Atkinson JP (1989) The regulators of complement activation (RCA) gene cluster. Adv Immunol 45:381–416
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden T (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5-9
Jones P, Binns D, Chang H, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn A, Sangrador-Vegas A, Scheremetjew M, Yong S, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Kozlov A, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35(21):4453–4455
Krushkal J, Bat O, Gigli I (2000) Evolutionary relationships among proteins encoded by the regulator of complement activation gene cluster. Mol Biol Evol 17:1718–1730
Lazzaro BP, Clark AG (2012) Rapid evolution of innate immune response genes. Rapidly evolving genes & genetic systems: PMC Exempt. Oxford University Press, Oxford
Lin YN, Roy A, Yan W, Burns KH, Matzuk MM (2007) Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol 27:6794–6805
Litscher ES, Williams Z, Wassarman PM (2009) Zona pellucida glycoprotein ZP3 and fertilization in mammals. Mol Reprod Dev 76:933–941
Miyata H, Castaneda JM, Fujihara Y, Yu Z, Archambeault DR, Isotani A, Kiyozumi D, Kriseman ML, Mashiko D, Matsumura T et al (2016) Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc Natl Acad Sci USA 113:7704–7710
Morgan CC, Hart MW (2019) Molecular evolution of mammalian genes with epistatic interactions in fertilization. BMC Evol Biol 19:154
Morgan CC, Loughran NB, Walsh TA, Harrison AJ, O’Connell MJ (2010) Positive selection neighboring functionally essential sites and disease-implicated regions of mammalian reproductive proteins. BMC Evol Biol 10:39
Morgan CC, Loughran NB, Walsh TA, Harrison AJ, O’Connell MJ (2017) Erratum to: Positive selection neighboring functionally essential sites and disease-implicated regions of mammalian reproductive proteins. BMC Evol Biol 17:170
Mori E, Baba T, Iwamatsu A, Mori T (1993) Purification and characterization of a 38-kDa protein, sp38, with zona pellucida-binding property from porcine epididymal sperm. Biochem Biophys Res Commun 196:196–202
Muro Y, Buffone MG, Okabe M, Gerton GL (2012) Function of the acrosomal matrix: zona pellucida 3 receptor (ZP3R/sp56) is not essential for mouse fertilization. Biol Reprod 86:1–6
Ojha H, Ghosh P, Singh Panwar H, Shende R, Gondane A, Mande SC, Sahu A (2019) Spatially conserved motifs in complement control protein domains determine functionality in regulators of complement activation-family proteins. Communications Biology 2(1):290
Okabe M (2018) Sperm-egg interaction and fertilization: past, present, and future. Biol Reprod 99:134–146
Okroj MBAM (2018) C4b-binding protein. In: Barnumb SST (ed) The complement handbook. Elsevier, New York
Pardo-Manuel de Villena F, Rodriguez S (1995) C4BPAL2: a second duplication of the C4BPA gene in the human RCA gene cluster. Immunogenetics 41:2–3
Rohlfs RV, Swanson WJ, Weir BS (2010) Detecting coevolution through allelic association between physically unlinked loci. Am J Hum Genet 86:674–685
Sanchez-Corral P, Pardo-Manuel de Villena F, Rey-Campos J, Rodriguez de Cordoba S (1993) C4BPAL1, a member of the human regulator of complement activation (RCA) gene cluster that resulted from the duplication of the gene coding for the alpha-chain of C4b-binding protein. Genomics 17:185–193
Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. https://doi.org/10.1186/1471-2105-6-31
Sievers F, Higgins D (2014) Clustal omega. Curr Protocols. https://doi.org/10.1002/0471250953.bi0313s48
Sutton KA, Jungnickel MK, Ward CJ, Harris PC, Florman HM (2006) Functional characterization of PKDREJ, a male germ cell-restricted polycystin. J Cell Physiol 209:493–500
Sutton KA, Jungnickel MK, Florman HM (2008) A polycystin-1 controls postcopulatory reproductive selection in mice. Proc Natl Acad Sci U S A 105:8661–8666
Swann CA, Cooper SJ, Breed WG (2007) Molecular evolution of the carboxy terminal region of the zona pellucida 3 glycoprotein in murine rodents. Reproduction 133:697–708
Swann CA, Cooper SJB, Breed WG (2017) The egg coat zona pellucida 3 glycoprotein - evolution of its putative sperm-binding region in Old World murine rodents (Rodentia: Muridae). Reprod Fertil Dev 29:2376–2386
Swanson WJ, Vacquier VD (2002) The rapid evolution of reproductive proteins. Nat Rev Genet 3:137–144
Swanson WJ, Yang Z, Wolfner MF, Aquadro CF (2001) Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci U S A 98:2509–2514
Swanson WJ, Nielsen R, Yang Q (2003) Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol 20:18–20
Velova H, Gutowska-Ding MW, Burt DW, Vinkler M (2018) Toll-Like Receptor Evolution in Birds: Gene Duplication, Pseudogenization, and Diversifying Selection. Mol Biol Evol 35:2170–2184
Wassarman PM (2009) Mammalian fertilization: the strange case of sperm protein 56. BioEssays 31:153–158
Wu J, Li H, Zhang S (2012) Regulator of complement activation (RCA) group 2 gene cluster in zebrafish: identification, expression, and evolution. Funct Integr Genomics 12:367–377
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Acknowledgements
This study was supported by NIH Grant HD076862 to Willie J. Swanson and a National Science Foundation Graduate Research Fellowship to Jolie A. Carlisle. Thank you to Dr. Damien Wilburn, Dr. Jan Aagaard, and Alberto Rivera for useful feedback. All research was performed on the traditional lands of the Duwamish Tribe. To learn more about the Duwamish Tribe and their continuing legacy, please visit https://www.duwamishtribe.org/.
Funding
Funding was supported by National Institute of Child Health and Human Development, HD076862.
Author information
Authors and Affiliations
Contributions
JAC and WJS designed the research. JAC and DHG performed the research. JAC wrote the paper.
Corresponding author
Additional information
Handling editor: David Liberles.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Carlisle, J.A., Gurbuz, D.H. & Swanson, W.J. Recurrent Independent Pseudogenization Events of the Sperm Fertilization Gene ZP3r in Apes and Monkeys. J Mol Evol (2024). https://doi.org/10.1007/s00239-024-10192-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00239-024-10192-x