Abstract
Adenosine-to-inosine (A-to-I) RNA editing recodes the genetic information. Apart from diversifying the proteome, another tempting advantage of RNA recoding is to correct deleterious DNA mutation and restore ancestral allele. Solid evidences for beneficial restorative editing are very rare in animals. By searching for “convergent recoding” under a phylogenetic context, we proposed this term for judging the potential restorative functions of particular editing site. For the well-known mammalian Gln>Arg (Q>R) recoding site, its ancestral state in vertebrate genomes was the pre-editing Gln, and all 470 available mammalian genomes strictly avoid other three equivalent ways to achieve Arg in protein. The absence of convergent recoding from His>Arg, or synonymous mutations on Gln codons, could be attributed to the strong maintenance on editing motif and structure, but the absence of direct A-to-G mutation is extremely unexpected. With similar ideas, we found cases of convergent recoding in Drosophila genus, reducing the possibility of their restorative function. In summary, we defined an interesting scenario of convergent recoding, the occurrence of which could be used as preliminary judgements for whether a recoding site has a sole restorative role. Our work provides novel insights to the natural selection and evolution of RNA editing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RNA Editing in All Kingdoms of Lives
RNA editing is the co-transcriptional or post-transcriptional alteration of RNA sequences. RNA editing is prevalent in all kingdoms of lives and largely diversifies the transcriptomes (Eisenberg and Levanon 2018). The two most abundant and well-studied types of RNA editing is the adenosine-to-inosine (A-to-I) RNA editing in animals, fungi, bacteria (Bar-Yaacov et al. 2018; Bian et al. 2019; Zhang et al. 2023) and the cytidine-to-uridine (C-to-U) RNA editing in plants (Duan et al. 2023a; Shikanai 2006; Takenaka et al. 2013). While animal A-to-I mRNA editing is mediated by ADAR (adenosine deaminase acting on RNA), the editing events in fungi and bacteria are mediated by ADAT (adenosine deaminase acting on tRNA) and TadA (tRNA-specific adenosine deaminase)-related protein complex ADAT2/3-Ame1 and Tad2/3-Ame1 (Duan et al. 2023b; Feng et al. 2024), and C-to-U editing in plants is mainly catalyzed by PPR (Pentatricopeptide repeat) families that possess a DYW deaminase domain (Hayes and Santibanez 2020; Yin et al. 2013). Since I is read as G by cellular machineries (Fig. 1A), both A-to-I and C-to-U RNA editing are able to change amino acid (AA) sequences and lead to nonsynonymous changes, and the process of nonsynonymous editing is usually referred to as “recoding” (Alon et al. 2015).
A general introduction on A-to-I RNA editing and the conserved Q>R recoding site in mammals. A The biogenesis of ADAR-mediated A-to-I RNA editing. I is recognized as G by cellular machineries. B Restorative RNA editing is further divided into compensatory editing and harm-permitting editing. C Mammalian GRIA2 gene encodes a glutamate receptor in brains. The Gln>Arg (Q>R) recoding site has a 100% editing level in all tested mammals and the absence of such editing, usually due to ADAR2-deficiency, leads to lethality
Molecular Error and Beneficial Recoding Theories
According to the “molecular error” theory (Zhang and Xu 2022), the majority of molecular outcomes of biological processes (e.g. RNA editing) are non-functional/neutral/deleterious, and only the RNA editing events especially the recoding sites conserved across different species might be the candidates of beneficial editing (Xu and Zhang 2015). Despite the rarity of beneficial recoding, there are indeed some clades of species exhibiting overrepresented RNA recoding events compared to neutral expectation, suggesting that at least part of the recoding sites was selectively favored (Liscovitch-Brauer et al. 2017; Yablonovitch et al. 2017). As a result, two theories on the advantage of recoding has been proposed, and in silico/experimental approaches to find such beneficial sites were developed.
The “proteomic diversifying hypothesis” (Shoshan et al. 2021) stresses that RNA editing can be controlled in a temporal–spatial manner, facilitating the adaptation of organisms to changeable conditions (Duan et al. 2024a, b). In contrast, the “restorative hypothesis” (Jiang and Zhang 2019) believes that RNA editing corrects deleterious DNA mutations and restores the ancestral allele. Notably, the restorative function of recoding could be further divided into two types. The first type is compensatory editing, which arises after a G-to-A mutation at the same site (Fig. 1B). In these cases, the fixation of the mutation is not dependent on RNA editing. The second type is harm-permitting editing, which allows for the fixation of otherwise deleterious G-to-A mutations (Jiang and Zhang 2019). Because the fixation of the G-to-A mutation is permitted by A-to-I editing, the mutation and editing could not be separated independently in evolution (Fig. 1B). If the G-to-A mutation is slightly deleterious, compensatory A-to-I editing can be considered adaptive, as having editing is more favorable than having no editing at that site. However, compensatory editing is expected to be less common since deleterious G-to-A mutations are likely to have been eliminated by purifying selection before the emergence of RNA editing (Gray 2012). In contrast, harm-permitting editing is supposed to be non-adaptive by the constructive neutral evolution (CNE) and harm-permitting models (Gray 2012; Jiang and Zhang 2019), as having a genomic A that is highly edited does not confer a fitness advantage over having the original genomic G intuitively.
Note that although the harm-permitting theories was original proposed as a non-adaptive explanation (Jiang and Zhang 2019), it likely referred to the entire “RNA editing mechanism”. It was the existence of the editing mechanism that allowed the fixation of the deleterious DNA mutation and thus the editing mechanism is overall non-adaptive. But for an individual restorative editing site, the editing event on this particular site did not exist before the occurrence of DNA mutation (because before the mutation, the genome was not adenosine at all). Therefore, a particular harm-permitting editing event itself can still be functional (beneficial) even the “DNA mutation coupled with RNA editing on it” or the global RNA editing mechanism was non-adaptive as a whole.
To generalize this notion, we further stress that the diversifying and restorative (compensatory or harm-permitting) hypotheses are making predictions on the genome-wide trend of recoding. The in silico approaches raised by the theories were applicable to global rather than individual editing site. Even it was demonstrated that the overall recoding sites in cephalopods were likely playing a diversifying role (Shoshan et al. 2021), it does not imply that every recoding site has a diversifying function. Given a particular recoding site (like the Q>R site mentioned below), there still lacks an in silico approach, especially under a large phylogenetic context, to help judge the confidence of restorative function.
Finding Beneficial Recoding from the Sea of Total RNA Editing Sites
Compelling evidences for both diversifying and restorative hypotheses exist. (1) For a particular A-to-I recoding site in fungi Fusarium graminearum (conserved missense editing, CME5), scientists constructed mutant strains and found that A-allele was fitter at asexual stage but G-allele was fitter at sexual stage, then in wild-type fungi RNA editing level could be flexibly regulated according to which allele was fitter under a given condition (Xin et al. 2023). This is the direct experimental evidence supporting the temporal–spatial flexibility of RNA recoding. Moreover, a more intuitive scenario predicted by the diversifying hypothesis is the heterozygote advantage, and accordingly, scientists found that the concurrent expression of both edited and unedited versions of site CME11 during the sexual stage is more advantageous than either version alone (Xin et al. 2023), supporting the diversifying advantage of RNA editing. The heterozygote advantage conferred by RNA editing in haploid fungi might be more remarkably than that in animals because diploids have an alternative way (heterozygous SNP in the genome) to achieve such advantage. (2) Furthermore, in silico analysis on hundreds of insect genomes revealed that several recoding sites in the extant insect species had an ancestral “uneditable” codon encoding the pre-editing AA, and this evolutionary trajectory suggests that there might be an advantage of being editable compared to uneditable (Duan et al. 2023c; Ma et al. 2023), supporting the diversifying hypothesis. One may argue that there might be sites that changed from editable to uneditable (Zhao et al. 2024), but what we focus here is the judgement on individual editing site rather than the global trend, thus there is no need to mention “other cases” of the opposite trajectory; and moreover, it is anti-intuitive to assume an editing event existed in ancestral node while the current sequence is an uneditable one.
The benefit of restorative RNA editing was also demonstrated in fungi F. graminearum (Qi et al. 2024). For several A-to-I(G) editing sites that turned a pre-mature stop codon (PSC) to a sense codon, the ancestral G-allele produced the extended protein isoform with “survival-reproduction trade-offs”, being more vulnerable under stress but being normal during sexual reproduction. This trade-off was resolved by G-to-A DNA mutation followed by restorative PSC editing in current species. The genomically encoded truncated isoform performs better for vegetative growth so at this stage RNA editing is absent. In contrast, the extended isoform functions well during sexual reproduction so accordingly RNA editing occurs to restore the ancestral full length version (Qi et al. 2024). This is the first experimental evidence for the advantage of restorative RNA editing since it demonstrates that harm-permitting A-to-I editing can also be adaptive when it resolves tradeoffs caused by antagonistic pleiotropy (meaning that the fitness of a flexibly editable A is higher than an uneditable A and the original genomic G). Note that there might be a hidden prerequisite for beneficial restorative RNA editing: in order to compensate for DNA mutation, the editing level has to be as high as possible, such as the ~ 80% editing levels observed in fungal PSC (Qi et al. 2024) and plant organelles (Duan et al. 2023a). Apart from fungi and plants, the solid examples of restorative RNA editing in animals are very rare. Moreover, in addition to simply looking at an ancestral G, a more informative in silico approach to help judge the confidence of restorative function was lacking.
Q>R Recoding in Mammalian GRIA2 Gene
Regarding extremely high editing level, a typical example is the Q>R site in mammalian GRIA2 gene. Mammalian GRIA2 (Glutamate Ionotropic Receptor AMPA Type Subunit 2), also known as GluR-2, is one of the four subunits (GRIA1-4) of the predominant excitatory neurotransmitter receptor in brains which is activated during many normal neurophysiologic processes. In mRNAs of gene GRIA2, a 100% level RNA editing event is observed in brains of mammals (Sommer et al. 1991), the position of which corresponds to the second transmembrane domain of GRIA2 protein, leading to a Gln>Arg change (a CAG>CGG change at codon level), and therefore this editing site is named as Q>R recoding site or simply Q>R site (Fig. 1C).
Among the three paralogs of editing enzyme ADAR in mammals (Savva et al. 2012; Zhan et al. 2023), ADAR2 is thought to be responsible for editing events in CDS (Tan et al. 2017). ADAR2-deficiency will cause the absence of Q>R recoding, leading to the flux of Ca2+ into the cell and cause lethality to the animal (Walkley and Li 2017) (Fig. 1C). The indispensability of this Q>R recoding event, together with its extremely high and robust editing level of 100%, leaves us an impression that this Q>R site is likely to be a restorative recoding site and that only the Arg version of GRIA2 protein is functionally necessary. However, the confidence of its restorative function and the conservation of codon sequence/editing events have not been studied under a large phylogenetic context.
Aims and Scope
In this study, we first defined a scenario termed “convergent A-to-I recoding”, abbreviated as “convergent recoding”, which means that the pre-editing AAs are different between species but the post-edited AA is the same (see the main text for detail). Based on this definition, we developed a new idea for judging whether a set of recoding sites are likely to exert a restorative function. Under the restorative hypothesis (which stresses the essentiality of the post-edited AA version regardless of compensatory or harm-permitting sites), the emergence of convergent recoding should be well tolerated. In other words, given a long-enough time during evolution, we should observe additional ways, including convergent recoding or direct genomic A-to-G substitution, to achieve post-edited AAs for particular restorative recoding sites.
We applied this notion to the mammalian Q>R site, finding several lines of observations that reduce the possibility of its restorative role. We then intended to perform this test in a wider range of recoding sites. As mammalian editing sites are poorly conserved and no cases of convergent recoding exist, we therefore look for an animal clade with highly conserved CDS editing sites plus a large set of species with documented genome sequences. Then the Drosophila genus was chosen and we indeed found a few cases of convergent recoding. However, population genomic analysis reveals that novel mutations leading to convergent recoding are drastically suppressed, which reduced the possibility that only the post-edited AA is essential, and thus reducing the probability that these recoding sites in Drosophila mainly exert restorative functions.
In conclusion, by defining convergent recoding sites, we have retrieved this previously ignored set of recoding sites and managed to uncover their evolutionary significance. We also used the occurrence of convergent recoding as a novel and preliminary criterion to test whether a particular set of recoding sites might have exerted a restorative function. Our work provides novel insights to the natural selection and evolution of RNA editing.
Methods
Data Availability
The genome accession IDs of all Drosophila species used in this study were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) and the accession links were listed in Supplementary Table S1. The lists of high-confidence A-to-I RNA editing sites in brains of Drosophila melanogaster, Drosophila simulans, and Drosophila pseudoobscura were retrieved from our previous study. The SNP data were downloaded from the D. melanogaster genetic reference panel (DGRP) (Mackay et al. 2012). The head transcriptomes of Drosophila ananassae were downloaded from NCBI with accession ID SRR7243210. The head transcriptomes of D. melanogaster were also downloaded from NCBI with accession IDs SRR7262144 (male) and SRR7262145 (female). The human adrenal (ERR3153450, ERR3153452, ERR3153392, ERR3153417, ERR3153385, and ERR3153335) and endometrium (ERR3153368, ERR3153491, ERR3153386, ERR3153495, ERR3153433, ERR3153361, and ERR3153438) transcriptomes were also downloaded from NCBI. The expression profile of human GRIA2 gene was seen from website https://www.ncbi.nlm.nih.gov/gene/2891. Adrenal and endometrium are the only non-brain tissues where GRIA2 is expressed.
Parsing the Evolution of Mammalian Q>R Site
We visited the UCSC genome browser (https://genome.ucsc.edu/) and entered the genomic coordinate of the human GRIA2 Q>R site (chr4:157336722-157336724). Then the alignments of 470 mammals and 100 vertebrates will be accessible. Since we already had the 470 mammalian sequences, we only focused on non-mammal species in the 100-vertebrate results. The topology of the major clades of the phylogenetic tree was confirmed by the TimeTree website (http://timetree.igem.temple.edu/). The phyloP scores of 470 mammals (https://hgdownload.soe.ucsc.edu/goldenPath/hg38/phyloP470way/) and 100 vertebrates (https://hgdownload.soe.ucsc.edu/goldenPath/hg38/phyloP100way/) were downloaded from UCSC genome browser (https://genome.ucsc.edu/). The GRIA2 nucleotide sequence of chimpanzee (Pan troglodytes) was downloaded from NCBI with accession ID NM_001184994.5. The dN and dS values between human and chimpanzee were calculated with codeml (Nei and Gojobori 1986).
RNA Structure Acquisition and Visualization
To obtain the pre-mRNA sequences of GRIA2, we downloaded the reference genome and annotation file of human (hg38, https://ftp.ensembl.org/pub/release-85/fasta/homo_sapiens/) and mouse (mm10, https://ftp.ensembl.org/pub/release-85/fasta/mus_musculus/). According to the genomic coordinates of the Q>R site (chr4:157336723 for human and chr3:80706912 for mouse), the sequence of its flanking ± 500 bp region was extracted using Bedtools getfasta (Quinlan and Hall 2010). Note that the genomic sequence represents the intron-containing pre-mRNA sequence rather than mature mRNA. Then, the 1001 bp sequence was folded and visualized using online tool RNAstructure (https://rna.urmc.rochester.edu/RNAstructureWeb/).
Phylogeny of Drosophila Genus
As mentioned above, we collected the reference genomes of all species in Drosophila genus (Supplementary Table S1). According to the established phylogeny of Drosophila genus provided by FlyBase (https://flybase.org/), there are 14 available species “between” D. pseudoobscura and the D. simulans–D. melanogaster branch. We only utilized the topology of the species tree as our results did not rely on the branch length.
Sequence Alignment to Extract Orthologous Codons
Our analyses entail extracting the orthologous codons from the alignment file. The positions of interest were based on the known RNA editing sites (or unedited sites as a control) in D. melanogaster. For all the ~ 13,000 coding genes in D. melanogaster, we selected the transcript with the longest CDS of each gene. We translated the CDS into protein, and aligned their protein sequences to those of other Drosophila species with blastp (Camacho et al. 2009). Default parameters were used. The hit with the lowest E value was regarded as the orthologous genes in each species. Then the orthologous sequences were aligned with mafft (Katoh and Standley 2013) with default parameters. CDSs were aligned according to the protein alignment. Since the edited genes in Drosophila generally have a high conservation level, the search for orthologs and the sequence alignment should be highly reliable and less sensitive to software, parameters, or cutoffs. The alignment of each codon/AA position was manually extracted from the sequence alignment file.
Transcriptome Mapping and Variant Visualization
BWA version 0.7.17 was used to map the RNA-Seq reads to the reference CDS sequence of the target species (Li and Durbin 2009). Default parameters were used. The sequence coverage and alignment at target region were visualized with IGV.
Annotation of Unedited Adenosines If They Are Edited
We split the reference genome of D. melanogaster into single bases. In gene region, we extracted the adenosines. If the gene is located in the positive strand of the reference genome, then we should extract A in the reference genome sequence; if the gene is located in the negative strand of the reference genome, then we should extract T in the reference genome sequence. Presume A-to-I RNA editing occurs, then A in the positive strand genes should be replaced with G, and T in the negative strand genes should be replaced with C. Then, software SnpEff (Cingolani et al. 2012) was used to annotate the change caused by A-to-G. Nonsynonymous and synonymous changes were counted.
Annotation of SNPs
The SNPs of D. melanogaster from DGRP project were also annotated by SnpEff (Cingolani et al. 2012). In coding region, the software will tell us which codon this SNP is located and thus we could infer the codon change and AA change based on this information. The nucleotide position on CDS and AA position on protein were also provided for each CDS SNP, and this enabled us to match the SNPs with the genome-wide unedited adenosines, consequently determining which SNPs are located in conserved codons with nonsynonymous adenosines.
Statistical Tests
Statistical tests were performed in R studio (R version 3.6.3). The graphical works were done in R environment.
Results
Q>R Recoding in Mammalian GRIA2 is Unlikely to Be Restorative Due to Several Paradoxes
The 100% editing level, the high conservation across mammals, and the strict indispensability of the GRIA2 Gln>Arg (Q>R) recoding site prompt us to believe that this editing event is used for restoring the ancestral allele and that only the Arg version is functionally essential. To systematically test the restorative function on this particular site, we retrieved the genome alignment of 470 mammals, 14 birds, 6 reptiles, 1 amphibian, 15 fishes, and 1 lamprey (Methods). According to the phylogeny, there is a clear trend that the ancestral state of all vertebrates was a CAG codon (Gln) and this codon has been maintained in the majority of extant vertebrates including mammals (Fig. 2A). Only in several fishes, different Arg codons are derived. Notably, six Arg codons can be divided into two groups: CGN and AG[A/G] (Fig. 2B), among which only CGG can be created by a single A-to-G mutation from the ancestral Gln codon CAG. The existence of various Arg codons in fishes indicates a multi-step mutation process on this codon (Fig. 2A). Nevertheless, the inference of ancestral Gln codon in vertebrates reduced the possibility of that mammalian Q>R site purely exerts a restorative function.
Potential restorative Q>R recoding in mammalian GRIA2 gene. A The evolution of Q>R site in vertebrate phylogeny. 470 mammalian species, 14 birds, 6 reptiles, 1 amphibian, 15 fishes, and 1 lamprey were used. Drosophila melanogaster was used as an outgroup to show that the CAG codon was conserved in invertebrates but there were no editing events. Female and male head transcriptomes were used. B Four panels in the codon table containing all two Gln codons and all six Arg codons. C Apart from Gln>Arg recoding, there are three other equivalent ways to achieve Arg in the final protein sequence: A-to-G DNA mutation, Gln>His DNA mutation (leading to convergent recoding), or synonymous mutation from CAG-to-CAA (Gln)
Moreover, since the Q>R recoding also existed in Xenopus (Nguyen et al. 2023), it prompts us to consider when did this CAG codon and recoding event originate. We retrieved the genome and head transcriptomes of representative invertebrate D. melanogaster (Methods) and found that Drosophila did have this gene (GluRIA, FBgn0004619) but the corresponding CAG codon was not edited in both male and female heads (Fig. 2A), presumably due to a bulge in the RNA structure near the CAG codon which is not optimal for Adar binding (Supplementary Fig. S1). The absence of Drosophila Q>R recoding is consistent with the fact that this site is not documented in the RADAR database who collected all the reported RNA editing sites in D. melanogaster (Ramaswami and Li 2014). This observation not only suggests that Q>R recoding might appear in the ancestor of vertebrates, but also indicates that at least some organisms can survive with the unedited allele or non-fully edited allele of Q>R site.
Next, the restorative function predicts that the AA produced from G-allele has higher fitness than the AA produced from A-allele, which means that Arg is fitter than Gln in this case of Q>R recoding. With further observation that the editing level is necessarily 100% in mammals, an intuitive implication is that only the Arg version is functional (at least in mammals). In theory, we ought to observe other ways to achieve Arg in the final protein sequence. Currently, we conceived the following three additional equivalent ways to obtain 100% Arg in protein (Fig. 2C).
-
(1)
Direct A-to-G DNA mutation from CAG (Gln) to CGG (Arg) will ensure 100% Arg in the protein.
-
(2)
DNA mutation from CAG (Gln) to CAC/T (His) and then His can also be recoded to Arg by RNA editing at the second codon position (Fig. 2B, C). We define this case as “convergent A-to-I recoding” or simply “convergent recoding”, which means pre-editing AAs are different between species (Gln and His) but the post-edited AA is converged to Arg. Note that our definition is based on DNA mutation near the conserved editing site, unless there is phylogenetic evidence suggesting that RNA editing events were independently gained at already-diverged codons.
-
(3)
Synonymous mutation from CAG-to-CAA (Gln). As long as the 100% editing exists, the Gln codons will always be converted to Arg codons.
However, it surprised us that none of the above three ways was utilized by the 470 mammals in our analysis as they all have the CAG codon (Fig. 2A). If only the Arg version is functional as predicted by the restorative role plus the 100% editing level seen in all mammals, then why should natural selection suppress the above three types of mutations? If those three ways were truly equivalent to the current Gln>Arg recoding, then it would be of very low probability to find that none of the 470 mammals took any of those ways. Without strong purifying selection, those three equivalent ways should have occurred and be maintained by chance.
Putative Explanations for the Absence of Three Additional Ways to Achieve Arg at the Q>R Site
A possibility raised by the harm-permitting model is that the ancestral CAG codon (Gln) first evolved into an Arg codon in the most recent common ancestor of mammals (or the common ancestor of vertebrates), and subsequently evolved back into an editable CAG codon (Gln) permitted by RNA editing. This hypothesis explains the emergence of Q>R recoding event but we still need to account for the absence of alternative ways to achieve Arg in mammals after RNA editing already emerged. A potential explanation is connected to the temporal–spatial nature of RNA editing. Gene GRIA2 encodes a glutamate receptor, if GRIA2 is not solely expressed in brain, then in other tissues, GRIA2 does not have to be 100% edited for the following reasons: (a) ADAR2 might be less efficient in other tissues; (b) the unedited Gln version protein might be functional and indispensable in other tissues as well. We name this as the “functional Gln in other tissues” theory. This assumption is supported by the observation that GRIA2 is indeed expressed in human adrenal and endometrium (Supplementary Fig. S2) but the majority of adrenal samples have editing levels lower than 0.5 (Supplementary Fig. S3) and in endometrium the editing levels are generally high and variable (Supplementary Fig. S4). Moreover, the absence of Q>R recoding in orthologous gene in Drosophila is a compelling evidence for the putative function of the pre-editing Gln (Fig. 2A). Then, under the “functional Gln” theory, way 1 (hardwired Arg codon in the genome) and way 2 (convergent recoding after a Gln>His substitution occurred) will not be allowed because these two cases do not produce any Gln version proteins in other tissues.
One may argue that the observation of Arg encoded by a few fish genomes contradicts the “functional Gln in other tissues” theory. First, the fraction of such hardwired Arg is very rare in fish and does not affect our inference of the function of Q>R in mammals. Second, the recent paper on restorative RNA editing in fungi (Qi et al. 2024) has just proposed a notion that different species are subjected to different environments and conditions, and the advantage of RNA editing might be unnecessary in some particular species so that the highly conserved and functional RNA editing sites can also be lost or hardwired in a few species (Qi et al. 2024). This notion perfectly explains the existence of hardwired Arg in the 13 fishes in our analysis.
However, “functional Gln in other tissues” theory does not explain the absence of CAG>CAA synonymous mutations in 470 mammalian genomes (Fig. 2A). It seems that not only Gln is indispensable, but also the CAG codon itself is unchangeable. The best-known selection constraint on synonymous mutation is the codon usage bias that the codons with higher frequency in the genome (like C/G-ending codons in human) might facilitate rapid translation of mRNAs, so that these optimal codons are favored and selectively maintained (Hanson and Coller 2018). This might account for the preference on CAG codon. Anyway, all these observations reduce the possibility of a pure restorative function of Q>R site.
The Strong Maintenance of Editing Motif and dsRNA Structure Around Q>R Site
Another crucial determinant of RNA editing is the cis elements. Regarding the absence of convergent recoding on Q>R site, one may come up with another explanation that compared to Arg and His codons, only the Gln CAG codon meets the editing motif. The editing motif in animals is mainly the tri-nucleotide context favored by ADAR where thE−1 position avoids G and the + 1 position favors G (Zhang and Duan 2023). The two His codons (CAC/T) and the synonymous Gln codon (CAA) lack the editing motif and this might explain the paucity of convergent recoding. However, (1) the ADAR motif in animals is much weaker than the editing motif in fungi (Bian et al. 2019), meaning that the sequence context has less impact on the occurrence of RNA editing events in animals; (2) the strongest determinants of RNA editing in animals should be the double-stranded RNA (dsRNA) structure. The intronic sequences complementary to the recoded exon usually facilitate the formation of dsRNA around editing sites. Given the fast evolution of intronic sequences, one should worry more about how could Q>R recoding be maintained under the fast evolution of introns. However, the fact is that all the mammals have robustly maintained the ability to edit the Q>R site, suggesting that there might be an unknown way to ensure the occurrence of Q>R site under the fluctuation of cis-elements; (3) Even if one insists that the editing motif matters, but it still cannot explain the absence of hardwired CAG (Gln) to CGG (Arg) mutations in mammals (we will quantify how unexpected it is to see this absence). In contrast, the “functional Gln in other tissues” might account for our observations.
However, although the sequence preference of RNA editing in animals is generally weak, it cannot be assumed that sequence preference has no effect on the editing level at specific sites. In fact, the robust maintenance of Q>R editing capability across all mammals implies that the occurrence and 100% editing level of Q>R recoding may be sensitive to the fluctuation of neighboring base sequences. We therefore set out to analyze the conservation of the flanking sequences and secondary structures around the Q>R sites in vertebrates.
We interrogated the phyloP score in the human genome, which represents the conservation level of each nucleotide. We obtained the phyloP scores generated from 470 mammals (https://hgdownload.soe.ucsc.edu/goldenPath/hg38/phyloP470way/) and 100 vertebrates (https://hgdownload.soe.ucsc.edu/goldenPath/hg38/phyloP100way/), respectively. We checked the 9 bp sequence centered by the GRIA2 Q>R site in human (Fig. 3A). In general, the 470-way score was higher than the 100-way score since the 100-vertebrate encompassed a much wider range of clades although the number of species was fewer. From the aspect of DNA mutation, any mutations at the first cytidine of the CAG codon would be nonsynonymous, and in contrast 1/3 of the mutations at the third guanosine of the CAG codon would be synonymous, this would intuitively imply that the third codon position should be less constrained. However, the opposite trend was observed in mammals where the third G is extremely conserved (Fig. 3A), raising a possibility that the third codon position was constrained by the need to maintain a 100% editing level at the second codon position. However, in the 100-vertebrate results, the phyloP score at the third codon position declined rapidly (Fig. 3A), presumably caused by the non-CAG codons in fishes.
Conservation of the CAG codon across vertebrate genomes. A PhyloP scores of 470 mammals and 100 vertebrates were shown for the 9 bp centered by the Q>R editing site. The third position of CAG is unexpectedly conserved across mammals, presumably due to the need to maintain 100% RNA editing level of the focal adenosine. B Calculation of the probability that a given site is identical across all 470 mammalian species. The neutral sites at the third codon position and the non-neutral sites at the second codon position were respectively used to infer the expected pairwise difference between species. Sites with phyloP score < 0 were defined as non-conserved sites
Regarding the second editing position of CAG, its conservation level across 470 mammals is undoubtedly high since we already know the site is conserved in all species. Next, we try to quantify how unexpected it is to see the absence of hardwired mutations from CAG (Gln) to CGG (Arg) (Fig. 3B). To estimate a neutral mutation rate on GRIA2 CDS (2634 bp not including stop codon), we looked at the third codon position (presumed neutral) and obtained 878 sites. 91 out of these 878 neutral sites were lowly conserved (mammalian phyloP score < 0), suggesting that on average there should be 91/878 = 10.36% of GRIA2 CDS to be different between two pairwise mammalian species. Given a particular nucleotide position (e.g. the Q>R editing site), the probability of seeing it being identical across all 470 mammals would be (1 − 10.36%)^470 = 4.6E−23 (Fig. 3B). This is an extremely unexpected case under neutral evolution by assuming that a genomically encoded Arg is equivalent. However, under our “functional Gln in other tissues” theory, the genomic mutation on this site is constrained and thus it will be unsurprising to observe the maintenance of genomic sequence. Then, even we used the non-neutral second codon position to estimate the pairwise sequence difference (26/878 = 2.96%, which is an underestimate of the real neutral divergence), the probability of observing a site being identical across all 470 mammals would be (1 − 2.96%)^470 = 7.3E−7 (Fig. 3B). This is still a low probability. To show the robustness of this low probability, we used the dS between human and chimpanzee (P. troglodytes) GRIA2 gene to represent the average divergence of 470 mammalian GRIA2 gene under neutral expectation (which is obviously an underestimation of the real average divergence among pairwise mammals). With dS = 0.037, we obtained P = (1 − 0.037)^470 = 2.02E−08 (and the real P value should be lower). If we replace dS with dN, then this P value would be 0.0305, which was still significantly skewed from an intuitive expectation.
For the conservation of dsRNA structure required for Q>R recoding, we made two parallel analyses. The first was to inspect the phyloP scores along the exons and introns of GRIA2 gene (Fig. 4A, B); and secondly, we folded the flanking ± 500 bp centered by Q>R site using human and mouse GRIA2 pre-mRNAs (Fig. 4C). The results supported a selective constraint that maintained the dsRNA structure around Q>R site. The human GRIA2 gene (hg38, https://asia.ensembl.org/) had 16 exons and the CDS region spanned 15 exons. The first intron was too long and due to space limitation, we only demonstrated the range from exon4 to exon16 (Fig. 4A). Q>R site (Chr4:157336723) was located in exon11 and it was only 24 bp away from the downstream exon–intron boundary. Thus, it is very likely that the local dsRNA structure required for Q>R editing was formed by exon11 and the downstream intron. Generally, exon sequences were much more conserved than introns (Fig. 4A) but there came one exception when we enlarged the region near the Q>R site. The intron sequence closely after exon11 was highly conserved as shown by extraordinarily high phyloP scores (Fig. 4B). The conserved intron region stretched for approximately 350 bp. We then folded the ± 500 bp around Q>R site in pre-mRNA (https://rna.urmc.rochester.edu/RNAstructureWeb/). The secondary structures were very similar between human and mouse (Fig. 4C). Interestingly, the sequence from the exon–intron boundary to the 3′ end of the displayed dsRNA structure was exactly 300 bp. This echoed our finding that the ~ 350 bp intron sequence after the boundary was highly conserved (Fig. 4B), which was presumably aimed at maintaining the RNA structure required for Q>R recoding (Fig. 4C).
Conservation of dsRNA structure near Q>R site in human and mouse pre-mRNAs. A PhyloP scores of 470 mammals along GRIA2 gene. As the gene model shows, Q>R recoding site is located in the 11th exon. B PhyloP score for the local region around Q>R site. The exon–intron boundary was labeled. The first 350 bp of the intron were highly conserved. C dsRNA structure required for Q>R recoding in human and mouse pre-mRNAs. The locations of Q>R site and the exon–intron boundary were labeled
Given that the Q>R site is a special case with 100% editing level and requires stringent editing motif and dsRNA structure, the nearby mutations leading to convergent recoding might be deleterious and eliminated, but we argue that this worry only exists for the Q>R site with obligated 100% editing level. For most harm-permitting RNA editing sites, as they already permitted a decrease of editing level from 100% (we mean, the ancestral G-allele) to the current level (mostly < 20% inferred from the median recoding level), then we naturally expect that they should permit new mutations that decrease the editing level again. Moreover, for Q>R site, CAG is an optimal motif for RNA editing, but for other potential cases of convergent recoding, the DNA mutations do not necessarily decrease editing level since it can also be the other way around. For compensatory editing events occurring after the deleterious DNA mutation (which should be very few), the decrease in editing level by nearby mutations might be deleterious. But here comes the same logic that new mutations do not necessarily decrease editing level. So far, the remaining unexplained observation for Q>R recoding site is the absence of direct Q>R genomic mutation, and accordingly, we propose that this might be potentially explained by the “functional Gln in other tissues” theory. Taken together, the Q>R site (CAG) is just a special case. We still need to expand the application of convergent recoding by finding other cases.
In Search of Convergent A-to-I Recoding Across Editomes
For an individual RNA recoding site, it might have a G-allele in the ancestral genome, but this RNA editing site might be non-functional, and therefore we consider the ancestral G-allele as a necessary but insufficient requirement for “exerting a restorative function”. Our convergent recoding methodology will then serve as an auxiliary judgement on this potential restorative function. Next, we try to extend this analysis to a wider range of sites and clades. We plan to: (i) systematically define all possible cases of convergent recoding from the codon table, not only the Gln>Arg and His>Arg pair; (ii) focus on the global editome instead of a single recoding site; (iii) use a proper control to quantify whether the observed occurrences of convergent recoding, or the mutations that lead to convergent recoding, are overrepresented or underrepresented.
First, let “X” and “Y” be the two AAs that can be “A-to-I(G) recoded” to become another AA “Z”, and X and Y themselves can be switched by a point mutation. Under this definition, from the codon table we found four groups of (X, Y, Z) meeting these criteria (Fig. 5). In all four groups, X and Y can be switched by a single DNA mutation at the third codon position. In groups 1 and 2, A-to-I RNA recoding takes place at the second codon position, while in groups 3 and 4, A-to-I RNA recoding occurs at the first codon position (Fig. 5).
Four groups of AAs that meet the prerequisite of convergent A-to-I recoding. Let X and Y be the two AAs that can be recoded to become another AA Z. The A-to-I recoding event takes place at a particular codon position, and X and Y can be switched by a point mutation at another codon position. Four groups of (X, Y, Z) are available
Next, to establish a pipeline for preliminarily judging the sole restorative function of RNA editing sites, we try to apply the searching of convergent recoding across the editome. For the Q>R recoding site in mammals, no convergent recoding is observed across 470 species. For the other known RNA editing sites in primates or rodents (Picardi et al. 2017; Ramaswami and Li 2014), the majority of sites are located in non-coding regions, leading to very few conserved editing sites in CDS (Xu and Zhang 2014) and no cases of convergent recoding exist. This fact cancels our further study in mammals. We therefore look for an animal clade with highly conserved CDS editing sites plus a large set of species with documented genome sequences, and promisingly with a global population data if available. Then we came to the Drosophila genus where a large fraction of editing sites are located in CDS and the recoding sites are highly conserved (Yablonovitch et al. 2017).
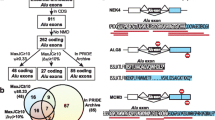
From our previous study (Zheng et al. 2024), we retrieved the known lists of A-to-I RNA editing sites in brains of three Drosophila species D. melanogaster, D. simulans, and D. pseudoobscura (Fig. 6A). Take group 1 convergent recoding (Gln>Arg and His>Arg) for instance, 258 conserved recoding sites between D. melanogaster and D. simulans were observed at the second codon position, meeting the criteria for group 1 (Fig. 5). Here we clarify that the 258 editing sites (which means the edited adenosines) are conserved but we did not restrict the 1st and 3rd nucleotide of the codon. Then, to obtain a fraction of group 1 convergent recoding, we checked how many of these 258 editing sites were found to have Gln>Arg recoding in D. melanogaster matching His>Arg recoding in D. simulans, or the other way around. The result showed that none of the 258 sites belonged to group 1 convergent recoding (Fig. 6A), and thus the occurrence was 0/258. In fact, since the two Drosophila species were close to each other, the recoding sites tended to be highly conserved so that all 258 editing sites led to the same types of AA changes in both species. Similarly, 153 conserved recoding sites between D. melanogaster and D. pseudoobscura were observed at the second codon position, and no cases of group 1 convergent recoding was found (Fig. 6A). The same attempts were done for groups 2, 3, and 4, but still no cases of convergent recoding were found.
Looking for convergent recoding in Drosophila. A Direct searching between D. melanogaster and D. simulans or D. pseudoobscura to find conserved A-to-I RNA editing on different types of codons but leads to the same post-edited AA. B Based on the highly conserved A-to-I editing sites across D. melanogaster, D. simulans, and D. pseudoobscura, we retrieved 14 Drosophila species “between” D. pseudoobscura and the D. melanogaster–D. simulans branch. In target species, we looked for a non-conserved codon that will lead to the same post-edited AA if edited
In addition to the direct searching for convergent recoding events between species, we also fully took advantage of the phylogenetic tree. We downloaded all available genomes in Drosophila genus and found that there were 14 species “between” D. pseudoobscura and the D. melanogaster–D. simulans branch (Fig. 6B). To clarify our scheme, we again take group 1 as an example. 132 recoding sites are conserved at the second codon position across D. melanogaster, D. simulans, and D. pseudoobscura. Since no convergent recoding was observed as shown in Fig. 6A, these 132 recoding sites will recode the same AA in three Drosophila species. This number will be 133 if we require conserved editing at the first codon position as defined in groups 3 and 4. Based on the phylogeny, it is very likely that the 14 target species also have RNA editing at the corresponding adenosine. Then, we delved into these 14 species to check whether we can find a different AA in the genomes (Fig. 6B). If the genomically encoded AA is different, then it might be a case of convergent recoding when subjected to editing. Interestingly, we found a position where a CAG>CGG (Gln>Arg) recoding is conserved across D. melanogaster, D. simulans, and D. pseudoobscura, but in two other species D. ananassae and D. bipectinata, the genome sequence is CAT (His). This would be a nice case of convergent recoding if the CAT codon was really edited at the second codon position (Fig. 6B). The confirmation of editing events requires additional transcriptome data and will be conducted later. Here, with similar workflow, we looked for potential convergent recoding belonging to groups 2, 3, and 4, and did not find such candidates based on genome sequence.
Then, we focused on the only case of potential convergent recoding of group 1. This site is located in gene Prosap encoding a synaptic scaffolding protein. It regulates synaptic growth and development, including the structure of the neuromuscular junction and synapses of the calyx, and olfactory acuity (FlyBase website https://flybase.org/). To know whether the CAT codon in D. ananassae and/or D. bipectinata is subjected to RNA editing, we searched for head/brain transcriptomes of the two Drosophila species and found RNA-Seq data of D. ananassae heads (Methods). We mapped the RNA-Seq reads to the orthologous gene sequence and identified A-to-G variation in RNAs, representing RNA editing events (Fig. 7). Moreover, the editing levels were highly consistent among D. melanogaster, D. simulans, and D. ananassae, suggesting the reliability of the detected editing signals (Fig. 7).
Observation of convergent recoding between D. ananassae and D. melanogaster/D. simulans/D. pseudoobscura. Phylogeny of the four species is displayed on the left. The IGV visualization plot shows the CAT>CGT (His>Arg) recoding event in heads of D. ananassae. Editing levels in heads of each species are shown. Drosophila bipectinata was not displayed due to the lack of head/brain transcriptome
In this part, we finally identified a bona fide case of convergent A-to-I recoding across different species. We estimated its occurrence rate to be two out of 14 × 265 = 3710, which is 1/1855. “Two” refers to the occurrence of convergent recoding in two target species D. ananassae and D. bipectinata, presuming that D. bipectinata is also edited as seen in D. ananassae; 14 refers to the total 14 target species we used; 265 stands for the number of conserved recoding sites (edited adenosines) between D. melanogaster, D. simulans, and D. pseudoobscura, with 133 adenosines at the first codon position and 132 adenosines at the second codon position. Given this observed low occurrence (< 1/1000) of convergent recoding fixed in different species, we wonder how can we obtain more such cases to increase the statistical power? What other angles can we investigate the evolution and adaptation of convergent recoding? How can we utilize convergent recoding to judge if a recoding site has a sole restorative function?
First, we need to conceptually distinguish the following two conditions. (1) Inter-species macro-evolutionary analysis discovered this single case of groups 1 convergent recoding in Drosophila genus. The rarity of its occurrence might represent the disadvantage of having such cases. But this potential deleteriousness has not been statistically tested; (2) at intra-species population level, we are able to directly test the selection force acting on the newly emerged mutations that lead to convergent recoding.
DNA Mutations in D. melanogaster Strongly Avoid Creating Convergent Recoding Between Different Populations
Given that convergent A-to-I recoding really exists between different Drosophila species, it is still unclear whether the DNA mutations leading to convergent recoding are favored or not (as we defined, convergent recoding is unlikely caused by independent gains of editing events on already-diverged codons unless phylogenetic evidence is provided). These puzzles might be answered in the light of population genomics. The micro-evolutionary analysis could reveal how the convergent recoding events emerged at the beginning.
Take group 1 (Gln>Arg and His>Arg) convergent recoding for instance (Fig. 8A). For a common CAG>CGG (Gln>Arg) recoding site in D. melanogaster, a novel DNA mutation might occur in the population. This mutation changes CAG to CAT (His) in part of the population but RNA editing at the second codon position still exists in the entire population. Then the two subpopulations will possess different types of recoding but the post-edited AA (Arg) is the same (Fig. 8A). This represents the initial stage of convergent recoding where the DNA sequences have just diverged between subpopulations, not between species yet. Under the restorative hypothesis, the post-edited Arg is fitter, then we should observe the tolerance of such DNA mutations leading to the switch between Gln and His (Fig. 8A). The same goes for the other three groups of convergent recoding AAs in Fig. 5. Note that the switch between X and Y is reciprocal so that even the editing motif is changed, the editing level does not necessarily decrease. In addition, the restorative function of recoding will also predict the tolerance of novel A-to-G DNA mutations in populations creating a hardwired G-allele (Fig. 8A). In summary, for a group of potential convergent recoding AAs (X, Y, Z), the “X ← → Y” or “ → Z” DNA mutations should be tolerated under the pure restorative role of the recoding sites. If not, then the sole restorative function of the recoding sites should be reconsidered.
Selection force acting on DNA mutations that create convergent recoding between populations. A Novel DNA mutations in population are labeled in purple. RNA editing sites conserved between D. melanogaster and D. simulans are in red. CAG (Gln) is shown in the plot but it can also be CAA (Gln) or CAT/CAC (His) in this case. Unedited adenosines are colored in cyan. DNA mutations can either make a switch between Gln and His codons, or can directly change Gln or His to the Arg codons. B Let “X” and “Y” be the two AAs that can be recoded to become another AA “Z”. We first counted the mutations occurring on all editable X and Y codons, and then counted how many of the mutations can make a switch between X and Y, or can lead to X > Z or Y > Z changes. The observed (obs.) and expected (exp.) fractions of such mutations (SNPs) were compared. Four groups of (X, Y, Z) are available. P values were calculated by one-sided Fisher’s exact tests. N.A. not applicable due to the occurrence of two zeros in the test (Color figure online)
We downloaded the single nucleotide polymorphism (SNP) data from the Drosophila melanogaster genetic reference panel (DGRP; Mackay et al. 2012). First, we searched for codons meeting the following criteria (Fig. 8A): (1) the codon is CAN (Gln or His, N = A/C/G/T); (2) the codon is identical between D. melanogaster and D. simulans; (3) the codon contains a conserved recoding event at the second codon position of D. melanogaster and D. simulans; (4) in D. melanogaster population, a derived DNA mutation is observed at this codon. It turns out that only five codons meet the above criteria (Fig. 8A). Then we interrogated how many of the five codons are potential convergent recoding or hardwired G, that is, contain a nonsynonymous SNP causing switches between Gln and His, or contain a nonsynonymous SNP leading to hardwired Arg. The beneficial restorative hypothesis should predict high tolerance of such cases, however, in real data no cases were observed. Thus, the fraction of mutations supporting beneficial restorative editing is 0/5 (Fig. 8A). Next, we need a negative control to judge whether this 0/5 occurrence is significantly underrepresented.
As a control, we retrieved the unedited codons meeting the same set of afore-mentioned criteria except for the requirement on RNA editing. Totally 13,637 CAN codons were obtained, among which 6200 (45.5%) of them contain a nonsynonymous SNP causing switches between Gln and His (4560 codons) or contain a nonsynonymous SNP leading to hardwired Arg (1640 codons). The difference between 0/5 and 6200/13637 is significant under Fisher’s exact test (Fig. 8B). Similarly, for potential convergent recoding codon groups 2, 3, and 4, we calculated the observed versus expected fractions of SNPs leading to the “X ← → Y” switch or hardwired “ → Z” mutation. In all comparisons, the observed fractions were remarkably lower than the expected ones, and in group 4 the difference is significant (Fig. 8B).
These results suggest that for the several edited codons we tested, mutations leading to convergent recoding or hardwired G-allele are suppressed. In other words, the alternative ways to achieve the post-edited AA are not favored by the organism. These observations reduce the possibility that those groups of recoding sites exert a sole restorative function. We re-emphasize that the potential change in editing motif is not an explanation for the paucity of convergent recoding because: (1) the switch between the two pre-editing codons is reciprocal and the editing level might increase, decrease, or keep intact; (2) the restorative function of a recoding site might not rely on a precise editing level as long as the site is edited. The mammalian Q>R recoding with obligated 100% editing level is only a special case, let alone its restorative function is still questionable.
Next, one may question that why only a handful of codons were used for this analysis, leading to a low statistical power. The reason is that the candidates for convergent evolution is rare given the specific requirements on codon properties. Nevertheless, our ideas could be utilized for determining whether a particular group of recoding sites conforms to the restorative hypothesis. Notably, as we have mentioned, most recoding sites might be neutral or deleterious and only a small fraction was selectively favored. Even for the favored ones, reducing their restorative role does not directly prove the proteomic diversifying role. Both hypotheses have their own predictions that can be computationally proved or disproved to facilitate our understanding of the adaptive nature of recoding events. We only claim that there is a tendency between “the existence of convergent recoding” and “the sole restorative function of an RNA editing site” due to the conceivable tolerance of additional equivalent ways.
Discussion
In this study, we defined and searched for a previously ignored type of RNA editing sites termed convergent recoding sites. At macro-evolution scale, it refers to a conserved editing site that make different AA alterations in different species. At micro-evolution scale, this case appears when novel DNA mutations occurs in an edited codon and produces a different genomically encoded AA but the post-edited AA remains the same.
The motivation for defining this type of RNA editing site originally came from the restorative hypothesis of recoding events but we added several restrictions. Under such hypothesis, the AA (Z) produced from post-edited G-allele has higher fitness than the pre-editing AA (X) version (Jiang and Zhang 2019). If this is true, then (1) the A-to-G DNA mutation leading to a hardwired post-edited AA (Z) should be more straightforward, and (2) if another pre-editing AA (Y) can also be recoded to the post-edited AA (Z), then the switch between X and Y in the genome sequence should also be inconsequential. The restrictions added by us include: (i) this recoding site solely exerts a restorative function so that G-allele is constantly fitter than A-allele; (ii) the restorative function of this particular recoding site does not rely on the precise editing level as long as editing could occur; (iii) take Qln>Arg recoding as an example, the unedited allele after genomic mutation e.g. His should not be harmful compared to the original unedited AA e.g. Gln. This issue will be erased if the editing level is constantly 100% where no unedited allele exists. Given all these potential limitations, the denial of a sole restorative role might have limited power in proving whether it belongs to proteomic diversifying site or other cases, but having this additional approach is better than nothing.
For the predicted situations under a pure restorative function, the situation of hardwired G-allele is always achievable by a single A-to-G point mutation. The second situation, what we call convergent recoding, is not applicable for every codon or AA because it only fits four groups of AAs (X, Y, Z) by definition (see Fig. 5). In order to determine whether a set of recoding sites solely exert a beneficial restorative function, we started to investigate whether convergent recoding or hardwired G-allele is tolerated at these codons at genome-wide level.
In Drosophila, we found an exact case of convergent recoding between species, and demonstrated that the novel DNA mutations leading to hardwired G or convergent recoding were disfavored. This suggests that, at least for the four groups of recoding sites in Drosophila, the benefit of recoding, if any, is not exerted purely by restoration. Intriguingly, note that in Drosophila, the overall recoding sites are beneficial because this statement is already well-established in this field (Yablonovitch et al. 2017), but we still did not claim that every Drosophila recoding site is beneficial and function. Otherwise, if an alternative non-adaptive hypothesis exists (e.g. purifying selection on global recoding sites), then more complicated analysis and additional preliminary work will be needed. Moreover, even within the scope of beneficial recoding, the classification of proteomic diversifying and restorative roles is not absolute. In theory, denying the possible restorative role does not directly prove the proteomic diversifying role. In fact, however, it is commonly believed that Drosophila A-to-I recoding sites mainly exert the proteomic diversifying role although no experimental evidences were provided, and our work further supported this notion by excluding a competing hypothesis at global level. This inference is completely based on that the recoding sites in Drosophila are selectively favored which indicates the beneficial function for at least part of the editing sites.
The traditional in silico analysis for a preliminary judgement for restorative hypothesis (particularly the harm-permitting model) was proposed earlier (Shoshan et al. 2021). If a restorative editing site, defined by the ancestral AA matching the edited state, has a solely restorative function, its editing should be primarily directed at restoring the preferred G allele. In this case, we would expect the nonsynonymous A>G mutation rate at this site to be at least as high as the neutral mutation rate (estimated by the mutation rate observed for synonymous sites). Conversely, if these restorative sites are not merely serving a harm-permitting role, the genomic G allele should be selected against, and a lower frequency of the nonsynonymous A>G mutation rate at this site should be observed. We used the two outgroups Drosophila virilis and Drosophila willistoni to infer the ancestral state of the D. melanogaster–D. pseudoobscura clade. We try to find the following situation: (1) If two outgroups have genomic G, and D. melanogaster and D. pseudoobscura have A-to-I RNA editing, then it suggests than the D. melanogaster–D. pseudoobscura ancestor have RNA editing and should be candidates for restorative sites. If D. simulans has a genomic G, then it implies that the editing site was replaced with genomic G (Fig. 9A). However, in real data, no D. melanogaster–D. pseudoobscura conserved editing sites were found on ancestral G positions, preventing us from testing whether recoding sites were less likely to be replaced with G compared to synonymous sites. (2) Nevertheless, we found 3 recoding and 6 synonymous editing sites in D. melanogaster with ancestral G-allele, then we checked how many of these editing sites have derived genomic A-to-G mutation in the global D. melanogaster populations. Note that to confirm a derived G in D. melanogaster population, we need to require RNA editing in sibling species D. simulans: because without D. simulans, the G allele in D. melanogaster population could be the ancestral state of this species (Fig. 9B). Again, no derived G was found in population. We surmise that the paucity of such cases was due to the overall less abundant CDS editing sites in Drosophila compared to cephalopods (Shoshan et al. 2021).
Attempt to see whether restorative recoding sites are likely to be replaced with genomic G during evolution. The numbers of sites found were given below each category. A Attempt to find derived genomic G in D. simulans at the conserved editing sites between D. melanogaster–D. pseudoobscura. The conserved editing site had an ancestral genomic G inferred from D. virilis and D. willistoni. B Attempt to find derived genomic G in D. melanogaster global populations at the site of conserved editing between D. melanogaster and D. simulans
For the Q>R recoding site in mammalian GRIA2 gene, the constant 100% editing level in brains and the indispensability of the G-allele made this site likely to be a pure restorative recoding site. However, we provided several lines of evidence to reduce the possibility of pure restoration. First, the A-allele (Gln) was the ancestral state in vertebrates and the Arg version was only derived in a handful of fishes; second, none of the 470 tested mammalian species chose alternative ways to achieve Arg, leaving us a feeling that the pre-editing AA or codon might be useful as well. The alternative ways included the direct A-to-G mutation in the genome and the convergent A-to-I recoding from His to Arg. If Arg has the highest fitness, then convergent recoding should be allowed and there were no needs to strongly maintain any of the ancestral forms. Even the convergent recoding of Q>R site might be unfeasible due to the constraint on editing motif and dsRNA structure, the absence of direct genomic A-to-G might lead to the denial of Q>R site being solely restorative.
Finally, one may come up with a question that even we do not consider the ancestral state of Q>R site at all, it is also very possible that the G-allele is constantly better than the A-allele. In other words, there might be a “G-better hypothesis” that only focuses on the relative fitness of A and G regardless of the ancestral state. First, since the definition of restorative hypothesis including the two subtypes (compensatory and harm-permitting) is already complicated, to avoid further confusion, we are not willing to propose a new hypothesis. Second, regarding the G-better or G-fitter scenario, there should be a reason where did the higher fitness come from? If the ancestral genome was C or T (for a long time) and then it suddenly mutated to A, it will be extremely unlikely that the G-allele could have the highest fitness because G-allele has never appeared at this particular site. Thus, a relatively reasonable situation for G-better scenario is still the restorative hypothesis where G was the ancestral allele, at least it demonstrated that the ancestors had lived well with G-allele for a long time. However, we do not try to distinguish which of the restorative types (compensatory or harm-permitting) does our methodology aim to test. The different between compensatory and harm-permitting editing is about the occurrence time of DNA mutation and RNA editing event. As mentioned, convergent recoding is actually achieved by genomic mutations after the conserved editing sites already existed, we think that the chance of such genomic mutation will be irrelevant to whether editing is compensatory or harm-permitting. It is only about the fact that if G-allele (e.g. Arg) is constantly better than A-allele (e.g. Gln), then convergent recoding should occur by chance because they do not have other unconfirmed harmful effects.
In conclusion, we defined an interesting scenario of convergent recoding between editomes of different species. The occurrence of such convergence events could be used as preliminary judgements for a pure restorative function. Our work provides novel insights to the natural selection and evolution of RNA editing.
Data availability
The genome accession IDs of all Drosophila species used in this study were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) and the accession links were listed in Supplementary Table S1. The lists of high-confidence A-to-I RNA editing sites in brains of Drosophila melanogaster, Drosophila simulans, and Drosophila pseudoobscura were retrieved from our previous study. The SNP data were downloaded from the Drosophila melanogaster genetic reference panel (DGRP) (Mackay et al. 2012). The head transcriptomes of D. ananassae were downloaded from NCBI with accession ID SRR7243210. The head transcriptomes of D. melanogaster were also downloaded from NCBI with accession IDs SRR7262144 (male) and SRR7262145 (female). The human adrenal (ERR3153450, ERR3153452, ERR3153392, ERR3153417, ERR3153385, and ERR3153335) and endometrium (ERR3153368, ERR3153491, ERR3153386, ERR3153495, ERR3153433, ERR3153361, and ERR3153438) transcriptomes were also downloaded from NCBI. The expression profile of human GRIA2 gene was seen from website https://www.ncbi.nlm.nih.gov/gene/2891.
Consent for Publication
Not applicable.
Abbreviations
- AA:
-
Amino acid
- A-to-I:
-
Adenosine-to-inosine
- ADAR:
-
Adenosine deaminase acting on RNA
- ADAT:
-
Adenosine deaminase acting on tRNA
- CDS:
-
Coding sequence
- CME:
-
Conserved missense editing
- DGRP:
-
Drosophila melanogaster Genetic reference panel
- dN:
-
Nonsynonymous substitution rate
- dS:
-
Synonymous substitution rate
- dsRNA:
-
Double-stranded RNA
- GRIA2:
-
Glutamate Ionotropic Receptor AMPA Type Subunit 2
- PPR:
-
Pentatricopeptide repeat
- PSC:
-
Pre-mature stop codon
- SNP:
-
Single nucleotide polymorphism
- TadA:
-
TRNA-specific adenosine deaminase
References
Alon S, Garrett SC, Levanon EY, Olson S, Graveley BR, Rosenthal JJ, Eisenberg E (2015) The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. Elife 4:e05198
Bar-Yaacov D, Pilpel Y, Dahan O (2018) RNA editing in bacteria: occurrence, regulation and significance. RNA Biol 15:863–867
Bian Z, Ni Y, Xu JR, Liu H (2019) A-to-I mRNA editing in fungi: occurrence, function, and evolution. Cell Mol Life Sci 76:329–340
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421
Cingolani P, Platts A, Le Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92
Duan Y, Cai W, Li H (2023a) Chloroplast C-to-U RNA editing in vascular plants is adaptive due to its restorative effect: testing the restorative hypothesis. RNA 29:141–152
Duan Y, Li H, Cai W (2023b) Adaptation of A-to-I RNA editing in bacteria, fungi, and animals. Front Microbiol 14:1204080
Duan Y, Ma L, Song F, Tian L, Cai W, Li H (2023c) Autorecoding A-to-I RNA editing sites in the Adar gene underwent compensatory gains and losses in major insect clades. RNA 29:1509–1519
Duan Y, Ma L, Liu J, Liu X, Song F, Tian L, Cai W, Li H (2024a) The first A-to-I RNA editome of hemipteran species Coridius chinensis reveals overrepresented recoding and prevalent intron editing in early-diverging insects. Cell Mol Life Sci 81:136
Duan Y, Ma L, Zhao T, Liu J, Zheng C, Song F, Tian L, Cai W, Li H (2024b) Conserved A-to-I RNA editing with non-conserved recoding expands the candidates of functional editing sites. Fly (Austin) 18:2367359
Eisenberg E, Levanon EY (2018) A-to-I RNA editing—immune protector and transcriptome diversifier. Nat Rev Genet 19:473–490
Feng C, Xin K, Du Y, Zou J, Xing X, Xiu Q, Zhang Y, Zhang R, Huang W, Wang Q et al (2024) Unveiling the A-to-I mRNA editing machinery and its regulation and evolution in fungi. Nat Commun 15:3934
Gray MW (2012) Evolutionary origin of RNA editing. Biochemistry 51:5235–5242
Hanson G, Coller J (2018) Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19:20–30
Hayes ML, Santibanez PI (2020) A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J Biol Chem 295:3497–3505
Jiang D, Zhang J (2019) The preponderance of nonsynonymous A-to-I RNA editing in coleoids is nonadaptive. Nat Commun 10:5411
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJC, Eisenberg E (2017) Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169(191–202):e111
Ma L, Zheng C, Xu S, Xu Y, Song F, Tian L, Cai W, Li H, Duan Y (2023) A full repertoire of Hemiptera genomes reveals a multi-step evolutionary trajectory of auto-RNA editing site in insect Adar gene. RNA Biol 20:703–714
Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM et al (2012) The Drosophila melanogaster genetic reference panel. Nature 482:173–178
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nguyen TA, Heng JWJ, Ng YT, Sun R, Fisher S, Oguz G, Kaewsapsak P, Xue S, Reversade B, Ramasamy A et al (2023) Deep transcriptome profiling reveals limited conservation of A-to-I RNA editing in Xenopus. BMC Biol 21:251
Picardi E, D’Erchia AM, Lo Giudice C, Pesole G (2017) REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res 45:D750–D757
Qi Z, Lu P, Long X, Cao X, Wu M, Xin K, Xue T, Gao X, Huang Y, Wang Q et al (2024) Adaptive advantages of restorative RNA editing in fungi for resolving survival–reproduction trade-offs. Sci Adv 10:eadk6130
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Ramaswami G, Li JB (2014) RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res 42:D109–D113
Savva YA, Rieder LE, Reenan RA (2012) The ADAR protein family. Genome Biol 13:252
Shikanai T (2006) RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol Life Sci 63:698–708
Shoshan Y, Liscovitch-Brauer N, Rosenthal JJC, Eisenberg E (2021) Adaptive proteome diversification by nonsynonymous A-to-I RNA editing in coleoid cephalopods. Mol Biol Evol 38:3775–3788
Sommer B, Kohler M, Sprengel R, Seeburg PH (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67:11–19
Takenaka M, Zehrmann A, Verbitskiy D, Hartel B, Brennicke A (2013) RNA editing in plants and its evolution. Annu Rev Genet 47:335–352
Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K et al (2017) Dynamic landscape and regulation of RNA editing in mammals. Nature 550:249–254
Walkley CR, Li JB (2017) Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs. Genome Biol 18:205
Xin K, Zhang Y, Fan L, Qi Z, Feng C, Wang Q, Jiang C, Xu JR, Liu H (2023) Experimental evidence for the functional importance and adaptive advantage of A-to-I RNA editing in fungi. Proc Natl Acad Sci U S A 120:e2219029120
Xu G, Zhang J (2014) Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci U S A 111:3769–3774
Xu G, Zhang J (2015) In search of beneficial coding RNA editing. Mol Biol Evol 32:536–541
Yablonovitch AL, Deng P, Jacobson D, Li JB (2017) The evolution and adaptation of A-to-I RNA editing. PLoS Genet 13:e1007064
Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang HW et al (2013) Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504:168–171
Zhan D, Zheng C, Cai W, Li H, Duan Y (2023) The many roles of A-to-I RNA editing in animals: functional or adaptive? Front Biosci Landmark 28:256
Zhang Y, Duan Y (2023) Genome-wide analysis on driver and passenger RNA editing sites suggests an underestimation of adaptive signals in insects. Genes (Basel) 14:1951
Zhang J, Xu C (2022) Gene product diversity: adaptive or not? Trends Genet 38:1112–1122
Zhang P, Zhu Y, Guo Q, Li J, Zhan X, Yu H, Xie N, Tan H, Lundholm N, Garcia-Cuetos L et al (2023) On the origin and evolution of RNA editing in metazoans. Cell Rep 42:112112
Zhao T, Ma L, Xu S, Cai W, Li H, Duan Y (2024) Narrowing down the candidates of beneficial A-to-I RNA editing by comparing the recoding sites with uneditable counterparts. Nucleus (Calcutta) 15:2304503
Zheng C, Ma L, Song F, Tian L, Cai W, Li H, Duan Y (2024) Comparative genomic analyses reveal evidence for adaptive A-to-I RNA editing in insect Adar gene. Epigenetics 19:2333665
Acknowledgements
We thank all the funders for their financial support.
Funding
This study is financially supported by the National Natural Science Foundation of China (No. 32300371), the Young Elite Scientist Sponsorship Program by CAST (No. 2023QNRC001), the Young Elite Scientist Sponsorship Program by BAST (No. BYESS2023160) and the 2115 Talent Development Program of China Agricultural University.
Author information
Authors and Affiliations
Contributions
Conceptualization and supervision: YD, WC, and HL. Data analysis: YD, LM, JL, CZ, FS, LT, WC, and HL. Writing—original draft: YD, LM, JL, CZ, FS, LT, WC, and HL. Writing—review & editing: YD, LM, JL, CZ, FS, LT, WC, and HL.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Additional information
Handling editor: Michelle Meyer.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, L., Zheng, C., Liu, J. et al. Learning from the Codon Table: Convergent Recoding Provides Novel Understanding on the Evolution of A-to-I RNA Editing. J Mol Evol 92, 488–504 (2024). https://doi.org/10.1007/s00239-024-10190-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-024-10190-z