Abstract
The obcell hypothesis is a proposed route for the RNA world to develop into a primitive cellular one. It posits that this transition began with the emergence of the proto-ribosome which enabled RNA to colonise the external surface of lipids by the synthesis of amphipathic peptidyl-RNAs. The obcell hypothesis also posits that the emergence of a predation-based ecosystem provided a selection mechanism for continued sophistication amongst early life forms. Here, I argue for this hypothesis owing to its significant explanatory power; it offers a rationale why a ribosome which initially was capable only of producing short non-coded peptides was advantageous and it forgoes issues related to maintaining a replicating RNA inside a lipid enclosure. I develop this model by proposing that the evolutionary selection for improved membrane anchors resulted in the emergence of primitive membrane pores which enabled obcells to gradually evolve into a cellular morphology. Moreover, I introduce a model of obcell production which advances that tRNAs developed from primers of the RNA world.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The RNA World hypothesis is a leading explanation for the origin of life. It proposes that RNA or RNA-like molecules emerged through Earth’s chemical processes, leading to an RNA sequence with the capacity to catalyse its own synthesis (Rich 1962; Visser 1984; Gilbert 1986; Bernhardt 2012; Goldman and Kacar 2021). RNA holds significant appeal as the precursor to complex life due to its dual characteristics as both a catalyst and a carrier of genetic information (Atkins et al. 2011; Bernhardt 2012).
The RNA World hypothesis has numerous unsolved challenges, including even its viability (Szostak 2012; Bregestovski 2015). Apart from these questions, the transition from the RNA world to a cellular state also requires elucidation. A model of this transition needs to provide an explanation for the acquisition of membranes and the emergence of the ribosome.
The concept of a self-assembling fatty acid vesicles enclosing an RNA replicator is a prevalent paradigm for the origin of the membrane (Bachmann et al. 1992; Segré et al. 2001; Szostak et al. 2001; Monnard and Deamer 2002; Chen et al. 2005; Albertsen et al. 2014; Monnard et al. 2015; Kurihara et al. 2015). The general thesis is that initially both the RNA and lipid components are self-assembling; however, with time, the RNA molecules developed catalytic functions that facilitated the replication of both the vesicle and RNA (Szostak et al. 2001). Contrary to being a simple lipid enclosure, the modern cell membrane functions as an active and dynamic barrier, playing a crucial role in regulating the movement of nutrients and waste within the cell. This regulation is essential for establishing and maintaining the complex network of enzyme-driven chemical processes of life. Therefore, for a lipid enclosure to sustain life, the first vesicles have been proposed to process a rather unusual set of properties, including spontaneous growth, permeability to nucleotide substrates (Szostak et al. 2001; Joyce and Szostak 2018), and a proto metabolism (Nunes Palmeira et al. 2022; Babajanyan et al. 2023).
Translation is a remarkably intricate and interdependent biological subsystem that has been conserved across all cellular life forms (Fox 2010). Although the evolutionary development of the ribosome has been modelled with high confidence (Fox and Naik 2004; Bokov and Steinberg 2009; Hsiao et al. 2009; Petrov et al. 2015), the reason for its emergence remains elusive (Wolf and Koonin 2007). The first proto-ribosome has been predicted to contain the peptidyl transferase centre (PTC) but lacks the ability to decode mRNA (Agmon et al. 2005; Bokov and Steinberg 2009). This proto-ribosome was estimated to be approximately 165 bases in length (Bokov and Steinberg 2009) and capable of synthesising short peptides comprised of either fixed or random amino acids. Whilst such peptides might not appear useful, the emergence of the complete translational apparatus implies that for a substantial period in the RNA world’s existence, the production of non-coded peptides served a significant purpose. It is generally proposed that these primitive peptides enhanced RNA’s functionality by serving as co-factors or structural influencers for RNA complexes (Szathmáry and Smith 1997; Noller 2012).
A lesser-known solution is the “inside-out” or obcell hypothesis. This posits that the proto-ribosome emerged in a pre-cellular RNA world in order to enable RNA life to colonise the outer surface of lipid membranes. This resolve concerns regarding lipid permeability and enables for the emergence of a relatively sophisticated lifeform, prior to its transition to a protocellular structure. In addition, by positing a high demand for peptides that all have a simply attained chemical property, the hypothesis provides a viable explanation for the early development of the ribosome. This hypothesis was originally introduced by Blobel (1980) and further elaborated by Cavalier-Smith (1987, 2001), with a further contribution from Griffiths (2007). Cavalier-Smith (1987) introduced the term obcell using the Latin prefix “ob-” meaning “towards”, “on”, and “against”.
The Emergence of a Predation-Based Ecosystem Offers an Explanation for Further Development of RNA World
With the RNA world scenario, i.e. a population of RNA replicators, one may expect that the population will proliferate until further growth is limited by the availability of a resource (Malthus 2018; Tang and Riley 2021). Reaching this limit could be extremely harmful to the RNA population. For example, consider an RNA-based world where the majority of replication efforts fail due to a lack of resources. In this scenario, the partial replication of the RNA replicators is an unproductive consumption of resources and would therefore predict a significant decrease in the carrying capacity of the environment. Regardless of the specific details, an ecosystem of competition, either direct or indirect, is likely to occur in an RNA world.
The availability of nucleotides is a possible candidate to be the limiting factor in the RNA world. Under such an environment one may imagine a significant evolutionary fitness gain for an RNA replicator that can acquire its nucleotides by the degradation of foreign RNA. Such an environment might lead to the emergence of a predation or decomposition-based ecosystem (Cavalier-Smith 2001).
Predation creates arms race dynamic and as such can have a major influence on the evolutionary landscape. For instance, it is cited as a potential driver of major transitions in evolution, including multicellularity and origin of eukaryotes (Bengtson 2002; de Nooijer et al. 2009; Ispolatov et al. 2023). Its emergence in the RNA world may be just as significant as it would favour the emergence of more sophisticated RNA lifeforms over time. It also offers a potential solution to the emergence of parasites; RNAs that selfishly increase their own replication at a cost of the fitness of the RNA lifeform (Smith 1979; Takeuchi and Hogeweg 2008; Bansho et al. 2012).
Evolutionary Selection for Increased RNA Cooperation May Result in the Emergence of Lipid-Based Compartmentation
Predation can help create conditions favouring the emergence of cooperative groups (Rubenstein 1978). This may occur if the reproductive success of the individuals is greater when acting in a cooperative manner and if the environment has capacity to sustain the groups (Rubenstein 1978). Cooperation also enables for functional specialisation. However, the evolution of cooperative behaviour in RNA necessitates a mechanism for RNA compartmentalization, a physical process that enables the production of inheritable groups of RNA.
Multiple means of compartmentation have been proposed, including encapsulation in fatty acid vesicles (Blain and Szostak 2014), coacervates (Koga et al. 2011), and aqueous two-phase systems (Andes-Koback and Keating 2011). Perhaps the simplest form of RNA compartmentation may be the complementary base pairing of RNA genes to produce multi-subunit RNA entities. This system of compartmentation is compatible as a predator; however, it is clearly limited in the number of RNA genes to which it could contain.
Life clearly eventually adopts a lipid membrane, and this would enable the increased sophistication of life. However, an immediate adoption of a lipid enclosed RNA lifeform appears to be incompatible with a predation-based RNA world. The replicating RNA within vesicles would be essentially incapable of interacting with foreign RNA.
The Obcell is a Lipid-Bound RNA Lifeform
The obcell offers a superior means compartmentation that is compatible as an RNA predator and that provides a means of adaption of life towards lipids. The tethering is proposed to be achieved by the production of peptidyl-RNAs polymers consisting of a hydrophobic peptide region and a hydrophilic RNA gene. Owing to hydrophobic interactions, the latter which would spontaneously enmesh itself into a lipid whilst the former would remain at the aqueous medium.
This RNA lifeform might be capable of colonising other lipids through environmental processes that facilitate the merging and dividing of lipid vesicles or obcells. This method of reproduction relies on the random segregation RNA genes, which may be viable once the parent obcell achieves a sufficiently high copy-number of each of its genes (Goldman and Landweber 2012). An independent modern analogue of this “statistical segregation” replication has been found to occur in Oxytricha, a genus of single-celled ciliated protists (Goldman and Landweber 2012). This segregation occurs during amitotic division of macronucleus of Oxytricha which contain approximately 1000 copies of each of 20,000 DNA nanochromosomes (Goldman and Landweber 2012; Swart et al. 2013). Unlike the obcell model, these nanochromosomes are not bound to a membrane.
An intuitive critique of the obcell model pertains to the potential dissociation of metabolites/nucleotides due to the non-enclosed structure. However, confinement of this life form could have been achieved within certain environments, such as rock cavities. Additionally, it may be that the cellular morphology is only viable for rather sophisticated lifeforms (Hutchison et al. 2016).
The hypothesis may also be received unfavourably because of its failure to provide a plausible path for the transition from the obcell to a cellular morphology. The subsequent section of this manuscript offers a new suggestion regarding this transition. A new model of the means by which obcells are formed is also presented.
On the Transition of Obcells to a Cellular Morphology
Positing a Gradual Transition to the Lipid Enclosed State
The transition from obcells to a cellular morphology has been previously suggested to occur through the inversion of the membrane, where the outer surface of an obcell becomes the inner surface of an enclosed protocell. A few different mechanisms have been proposed, such as the merger of curved obcells (Blobel 1980; Cavalier-Smith 2001) or an invagination of the membrane facilitated by an early cytoskeletal system, ultimately resulting in complete encapsulation (Griffiths 2007). These models however are not particularly satisfactory. The evolutionary advantage granted by the curved obcell or invagination of membrane is not apparent yet to maintain such a conformation would require a significant energetic expenditure. Furthermore, the model demands that the RNA that is specialised to colonise the outer membrane to immediately adapt to lipid-based encapsulation.
I propose an alternative scenario in which obcells transition to cellular morphology through the emergence of a permeable membrane that enabled RNA life to spread into the internal lumen. In this model, the emergence of membrane pores enables free diffusion of RNAs and ions across the membrane. This enables RNA to colonise the vesicle’s lumen and for this to become an organelle of the obcell. The lumen could confer evolutionary advantages by compartmentalising activities, promoting reactions by molecular crowding (Ichihashi and Yomo 2014; Saha et al. 2014) and reducing the loss of metabolites to the environment. Over time, as life becomes more biochemically capable and less reliant on the external environment, the lumen or cytosol becomes the principal compartment of life.
Emergence of Both Template Decoding Proto-Ribosome and a Porous Membrane Owing to Selection for Improved Anchors
It is reasonable to expect that early development of the proto-ribosome including of template decoding occurred as it enabled for improvements of its original function. In other words, the early development of the proto-ribosome occurred because of the benefits conferred by enhanced membrane anchors. I propose that the evolutionary pressure that favoured this development eventually led to the incidental production of pores with the membrane anchors.
Several hypotheses can explain this evolutionary path. For example, it is plausible that the advancements resulted in more hydrophobically stable membrane anchors. This stability could permit a more concentrated placement of these anchors on the obcell membrane without undermining its structural integrity. It also might have supported the obcells’ adaptation to varied environments, like vesicles with distinct lipid compositions or areas with elevated temperatures. Furthermore, the advancements in proto-ribosome functionality, including template decoding, could have allowed for a more effective utilisation of the limited, highly sought-after hydrophobic residues and it could have facilitated the incorporation of residues that were previously unsuitable for anchor formation.
Regardless of the exact selection process, obcell membranes become sufficiently permeable that there is sufficiently free diffusion of RNA across the membranes to support colonisation. This may have required that this surface of obcells to be saturated with pores. The colonisation of the inside lumen would likely incentivize the further development of membrane pores (Pohorille et al. 2003). This model aligns with prior predictions that the membranes of early life were leaky and gradually became less permeable over time due to advantages related to maintaining low sodium levels initially and later evolving membrane bioenergetics (Mulkidjanian et al. 2007, 2009).
Model of Obcell Structure and Synthesis
Production of Obcell by a Proto-Ribosome and a tRNA-Derived Primer
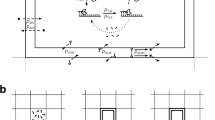
A new model by which the obcell creates a membrane-bound RNA is illustrated in Fig. 1. This model posits that (proto-)tRNAs, which here I assume are essentially the same as contemporary tRNAs, are charged with a hydrophobic residue by an RNA gene. Next, in a fashion similar to modern ribosomes, the proto-ribosome utilises these charged tRNAs to form a hydrophobic peptidyl-tRNA. Once the peptidyl group becomes of sufficient length the peptidyl-tRNA separates from the proto-ribosome and the peptidyl group rapidly integrates into a lipid vesicle, whilst the tRNA group remains in the aqueous medium. A segment of the tRNA then hybridises at the 5′ end to the template/reverse strand of an RNA gene. Following this, the hybridised tRNA acts as an RNA primer, initiating template-based replication of the catalytic/forward stand. Once the synthesis of the complementary strand is complete, the strands separate.
Model of an obcell. Catalytic/forward strand RNAs genes are tethered to lipid, whilst template strand RNA and (proto-)tRNAs are free floating. The steps to add a new gene are (1) tRNA are charged with hydrophobic amino acids. (2) A proto-ribosome uses the aminoacylated tRNAs to make a peptidyl charged tRNAs. (3) These peptidyl-tRNAs dissociate from the proto-ribosome and become bound to the membrane. The tRNA is then cleaved, which enables remaining part (the minihelix) to become accessible to base-pair to RNA genes. (4) This tethered minihelix hybridises to a complementary sequence at the 5′ end of a RNA gene and it promotes the synthesis of RNA strand (either enzymatically or otherwise). A different primer is used to produce the reverse strand non-membrane-bound strands. (6) One of the non-membrane-bound (reverse strand) RNAs includes a primer source gene, which is spliced to excise tRNAs and reverse primers
Whilst a ~ 76-nucleotide long tRNA might seem an unlikely candidate to be an RNA primer, it has previously been proposed the tRNA evolved from a simpler stem-loop structure approximately half the size of a full tRNA (Di Giulio 1992; Tamura 2015; Burton 2020). I suggest that this 3′ half of a tRNA, often referred to as the “minihelix”, served as a primer.
As has previously been suggested by Robertson (1994) this hypothesis proposes template-based RNA replication in a manner opposite to that of modern cellular life; RNA replication in extant life consists of single-nucleotide extension in the 3′ direction, where a phosphate group attached carbon 5 of an activated nucleotide bonds to hydroxyl group in carbon 3 of the growing RNA polymer. This difference should not diminish its plausibility given that the process of RNA replication in the RNA world is predicted to be very different to current mechanisms (Szostak 2012). In addition, template directed replication of oligonucleotides in 5′ direction has been previously demonstrated using a non-enzymatic process (Kaiser et al. 2012). Ribozyme-catalysed template-based extension in the 5′ direction have also been reported although it was capable only of an extension by 3 successive nucleotides (McGinness et al. 2002).
The model proposed here differs from (Cavalier-Smith 2001) model which proposed the transfer of the hydrophobic peptidyl group from the tRNA to RNA genes. I argue that the model presented here is more plausible in part as it avoids this extra processing step. A second, more significant, advantage however is that it proposes a function for tRNAs prior to the emergence of the proto-ribosome thereby simplifying the emergence of the early translational apparatus.
The placement of the tRNA has some similarity with the genomic tag hypothesis which proposed that a minihelix was present at the 3′ end of each RNAs genome (Weiner and Maizels 1987; Maizels and Weiner 1993). Weiner and Maizels however did not propose the minihelixes to be primers.
Obcells Requires a Source of RNA Primers
To ensure a steady supply of primers, both for catalytic and template strands, the obcell requires an RNA gene that contains multiple primer sequences. These sequences could be excised by a ribozyme similar to RNAse P (Guerrier-Takada et al. 1983) in order to generate the primers. As these primer source genes are likely to be quickly degraded after their synthesis it appears unnecessary for them to be membrane bound. In addition, given that a single template strand can be used to create multiple catalytic strands, it stands to reason that using different primers for each strand could optimise the utilisation of available nucleotides.
Typically, one would not expect a sequence that acts as a primer to contain an RNA structure. However, the requirement for a ribozyme to recognise the sequences to excise it may explain why the minihelix (RNA primer) has a stem-loop structure. The large ribosome subunit RNA has been proposed to emerge from a gene containing a multiple tRNAs sequences (Tamura 2011; Farias et al. 2014; de Farias et al. 2016). This has previously been explained by a fusion of tRNAs; however, a remarkable possibility is that the proto-ribosome first emerged from such a primer source gene.
Discussion
The evolutionary transition from the RNA world to the cellular world, as outlined here, can be condensed into six key stages, as illustrated in Fig. 2. These stages encompass the following: (1) the emergence of the replicator, (2) the development of predation, (3) the emergence of a non-template decoding proto-ribosome, (4) its development to a template decoding proto-ribosome, (5) the introduction of membranes pores, and (6) the colonisation of the internal lumen.
Proposed stages of evolutionary transition from the RNA world to the cellular one. RNA genes are shown in orange. Peptidyl groups are represented with square boxes with hydrophobic domains in cyan and hydrophilic domains in red. Nucleotides are shown in light green circles, whilst tRNA-derived primer are shown in dark green. For clarity, the RNA genes are not always shown
The first stage is the emergence of an RNA (or RNA network) that catalyses its own replication. Given the utility of primers for RNA replication, it is intriguing to consider that this RNA replicator was a nuclease that excised RNA motifs, available in a randomly synthesised pool, to serve as primers for its own replication. This mechanism could have been effective in catalysing its own replication if the template-based extension were a non-enzymatic reaction (Szostak 2012). Although an RNA polymerases may also be viable as a RNA replicator (Tjhung et al. 2020), this alternative would require cooperation of a two polymerases to achieve replication.
The emergence of a predation-based ecosystem is proposed to be the major driver of selection in this model and a reason for increase of complexity with time. The emergence of predation would seem necessary if the RNA replication rate is limited by the availability of nucleotides. I argue that its emergence creates an ecosystem that prevents the RNA world from undergoing decay (Boltzmann 1974) and instead drives increasing RNA cooperation and compartmentation. It is likely that the importance of nucleotide availability would only wane after the emergence of nucleotide biosynthesis in protocells. Owing to its importance to the model proposed here, the examination of a ribozymes capability to degrade RNA is a significant means to evaluate it.
The predation of RNA-by-RNA lifeforms was unlikely to be rapid process as such an event would necessitate the existence of extremely effective, indiscriminate RNA nucleases which likely would result in deleterious exchanges between predators. Instead, it may be a decomposition like process, i.e. a slow consumption process that may be principally focused on RNAs transcripts that were already partly hydrolysed.
The latter stages consist of various stages in the development of the ribosome and essentially posits that this occurred to achieved superior compartmentation. The proposal that the template decoding proto-ribosome emerged to better fulfil the role of the non-template decoding proto-ribosome, as opposed to production true proteins is reasonable. Although the evolutionary selection behind the emergence of membrane pores is not immediately obvious, if offers a plausible pathway towards cellularisation.
It is apparent that DNA emerged relatively late in the development of the protocell, for instance, it is a genuine possibility the genome of the Last Universal Common Ancestors (LUCA) was composed of RNA instead of DNA (Mushegian and Koonin 1996; Forterre 2006). Owing to the relative instability of RNA (Koonin 2014) argued that this would require that the genome of protocells to consist of numerous RNA segments and that cell division is achieved by the statistical segregation approach. In order to reduce the stochasticity with the segregation of genes during cell division it is possible that some form tethering of RNA to membranes persisted after the adoption of the cell-like structure perhaps until the emergence of DNA genomes (Koonin 2014). For this to be achieved with peptidyl-tRNAs however would require two distinct modes of translational termination during this protocellular stage: one that maintained the peptidyl-tRNA bond to create membrane anchors and another that broke this bond to produce the other proteins. The potential existence of distinct translation termination mechanisms is corroborated by the appearance of release factors (the proteins that break the peptidyl-tRNA bond) only after LUCA (Maxwell Burroughs and Aravind 2019).
Support for the obcell hypothesis comes from the structure of the oldest (core) ribosome proteins. Based in part on its structure and characteristics, (Smith et al. 2008; Hartman and Smith 2010) have proposed that the original function of the core ribosome proteins was to anchor the primitive ribosomal RNA to the membrane.
The obcell hypothesis also aligns well with the “adaptive” model of genetic code. This proposes that the code is optimised to reduce the impact of translational errors (Knight et al. 1999). Haig and Hurst (1991) argued that the code maximises the correct placement of hydrophobic and hydrophilic residues (Haig and Hurst 1991; Chiusano et al. 2000). The authors even speculated that translated peptides produced during the optimisation of the genetic code had a role as membrane anchors (Haig and Hurst 1991). This optimisation is possible with the obcell hypothesis given that it proposes a stage of development where the only proteins produced were membrane anchors and as such the code would not be frozen (Crick 1968). This does not imply, however, that the obcell had developed the universal genetic code. For example, it suggests that whilst the codon GUU coded for a hydrophobic amino acid, but it was not necessarily valine. The universal genetic code specification may have emerged at a later stage by substitution of residues with those of similar properties. The evidential value of this aspect of the genetic code for the obcell hypothesis is also tempered however by other hypothesis that have been proposed to explain its nature (Trifonov 2000; Crick 1968; Szathmáry 1999; Baranov et al. 2009).
The obcell hypothesis offers a framework for understanding multiple facets of cell origin. Notably, it avoids proposing singular transformational events or requiring major environmental demands. Instead, it emphasises gradual transitions and the emergence of an ecosystem that continually selects for greater cooperation.
Availability of data and materials
Not applicable.
References
Agmon I, Bashan A, Zarivach R, Yonath A (2005) Symmetry at the active site of the ribosome: structural and functional implications. Biol Chem 386:833–844. https://doi.org/10.1515/BC.2005.098
Albertsen AN, Maurer SE, Nielsen KA, Monnard PA (2014) Transmission of photo-catalytic function in a self-replicating chemical system: in situ amphiphile production over two protocell generations. Chem Commun 50:8989–8992. https://doi.org/10.1039/C4CC01543F
Andes-Koback M, Keating CD (2011) Complete budding and asymmetric division of primitive model cells to produce daughter vesicles with different interior and membrane compositions. J Am Chem Soc 133:9545–9555. https://doi.org/10.1021/JA202406V
Akins JF, Gesteland RF, Cech TR (eds) (2011) RNA worlds: from life’s origins to diversity in gene regulation. Cold Spring Harbor Laboratory Press, New York
Babajanyan SG, Wolf YI, Khachatryan A et al (2023) Coevolution of reproducers and replicators at the origin of life and the conditions for the origin of genomes. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.2301522120
Bachmann PA, Luisi PL, Lang J (1992) Autocatalytic self-replicating micelles as models for prebiotic structures. Nature 357(6373):57–59. https://doi.org/10.1038/357057a0
Bansho Y, Ichihashi N, Kazuta Y et al (2012) Importance of parasite RNA species repression for prolonged translation-coupled RNA self-replication. Chem Biol 19:478–487. https://doi.org/10.1016/J.CHEMBIOL.2012.01.019
Baranov PV, Venin M, Provan G (2009) Codon size reduction as the origin of the triplet genetic code. PLoS One. https://doi.org/10.1371/journal.pone.0005708
Bengtson S (2002) Origins and early evolution of predation. Paleontol Soc Pap 8:289–318. https://doi.org/10.1017/S1089332600001133
Bernhardt HS (2012) The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others)a. Biol Direct 7:23. https://doi.org/10.1186/1745-6150-7-23
Blain JC, Szostak JW (2014) Progress toward synthetic cells. Ann Rev Biochem 83:615–640. https://doi.org/10.1146/annurev-biochem-080411-124036
Blobel G (1980) Intracellular protein topogenesis. Proc Natl Acad Sci USA 77:1496–1500. https://doi.org/10.1073/PNAS.77.3.1496
Bokov K, Steinberg SV (2009) A hierarchical model for evolution of 23S ribosomal RNA. Nature 457(7232):977–980. https://doi.org/10.1038/nature07749
Boltzmann L (1974) Theoretical Physics and Philosophical Problems. Theor Phys Philos Probl. https://doi.org/10.1007/978-94-010-2091-6
Bregestovski PD (2015) “RNA World”, a highly improbable scenario of the origin and early evolution of life on earth. J Evol Biochem Physiol 51:72–84. https://doi.org/10.1134/S0022093015010111
Burton ZF (2020) The 3-Minihelix tRNA evolution theorem. J Mol Evol 88:234–242. https://doi.org/10.1007/S00239-020-09928-2
Cavalier-Smith T (1987) The origin of cells: a symbiosis between genes, catalysts, and membranes. Cold Spring Harb Symp Quant Biol 52:805–824. https://doi.org/10.1101/SQB.1987.052.01.089
Cavalier-Smith T (2001) Obcells as proto-organisms: membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J Mol Evol 53:555–595. https://doi.org/10.1007/S002390010245
Chen IA, Salehi-Ashtiani K, Szostak JW (2005) RNA catalysis in model protocell vesicles. J Am Chem Soc 127:13213–13219. https://doi.org/10.1021/JA051784P
Chiusano ML, Alvarez-Valin F, Di Giulio M et al (2000) Second codon positions of genes and the secondary structures of proteins. Relationships and implications for the origin of the genetic code. Gene 261:63–69. https://doi.org/10.1016/S0378-1119(00)00521-7
Crick FHC (1968) The origin of the genetic code. J Mol Biol 38:367–379. https://doi.org/10.1016/0022-2836(68)90392-6
de Farias ST, Rêgo TG, José MV (2016) tRNA Core Hypothesis for the transition from the RNA world to the ribonucleoprotein world. Life. https://doi.org/10.3390/LIFE6020015
de Nooijer S, Holland BR, Penny D (2009) The Emergence of predators in early life: there was no garden of Eden. PLoS One 4:e5507. https://doi.org/10.1371/JOURNAL.PONE.0005507
Di Giulio M (1992) On the origin of the transfer RNA molecule. J Theor Biol 159:199–214. https://doi.org/10.1016/S0022-5193(05)80702-7
Farias ST, Rêgo TG, José MV (2014) Origin and evolution of the peptidyl transferase Center from proto-tRNAs. FEBS Open Bio 4:175–178. https://doi.org/10.1016/j.fob.2014.01.010
Forterre P (2006) Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc Natl Acad Sci 103:3669–3674. https://doi.org/10.1073/PNAS.0510333103
Fox GE (2010) Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol 2:a003483. https://doi.org/10.1101/CSHPERSPECT.A003483
Fox GE, Naik AK (2004) The evolutionary history of the translation machinery. Genet Code Orig Life. https://doi.org/10.1007/0-387-26887-1_6
Gilbert W (1986) Origin of life: the RNA world. Nature 319(6055):618–618. https://doi.org/10.1038/319618a0
Goldman AD, Kacar B (2021) Cofactors are remnants of life’s origin and early evolution. J Mol Evol 89:127–133. https://doi.org/10.1007/S00239-020-09988-4
Goldman AD, Landweber LF (2012) Oxytricha as a modern analog of ancient genome evolution. Trends Genet 28:382–388. https://doi.org/10.1016/j.tig.2012.03.010
Griffiths G (2007) Cell evolution and the problem of membrane topology. Nat Rev Mol Cell Biol 8:1018–1024. https://doi.org/10.1038/NRM2287
Guerrier-Takada C, Gardiner K, Marsh T et al (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–857. https://doi.org/10.1016/0092-8674(83)90117-4
Haig D, Hurst LD (1991) A quantitative measure of error minimization in the genetic code. J Mol Evol 33:412–417. https://doi.org/10.1007/BF02103132
Hartman H, Smith TF (2010) GTPases and the origin of the ribosome. Biol Direct 5:36. https://doi.org/10.1186/1745-6150-5-36
Hsiao C, Mohan S, Kalahar BK, Williams LD (2009) Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol 26:2415–2425. https://doi.org/10.1093/MOLBEV/MSP163
Hutchison CA, Chuang RY, Noskov VN et al (2016) Design and synthesis of a minimal bacterial genome. Science. https://doi.org/10.1126/SCIENCE.AAD6253
Ichihashi N, Yomo T (2014) Positive roles of compartmentalization in internal reactions. Curr Opin Chem Biol 22:12–17. https://doi.org/10.1016/J.CBPA.2014.06.011
Ispolatov Y, Doebeli C, Doebeli M (2023) On the evolutionary emergence of predation. J Theor Biol 572:111578. https://doi.org/10.1016/J.JTBI.2023.111578
Joyce GF, Szostak JW (2018) Protocells and RNA self-replication. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/CSHPERSPECT.A034801
Kaiser A, Spies S, Lommel T, Richert C (2012) Template-directed synthesis in 3′- and 5′-direction with reversible termination. Angew Chem Int Ed 51:8299–8303. https://doi.org/10.1002/ANIE.201203859
Knight RD, Freeland SJ, Landweber LF (1999) Selection, history and chemistry: the three faces of the genetic code. Trends Biochem Sci 24:241–247. https://doi.org/10.1016/S0968-0004(99)01392-4
Koga S, Williams DS, Perriman AW, Mann S (2011) Peptide–nucleotide microdroplets as a step towards a membrane-free protocell model. Nat Chem 3(9):720–724. https://doi.org/10.1038/nchem.1110
Koonin EV (2014) The origins of cellular life. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol 106:27–41. https://doi.org/10.1007/S10482-014-0169-5
Kurihara K, Okura Y, Matsuo M et al (2015) A recursive vesicle-based model protocell with a primitive model cell cycle. Nat Commun. https://doi.org/10.1038/NCOMMS9352
Maizels N, Weiner AM (1993) The genomic tag hypothesis: modern viruses as molecular fossils of ancient strategies for genomic replication
Malthus TR (2018) An essay on the principle of population, 1803 edn. Yale University Press
Maxwell BA, Aravind L (2019) The origin and evolution of release factors: implications for translation termination, ribosome rescue, and quality control pathways. Int J Mol Sci. https://doi.org/10.3390/IJMS20081981
McGinness KE, Wright MC, Joyce GF (2002) Continuous in vitro evolution of a ribozyme that catalyzes three successive nucleotidyl addition reactions. Chem Biol 9:585–596. https://doi.org/10.1016/S1074-5521(02)00136-9
Monnard PA, Deamer DW (2002) Membrane self-assembly processes: steps toward the first cellular life. Anat Rec 268:196–207. https://doi.org/10.1002/AR.10154
Monnard P-A, Walde P, Mavelli F, Stano P (2015) Current ideas about prebiological compartmentalization. Life 5:1239–1263. https://doi.org/10.3390/LIFE5021239
Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV (2007) Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat Rev Microbiol 5:892–899. https://doi.org/10.1038/NRMICRO1767
Mulkidjanian AY, Galperin MY, Koonin EV (2009) Co-evolution of primordial membranes and membrane proteins. Trends Biochem Sci 34:206–215. https://doi.org/10.1016/J.TIBS.2009.01.005
Mushegian AR, Koonin EV (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA 93:10268–10273. https://doi.org/10.1073/PNAS.93.19.10268
Noller HF (2012) Evolution of protein synthesis from an RNA world. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/CSHPERSPECT.A003681
Nunes Palmeira R, Colnaghi M, Harrison SA et al (2022) The limits of metabolic heredity in protocells. Proc R Soc B Biol Sci. https://doi.org/10.1098/rspb.2022.1469
Petrov AS, Gulen B, Norris AM et al (2015) History of the ribosome and the origin of translation. Proc Natl Acad Sci USA 112:15396–15401. https://doi.org/10.1073/PNAS.1509761112
Pohorille A, Wilson MA, Chipot C (2003) Membrane peptides and their role in protobiological evolution. Orig Life Evol Biosph 33:173–197. https://doi.org/10.1023/A:1024627726231
Rich A (1962) On the problems of evolution and biochemical information transfer. In: Kasha M, Pullman B (eds) Horizons in biochemistry. Academic Press, New York, pp 103−126
Robertson HD (1994) Life Before DNA: the RNA World. Science 264:1479–1480. https://doi.org/10.1126/SCIENCE.264.5164.1479
Rubenstein DI (1978) On predation, competition, and the advantages of group living. pp 205–231. https://doi.org/10.1007/978-1-4684-2901-5_9
Saha R, Pohorille A, Chen IA (2014) Molecular crowding and early evolution. Orig Life Evol Biosph 44:319–324. https://doi.org/10.1007/s11084-014-9392-3
Segré D, Ben-Eli D, Deamer DW, Lancet D (2001) The lipid world. Orig Life Evol Biosph 31:119–145. https://doi.org/10.1023/A:1006746807104
Smith JM (1979) Hypercycles and the origin of life. Nature 280(5722):445–446. https://doi.org/10.1038/280445a0
Smith TF, Lee JC, Gutell RR, Hartman H (2008) The origin and evolution of the ribosome. Biol Direct 3:16. https://doi.org/10.1186/1745-6150-3-16
Swart EC, Bracht JR, Magrini V et al (2013) The oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 11:e1001473. https://doi.org/10.1371/JOURNAL.PBIO.1001473
Szathmáry E (1999) The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet 15:223–229. https://doi.org/10.1016/S0168-9525(99)01730-8
Szathmáry E, Smith JM (1997) From replicators to reproducers: the first major transitions leading to life. J Theor Biol 187:555–571. https://doi.org/10.1006/JTBI.1996.0389
Szostak JW (2012) The eightfold path to non-enzymatic RNA replication. J Syst Chem 3:2. https://doi.org/10.1186/1759-2208-3-2
Szostak JW, Bartel DP, Luisi PL (2001) Synthesizing life. Nature 409:387–390. https://doi.org/10.1038/35053176
Takeuchi N, Hogeweg P (2008) Evolution of complexity in RNA-like replicator systems. Biol Direct. https://doi.org/10.1186/1745-6150-3-11
Tamura K (2011) Ribosome evolution: emergence of peptide synthesis machinery. J Biosci 36:921–928. https://doi.org/10.1007/S12038-011-9158-2
Tamura K (2015) Origins and early evolution of the tRNA molecule. Life 5:1687–1699. https://doi.org/10.3390/LIFE5041687
Tang J, Riley WJ (2021) Finding Liebig’s law of the minimum. Ecol Appl 31:e02458. https://doi.org/10.1002/EAP.2458
Tjhung KF, Shokhirev MN, Horning DP, Joyce GF (2020) An RNA polymerase ribozyme that synthesizes its own ancestor. Proc Natl Acad Sci USA 117:2906–2913. https://doi.org/10.1073/PNAS.1914282117
Trifonov EN (2000) Consensus temporal order of amino acids and evolution of the triplet code. Gene 261(1):139–151. https://doi.org/10.1016/s0378-1119(00)00476-5
Visser CM (1984) Evolution of biocatalysis 1. Possible pre-genetic-code RNA catalysts which are their own replicase. Orig Life 14:291–300. https://doi.org/10.1007/BF00933670
Weiner AM, Maizels N (1987) tRNA-like structures tag the 3’ ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci 84:7383–7387. https://doi.org/10.1073/PNAS.84.21.7383
Wolf YI, Koonin EV (2007) On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct 2:14. https://doi.org/10.1186/1745-6150-2-14
Acknowledgements
I thank Pasha Baranov and Jack Tierney for discussions and help with preparation of the manuscript.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no competing interests.
Ethical approval
Not applicable.
Additional information
Handling editor: Aaron Goldman.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
O’Connor, P.B.F. The Evolutionary Transition of the RNA World to Obcells to Cellular-Based Life. J Mol Evol 92, 278–285 (2024). https://doi.org/10.1007/s00239-024-10171-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-024-10171-2