Abstract
Autoimmune Regulator 1 (AIRE1) and Forebrain Embryonic Zinc Finger-Like Protein 2 (FEZF2) play pivotal roles in orchestrating the expression of tissue-restricted antigens (TRA) to facilitate the elimination of autoreactive T cells. AIRE1’s presence in the gonads of various vertebrates has raised questions about its potential involvement in gene expression control for germline cell selection. Nevertheless, the evolutionary history of these genes has remained enigmatic, as has the rationale behind their apparent redundancy in vertebrates. Furthermore, the origin of the elimination process itself has remained elusive. To shed light on these mysteries, we conducted a comprehensive evolutionary analysis employing a range of tools, including multiple sequence alignment, phylogenetic tree construction, ancestral sequence reconstruction, and positive selection assessment. Our investigations revealed intriguing insights. AIRE1 homologs emerged during the divergence of T cells in higher vertebrates, signifying its role in this context. Conversely, FEZF2 exhibited multiple homologs spanning invertebrates, lampreys, and higher vertebrates. Ancestral sequence reconstruction demonstrated distinct origins for AIRE1 and FEZF2, underscoring that their roles in regulating TRA have evolved through disparate pathways. Furthermore, it became evident that both FEZF2 and AIRE1 govern a diverse repertoire of genes, encompassing ancient and more recently diverged targets. Notably, FEZF2 demonstrates expression in both vertebrate and invertebrate embryos and germlines, accentuating its widespread role. Intriguingly, FEZF2 harbors motifs associated with autophagy, such as DKFPHP, SYSELWKSSL, and SYSEL, a process integral to cell selection in invertebrates. Our findings suggest that FEZF2 initially emerged to regulate self-elimination in the gonads of invertebrates. As organisms evolved toward greater complexity, AIRE1 likely emerged to complement FEZF2’s role, participating in the regulation of cell selection for elimination in both gonads and the thymus. This dynamic interplay between AIRE1 and FEZF2 underscores their multifaceted contributions to TRA expression regulation across diverse evolutionary contexts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Compared to the innate immune system, the adaptive immune system is highly diverse and specific. The remarkable diversity and specificity of the adaptive immune system stem from the unique properties of its key players, particularly the T cells. The adaptive immune system can mount a tailored response to a wide range of pathogens due to the diverse repertoire of T-cell receptors (TCRs) expressed on the surface of T cells (Abbas et al. 2018). Each TCR is specifically designed to recognize a particular antigen presented by the major histocompatibility complex (MHC) molecules on the surface of antigen-presenting cells. This paradigm allows T cells to recognize and respond to millions of different viruses, bacteria, and other pathogens effectively. CD4-expressing T cells specifically bind to MHC class II molecules, while CD8-expressing T cells bind to MHC class I molecules, broadening the range of antigens they can detect. Additionally, each TCR is only able to bind to a limited number of unique antigens, ensuring a highly specific response to particular pathogens. To achieve this high level of diversity and specificity, T cells undergo three critical developmental stages in the thymus, where their TCRs are generated through a process of genetic rearrangement. The first stage takes place during early T-cell development, and it is known as the variability, diversity, and joining (VDJ) recombination process. This process ensures the uniqueness of each TCR complex, rendering it exclusive in its ability to bind to a very limited number of antigens (Abbas et al. 2018). The second stage is the positive selection process, and it takes place in the cortex of the thymus. In this process, T cells expressing both CD4 and CD8 markers are filtered based on their affinity to either MHCII or MHCI (Xing and Hogquist 2012; Takaba and Takayanagi 2017). Cells that bind to an antigen or MHC with an appropriate affinity survive, whereas cells that interact with a weaker affinity die by apoptosis. The third stage is the regulatory process of eliminating autoreactive T cells, which is also known as the negative selection process. In this process, T cells are recruited to the medulla of the thymus. This migration process is controlled by the interaction between CCR7 expressed on T cells and its ligands (e.g., CCL19 and CCL21) expressed by medullary thymic epithelial cells (mTECs) and dendritic cells. mTECs express various tissue-restricted antigens (TRA) to allow deletions of T cells specific for antigens that otherwise would only be encountered in the periphery. Almost all T cells that recognize self-antigen—the MHC complexes—are deleted by mTECs and thymic dendritic cells (Xing and Hogquist 2012). T cells expressing a functional TCR without significant reactivity to self-antigens migrate to secondary lymphoid organs (e.g., the spleen and lymph nodes) and circulate throughout the body.
The process of regulating the expression of TRA is controlled by two main proteins, namely Autoimmune Regulator 1 (AIRE1) and Forebrain Embryonic Zinc Finger-Like Protein 2 (FEZF2) (Takaba and Takayanagi 2017). In the mouse thymus, AIRE1 controls 40% of the TRA expression (Peterson et al. 2008; St-Pierre et al. 2015). AIRE1 does not seem to be a transcription factor (Takaba and Takayanagi 2017). Rather, AIRE1 induces the transcription of TRA genes through interactions with various transcriptional factors. These interactions include enrichment of repressive markers in its promoter region (e.g., H3K27) (Takaba and Takayanagi 2017). It also supports transcriptional elongation through facilitating p-TEFb recruitment. Additionally, AIRE1 interacts with BRD4, which is a transcriptional and epigenetic regulator (Yoshida et al. 2015). Similarly, AIRE1 interacts with TOP1 and TOP2, which are known for their role in regulating the topologic states of DNA during transcription (Pommier et al. 2016). AIRE1 expression is controlled by various gene networks, such as the TNFR family network. This network includes RANK, CD40, RANK-ligand, and CD40-ligand, as well as NF-κB pathways (Akiyama et al. 2008). It was also shown that estrogen and androgen perform a crucial role in controlling AIRE1 expression (Dragin et al. 2016). Recently, strong evidence emerged supporting a crucial role of FEZF2 in central tolerance, probably through acting as a transcription factor. A difference in the usage of TCR Vβ chains in CD4 or CD8 T cells between WT and FEZF2-deficient thymus demonstrated that FEZF2 regulates the negative selection of T cells (Takaba et al. 2015). Notably, some of the genes controlled by FEZF2 are not controlled by AIRE1. However, some exceptions exist, such as FABP2, SAA2, and CDKN1C. FEZF2 expression itself is controlled by the LTBR signaling pathway. Whether other genes are fundamental for the process of expression of TRA is not currently known (Takaba and Takayanagi 2017).
The natural history of the process of expressing TRA to eliminate autoreactive T cells remains unclear. Additionally, the evolutionary advantage of having both AIRE1 and FEZF2 control the TRA expression process has not yet been identified. There is compelling evidence suggesting that invertebrates possess effective mechanisms for eliminating self-reactive cells. For instance, studies show that cells in sponges aggressively attack grafts from other sponges, indicating a recognition of non-self-elements (Beck and Habicht 1996). Phagocytosis, a prevalent mechanism in invertebrates, facilitates the elimination of foreign elements and is observed in a wide range of animals, from starfish to humans (Beck and Habicht 1996). Moreover, invertebrates exhibit systems like prophenoloxidase (proPO) and lectins, which bear striking similarities to the vertebrate complement system and antibodies, respectively. Notably, Drosophila has a mechanism to eliminate miss-migrating embryonic cells (Sano et al. 2005). The presence of such selective processes in invertebrates suggests the existence of mechanisms distinguishing self from non-self-proteins, which could potentially involve the expression of specific proteins to target cells reacting to those proteins. Given the differences in structure and function between AIRE1 and FEZF2, the need for more than one mechanism performing the same function is intriguing. FEZF2 belongs to the FEZF family which functions as a transcriptional repressor, and it is known to contain six C2H2 zinc fingers and an EH1 repressor motif (Copley 2005; Shimizu et al. 2010). Mammalian AIRE1 contains four crucial motifs that support its function in controlling the function of various TFs (Takaba and Takayanagi 2017). These motifs are, namely, CARD (caspase recruitment domain), SAND, PHD1 (plant homeodomain 1), and PHD2. There are two opposing arguments concerning the evolutionary relationship between AIRE1 and FEZF2 in the context of TRA regulation. The first argument proposes that AIRE1 and FEZF2 share a common ancestral origin but have diverged over time due to accumulating differences, eventually leading to distinct roles in TRA regulation. On the other hand, the second argument posits that AIRE1 and FEZF2 are not related and are products of different gene families from the outset. Despite their different origins, they have independently evolved to perform similar functions in TRA regulation through convergent evolution. To evaluate these contrasting perspectives, our hypothesis suggests that AIRE1 and FEZF2 are not ancestrally related and have independently evolved to serve similar functions in TRA regulation, which can be attributed to convergent evolution mechanisms rather than a common ancestral pathway.
In this investigation, we studied the evolutionary history of the mechanisms responsible for the regulation of TRA expression, focusing primarily on the roles of AIRE1 and FEZF2. To test our hypothesis, we conducted a range of analyses, including multiple sequence alignment followed by phylogenetic analysis. Additionally, to determine whether the FEZF2 and AIRE1 families were structurally related, we constructed putative homologs of their ancestral sequences using ancestral sequence reconstruction. We also explored whether they evolved under positive selection pressure using Phylogenetic Analysis by Maximum Likelihood (PAML). To compare their functional specificity, we employed type II functional divergence analysis, linear functional motif searches, and gene enrichment analysis for their downstream targets. Furthermore, we utilized non-homology-based artificial intelligence methods to predict the function of AIRE1 and FEZF2 in earlier diverging species. The results of our research allowed us to infer the earliest diverging homologs and the evolutionary history of the regulatory mechanisms responsible for controlling the expression of TRA, while excluding PAML from the analysis.
Materials and Methods
Workflow
To investigate the evolutionary history of the proteins responsible for the negative selection pathway, we divided these proteins into three groups (i) main regulators (AIRE1 and FEZF2), (ii) upstream of the main regulators (e.g., regulators of regulators), and (iii) downstream targets of the regulatory elements (Table 1). For the first group, we drew the phylogenetic history, calculated the ancestral sequence, investigated positive selection, functional divergence, and functional specificity, and then identified functional motifs. For the other two groups, we only investigated the evolutionary history and gene ontology.
Database Search
In this study, our primary focus was investigating the evolutionary origins of the FEZF2 and AIRE1 pathways. To provide a comprehensive understanding of these pathways, we thoroughly examined not only FEZF2 and AIRE1 themselves but also their associated regulators and downstream targets (Table 1). Given the diverse nature and extensive evolutionary history of the genes under investigation, we employed a strategy based on sequence alignment of their respective proteins (Mickael et al. 2016). We conducted a detailed analysis of FEZF2 and AIRE1 presence across various taxonomic classes, spanning over 500 million years and encompassing more than 200 protein sequences, including Mammalia, Actinopterygii, Insecta, Arachnida, Gastropoda, Bivalvia, Cephalopoda, Anthozoa, and Placozoa. To maintain consistency and comparability across our analysis, we conducted BLASTP searches utilizing human protein families against the aforementioned proteomes. In order to ascertain robust results, we employed the longest transcript homolog for each species during our investigation (Supplementary file 1). To establish candidate proteins, we set a stringent threshold, only accepting sequences with E values below 1e–10 (Wiemerslage et al. 2016). Additionally, we implemented a filtering step by comparing conserved domains within each identified protein against the query human protein sequences. By adopting this comprehensive approach and standardizing our taxonomic categorization, we aim to provide a clear basis for comparison across different evolutionary time frames. This ensures that our analysis accurately captures the evolutionary trajectories of the FEZF2 and AIRE1 pathways within the context of their respective taxa.
Alignment and Phylogenetic Analysis
The phylogenetic investigation was done in two stages (Kubick et al. 2018; 2021a, b). First, amino acid sequences were aligned using MAFFT by utilizing the iterative refinement method (FFT-NS-i) (Huson and Bryant 2006; Katoh and Standley 2013; Tamura et al. 2013). To ensure the utmost accuracy in capturing the complex evolutionary relationships within our dataset, we harnessed the powerful capabilities of IQ-Tree (Nguyen et al. 2015). This advanced tool enabled us to investigate a diverse range of substitution models, including but not limited to GTR, HKY, and JC. Through the comprehensive analysis offered by IQ-Tree, we employed sophisticated techniques such as the Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) to rigorously compare and evaluate different models’ performance in representing our data's intricacies. Through IQ-Tree’s meticulous examination, we were able to pinpoint the most suitable model for tree investigated. This selection was grounded in the model’s ability to effectively explain the observed sequence variations and reflect the underlying evolutionary processes accurately.
Positive Selection Analysis
We employed a maximum likelihood approach to explore whether FEZF2 or AIRE1 underwent positive selection during evolution (Kubick et al. 2021a, b). First, we back translated the respective complementary DNAs (cDNAs) using the EMBOSS Backtranseq tool and aligned them based on their codon arrangement (Madeira et al. 2019). Next, we examined patterns of positive selection in both genes using CODEML (PAML, Version 4) (Yang 2007). To investigate selection, we calculated the substitution rate ratio (ω) as the ratio of nonsynonymous (dN) to synonymous (dS) mutations. We conducted three levels of analysis: (i) basic (global) selection, (ii) branch-specific selection, (iii) branch-site-specific selection, and (iv) site-specific selection models (Kubick et al. 2018). Statistical significance was determined through a likelihood ratio test (LRT), calculated using the following equation: \(p{\text{ - value}} = \chi^{{2}} \left( {{2}*\Delta \left( {{\text{ln}}\left( {{\text{LRTmodel}}} \right) - {\text{ln}}\left( {{\text{LRTneutral}}} \right)} \right),{\text{ number of degrees of freedom}}} \right).\).
Functional Divergence Estimation
We conducted an analysis of Type II functional divergence between the FEZF2 and AIRE1 proteins using the DIVERGE software. This analysis aimed to identify any shifts in cluster-specific amino acid properties. Type II functional divergence represents a change in amino acid properties, such as charge, size, or hydropathy (Gu and Velden 2002). In this analysis, AIRE1 was grouped into higher and lower vertebrates, while FEZF2 was classified in both vertebrates and invertebrates (Gu and Velden 2002; Kubick et al. 2018).
Linear Motifs Search
To investigate the distinction in the evolution–function relationship between FEZF2 and AIRE1, we conducted a search for linear motifs within the protein sequences of both. Linear motifs are short sequences of amino acids that serve as potential protein interaction sites. We performed this search using the ELM server (http://elm.eu.org/) with a motif significance threshold set to 100 (Kumar et al. 2019).
Non-Homology Functional Prediction
To substantiate our findings, we utilized two non-homology-based techniques for predicting the functions of FEZF2 and AIRE1. Initially, we employed DeepGO, a method that predicts protein function by leveraging neural networks and gene network linkages (Kulmanov et al. 2018). Additionally, we utilized a weighted K-nearest neighbor classifier as implemented in Pannzer (Törönen et al. 2018).
Functional Ontologies
To complement our study and address the question of why two different genes perform the same function, we employed functional ontologies. We aimed to determine whether the reason behind this redundancy was based on the unique functions of the genes they controlled. To achieve this, we compared the gene enrichment profiles of AIRE1 and FEZF2 downstream targets using three methods. First, we analyzed the microarray gene sets GSE69105 and GSE2585, which consist of groups of WT and knockout mice for AIRE1 and FEZF2, respectively, using GeneSpring©. We considered downstream gene candidates that exhibited a fold change greater than 1.4 and a p-value less than 0.05. Subsequently, we utilized the Gene Ontology (GO) Gorilla server to identify gene enrichment in both datasets. Additionally, we employed the list of downstream targets as input for Metascape©. To support the results of our functional ontology investigation, we examined the expression of FEZF homologs in invertebrates by referencing FlyBase (https://flybase.org/) and WormBase (https://wormbase.org/) (accessed on 21 February 2023).
Results
Evolutionary History of AIRE1 and FEZF2
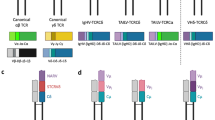
FEZF2 and AIRE1 differ considerably in their evolutionary history. In addition to FEZF2, the FEZF family possesses another member, namely FEZF1. We only found homologs for FEZF1 in vertebrates (Fig. 1A). Conversely, FEZF2 seems to have homologs in higher vertebrates (mammals and fish) as well as lampreys. Additionally, BLASTP analysis revealed that FEZF2 has multiple homologs in various invertebrates investigated, including Spiralia (Crassostrea virginica, E-value < 3e-112), Cnidaria (Nematostella vectensis, E-value < 1e-110), and Arthropoda (Drosophila melanogaster, E-value < 2e-114). Thus, the FEZF family seems to have undergone one round of duplication that occurred during the Cambrian explosion between 541 and 530 million years ago. FEZF2 from invertebrates are orthologs of FEZF2 from vertebrates. FEZF2 from invertebrates and vertebrates are paralogs of FEZF1 from vertebrates. The duplication event occurred in the branch before all vertebrates (Fig. 1A, branch with bootstrap 76). FEZF2 is most likely the ancestral version of this protein/gene. Interestingly, we were not able to locate homologs for AIRE1 beyond vertebrates. On the structural level, multiple sequence alignment and conserved sequence investigations revealed that both FEZF1 and FEZF2 share a structural domain consisting of six C2H2 zinc finger domains (Fig. 1B). This domain appears to be conserved in all species investigated. In the case of AIRE1, four conserved domains were found among the investigated species, namely the homogeneous staining region (HSR), the SAND domain (Sp100, AIRE1, NucP41/75, and DEAF-1), as well as two Plant Homeodomain (PHD) subunits.
FEZF2 and AIRE1 evolution histories. A The FEZF2 family appears to have homologs as early as Trichoplax adhaerens, while the oldest homolog of the AIRE1 family can be found in bony fish. CD4 protein sequences from humans were used as an outgroup. Internal node labels represent bootstrap values. The putative occurrence of a round of duplication is evident in FEZF2 but not in AIRE1. B On a structural level, AIRE1 contains four main functional domains, namely HSR, SAND, and two PHD units. In contrast, the FEZF family is characterized by having a single conserved domain consisting of six zinc fingers
Evolution of the Process of TRA Expression in the Thymus
The evolutionary history of TRA is intricate. While AIRE1 seem to have first emerged during the divergence of bony fish, its regulators do not share a specific emergence period (Supplementary file 1 and Fig. 2). Notably, RANK first emerged during the divergence of lampreys, and CD40 has homologs in both lampreys and tunicates. Homologs of CD40L, on the other hand, first appeared in fish. Estrogen receptor homologs (ER1 and ER2) are both present in lampreys, but ER1 seems to be more ancient, with a homolog found in Spiralia and in Cnidaria. The downstream targets of AIRE1 also exhibit a diverse evolutionary history. Various genes controlled by AIRE1 first emerged in bony fish (e.g., CAMK2B, C4BP, KRT2) or rodents (e.g., AMPLEX). Similarly, various proteins controlled by AIRE1 have homologs in cnidarians, such as GSTA2 and IGF2 (Supplementary file 1). The evolution of the FEZF2 pathways appears to follow a similar pattern to AIRE1, with no apparent common emergence period. Furthermore, several proteins regulated by FEZF2 appeared in invertebrates (Fig. 2). For example, BHMT has homologs in both Cnidaria and Trichoplax, while F2 and MAOA homologs exist in Cnidaria, and KCNJ5 in Spiralia. Interestingly, known regulators of FEZF2 seem to have emerged much later, with LTA, LTB, and LIGHT homologs first diverging in bony fish, and LTBR homologs first appearing in lampreys.
Investigating the evolution of AIRE1 regulators, and downstream targets reveals a complex picture. A Several of AIRE1 known regulatory proteins and genes, appear to have earlier homologs. For example, RANK, CD40, and ESR1 all have homologs in Spiralia. Since mollusks lack T cells, this observation indicates that these regulators might have other functions. B We noticed that more than half of the genes regulated by AIRE1 seem to have homologs that predate the earliest divergence of AIRE1 homologs. This observation cannot be extended toward FEZF2-regulated pathways, as FEZF2 possesses homologs as ancient as Trichoplax. Overall, these observations indicate that controlling one gene by another is not necessarily connected to the time of divergence of either of these two genes. Numbers in parentheses are SH-aLRT support (%)/ultrafast bootstrap support (%)
AIRE1 and FEZF2 Families Do Not Share a Recent Common Origin
Regulatory elements controlling the process of TRA expression have a divergent evolutionary history (Table 2). The FEZ family contains two main genes, namely FEZ1 and FEZ2. The nearest homolog for the constructed ancestral sequence of the FEZF family is a protein that contains a zinc finger (C2H2 type) and is expressed in Metschnikowia aff. Pulcherrima (Fig. 1B and Table 2). The nearest homologs to the reconstructed ancestral sequence of the AIRE1 family include three proteins, namely PHD finger protein 12 (Drosophila busckii), E3 ubiquitin-protein ligase TRIM33 like (Actinia tenebrosa), and Chromodomain-helicase-DNA-binding protein 4-like isoform X3 (Orbicella faveolata). We constructed an evolutionary a phylogenetic tree for these sequence. Our results indicate that Chromodomain-helicase-DNA-binding protein 4-like isoform X3 (Orbicella faveolata) could be the nearest homolog of the reconstructed ancestral protein sequence of AIRE1. Taken together, our results do not support the hypothesis that the FEZF family and AIRE1 are evolutionarily related (Fig. 3).
Positive Selection
We conducted a comprehensive analysis of positive selection to assess the evolutionary dynamics of AIRE1 and FEZF2. Notably, our analysis revealed distinct patterns in the conservation of these two genes across various taxa. In the case of FEZF2, we observed a high degree of conservation between vertebrates and invertebrates, as evidenced by an ω value below 1 (0.3, p-value < 0.01) in both the global and branch evolution patterns. Our investigation at the branch-site and site levels, using the phylogenetic tree (Fig. 1), did not uncover any amino acids subject to positive evolution within the vertebrates and invertebrates divide. These results were based on the Bayes Empirical Bayes (BEB) method (Table 3). However, for AIRE1, we detected a more relaxed conservation evolutionary pattern compared to FEZF2. Globally, AIRE1 appeared to have evolved under neutral selection, reflected by an ω value of 1. Similarly, we did not find conclusive evidence of positive selection on the primates branch. Moving to the site-branch analysis, we identified only two amino acids subjected to positive selection: 147 P (probability: 0.998**, BEB) and 315 V (probability: 0.992**, BEB).
Functional Divergence
Overall, we detected a low degree of functional divergence. We utilized DIVERGE 3.0 to identify putative functional divergence type II sites using a cutoff value of 0.5 (Supplementary file 2). We found only two functional divergent sites in the AIRE1 sequence, particularly at site 290 (where higher vertebrates have Histidine, while bony fish have Tyrosine). Additionally, at amino acids 320 and 321, higher vertebrates have HA, while fish have YS. Although the FEZ2 family has diverged much earlier than AIRE1, we were able to locate only one single site that could represent a candidate for functional divergence (site 454). There is a strong variation at this site, with vertebrates having glutamine, insects having threonine, Cnidaria having isoleucine, and Trichoplax having valine. These results suggest that both AIRE1 and FEZ2 exhibit a limited number of functional divergent sites.
Motifs Search
Our linear motif search revealed a complex picture for AIRE1 and FEZF2. We found that AIRE1 contains an LXXLL motif, known to play a role in binding nuclear receptors. Conversely, FEZF2 has several motifs related to autophagy, such as DKFPHP, SYSELWKSSL, and SYSEL, as well as motifs involved in the regulation of actin and cytoskeleton dynamics, such as PPACPR. However, both genes share various similar motifs related to MAPK regulation, such as AGASPAT (see supplementary file 3).
Non-Homolog-Based Functional Ontology of FEZF2 and AIRE1
In order to reinforce our findings, we employed non-homology-based methods for functional ontology prediction of FEZF2 and AIRE1. Specifically, we utilized DeepGO to examine the functional conservation of these proteins (Fig. 4). This approach is AI driven and not reliant on homology. The results revealed a high degree of functional conservation for both FEZF2 and AIRE1 among the species in which they are expressed. This suggests that FEZF2 may serve a similar function in invertebrates.
Non-homology-based Functional Prediction. We employed AI-driven methods to predict functions based on protein sequences. This approach was chosen because homology-based assumptions may not provide a complete representation of the evolutionary history of these distant genes (Bhaumik et al. 2023). Our results indicate that AIRE1 oldest homologs appear in fish, while FEZF2 has homologs in more ancient divergent species, such as Spiralia and Cnidaria
AIRE1 and FEZF2 Have Distinctive Functional Ontologies
We explored the functions of the top genes identified through microarray data analysis (GSE69105 and GSE2585) in GeneSpring, with a fold change > 1.4 and a p-value < 0.05. In these two datasets, FEZF2 and AIRE1 were knocked out in mice mTECs. We employed several gene enrichment approaches, including Gorilla (Supplementary file 4), Gene cards (Supplementary file 5, Table 1), the Shiny server (Supplementary file 1, Table 2), and Metascape analysis (Fig. 5). Our results indicate that FEZF2 and AIRE1 control multiple genes in a complementary manner. It is noteworthy that the knockout of AIRE1 affected the expression of more than 1000 genes, whereas knocking out FEZF2 only affected 100 genes (Supplementary file 5). Importantly, when examining the expression patterns of FEZF2 homologs in invertebrates, we found that one FEZF2 homolog (e.g., erm) is expressed in the male and female germline of Drosophila Melanogaster, as well as in the embryos of both Drosophila and C. elegans.
Functional Ontologies of FEZF2 versus AIRE1 Downstream Targets. A FEZF2 downstream targets exhibit a high degree of interconnectedness and form a single cluster, whereas genes regulated by AIRE1 display greater diversity. B Examination of the expression of a FEZF2 homolog (e.g., erm) in Drosophila melanogaster via scRNA-seq and manual literature curation reveals erm expression in both male and female germlines, as well as in Drosophila embryos (darker color indicates higher expression). C Similar expression patterns are observed in C. elegans embryos
Discussion
FEZF2 is a member of the FEZ family and its closest known ancestor is a protein containing a zinc finger domain of the C2H2 type found in Metschnikowia aff. Pulcherrima (E-value < 9e-32) (Table 2). In mammals, FEZF2 still retains these zinc finger protein domains (Fig. 1B). Interestingly, zinc finger domains are known for their role as DNA-binding motifs in transcription factors, like TFIIIA. They have also been shown to have the ability to interact with DNA, RNA, and proteins (Negi et al. 2008). As a result of various evolutionary processes C2H2 zinc finger domains contain only four highly conserved residues, while the rest of the residues are highly variable (Albà 2017). C2H2 zinc finger domains are extremely diverse, with the ability to recognize the complete range of possible DNA triplets (Albà 2017). Interestingly, MCPIP1, which also contains a zinc finger, plays a role in the elimination of autoantibodies (Dobosz et al. 2021; Rakhra and Rakhra 2021). Taken together, these results confirm our hypothesis that FEZF2 can regulate the expression of a multitude of TRAs and extend it to suggest that zinc finger proteins could play a fundamental role in the process of eliminating harmful self-proteins in an immune context.
The nearest known ancestor for the reconstructed ancestral sequence of AIRE1 appears to be CHD4 LIKE (Orbicella Faveolata) (Fig. 3). CHD4 is a protein involved in chromatin remodeling and belongs to the CHD (Chromodomain-Helicase-DNA binding) family of proteins, which are ATP-dependent chromatin remodeling factors. CHD4 plays a vital role in the regulation of gene expression by modifying chromatin structure to either activate or repress gene transcription. Dysregulation of CHD4 function has been linked to diseases, such as cancer and developmental disorders. Importantly, it has been recently reported that Chd4 might be involved in regulating the thymus’s TRA expression, where it is responsible for organizing the promoter regions of Fezf2-dependent genes and contributing to the AIRE1-mediated induction of self-antigens via super-enhancers (Tomofuji et al. 2020; Benlaribi et al. 2022). These findings support the notion that AIRE1 may regulate transcription factors both directly through protein–protein interactions and indirectly through epigenetic mechanisms. Although these results contradict the hypothesis of a recent common evolutionary origin for the regulation of TRA expression through FEZF2 and AIRE1, nevertheless it indicates that the regulation of these two regulators might have been mediated by a common mechanism.
FEZF2 and AIRE1 have evolved through a finely tuned and controlled evolutionary process. The elimination of autoreactive T cells is a crucial requirement for the body's survival. Our study reveals a significant level of complementarity between FEZF2 and AIRE1 on various levels, including the evolutionary history of their downstream targets and the functional characteristics of these targets. Regarding the evolutionary history of downstream targets, we observed that the earliest homologs of FEZF2 diverged before the emergence of the earliest homologs of AIRE1 (Figs. 1A and 2). However, both AIRE1 and FEZF2 regulate genes that can be classified as ancient, existing in invertebrates. Examples of such genes include BHMT, F2, ITIH3, KCNJ5, LGALS7, MAOA, MYO15B, PLAG1, and ZP2 in the case of FEZF2, and IGF2 and SPT1 in the case of AIRE1 (Supplementary file 1). We also found multiple downstream targets for both pathways, with homologs emerging as late as in bony fish, such as C4BP and KRT2 (Fig. 2). When considering the functional characteristics of the downstream targets, we discovered that both pathways control genes with shared functional annotations, exemplified by genes, like ZP2 and ZP3 (Fig. 4, Supplementary file 2, and Supplementary file 4). However, it is worth noting that based on the analysis of knockout microarray studies (GSE69105 and GSE2585), AIRE1 appears to control the expression of approximately 1000 genes, while FEZF2 regulates around 100 genes (Supplementary file 5). Consequently, genes controlled by AIRE1 exhibit greater diversity (Fig. 4). Overall, our results seem to confirm the convergence of the functions of these two genes. Convergence evolution can be classified into two types: (i) parallel evolution and (ii) collateral evolution based on the existence of identical mutations taking part in independent lineages or the occurrence of a hybridization process between entities, respectively (Stern 2013). As AIRE1 and FEZF2, and possibly other genes, regulate the levels of TRA in mTECs, it is safe to assume that the evolution of AIRE1 and FEZF2 followed a parallel evolution scheme. The main reason behind the need for this particular approach seems to be minimizing collateral damage by reducing possible pleiotropic effects and maximizing adaptation.
Our findings suggest a potential connection between the functions of both FEZF2 and AIRE1 and the autophagy mechanism. Macroautophagy mediates endogenous MHC class II-loading in mTEC (Nedjic et al. 2008; Klein et al. 2014). Notably, an elevated level of AIRE expression in mTECs has been correlated with the enhancement of autophagy processes (Shevyrev et al. 2022). AIRE1’s ability to regulate the expression of various proteins in the gonads has also been well established (Adamson et al. 2004; Radhakrishnan et al. 2016). AIRE1 likely facilitated the auditioning of gonadal and perhaps early embryonic cells (Forsdyke 2020). The gonadal environment is rich in mechanisms that facilitate the selection of offspring. Processes related to ovarian follicle survival and death are under the regulation of autophagy (Yadav et al. 2018). Autophagy involves the modification of internal membranes and an increase in the number of vesicles that engulf both bulk cytoplasm and organelles. The initial stage of autophagy includes the encapsulation of cytoplasmic components within membrane sacs known as autophagosomes. Ovarian follicular atresia results from the degradation of autophagic vesicles by lysosomal enzymes. Therefore, it has been suggested that AIRE1’s role in the thymus may be an adaptation of earlier evolving mechanisms. The apparent conservation of FEZF2 function between vertebrates and invertebrates hints at a potential role for FEZF2 in regulating simpler mechanisms that may have existed in invertebrates (Buss et al. 1985). Our investigations into positive selection using PAML and non-homology-based functional prediction of FEZF2 homologs found in invertebrates indicate that FEZF2 might have the ability to function as a transcription factor (Table 2 and Supplementary 3). Furthermore, FEZF2 has been reported to be expressed in primordial germ cells, which are precursors to embryonic sperm and egg cells (Jean et al. 2015). A FEZF2 homolog is also expressed in the germline and embryos of Drosophila (Figs. 4B and C). Autophagy plays a fundamental role in the Drosophila germline and has been reported in organisms, like Hydra (Chera et al. 2009). Interestingly, FEZF2 contains motifs associated with autophagy regulation, such as DKFPHP, SYSELWKSSL, and SYSEL. These findings suggest the possibility that FEZF2 may contribute to an invertebrate auto-elimination mechanism through the regulation of genes involved in autophagy.
Our research reveals valuable insights into the negative selection of immune cells in lampreys. Lampreys exhibit unique immune cell phenotypes, including VRLA (akin to α/β T cells), VRLB (similar to B cells), and VRLC (resembling γ/δ T cells). These specialized immune cell types have evolved specific mechanisms for immune cell elimination. Notably, lampreys possess a thymoid structure that expresses VRL, indicating the presence of a mechanism to eliminate autoreactive immune cells (Bajoghli et al. 2011). Interestingly, while we found a homolog of FEZF2 (XP_032822733.1) in lampreys, we could not locate a homolog of AIRE1. Moreover, we identified the presence of LTBR (XP_032823234.1), a gene known to regulate FEZF2 expression in lampreys (Takaba et al. 2015). Additionally, we observed the expression of downstream target genes that are regulated by FEZF2 in lampreys. These genes include BHMT, CALCA, COL17A1, CRISP 1, CYP24A1, F2, FABP7, FABP9, GC, ITIH3, KRT10, KCNJ5, LGALS7, LYPD1, MAOA, MUC3, MYO15B, PLAGL1, PLD1, Resp18, and ZP2. In summary, our findings suggest that lampreys may have evolved a mechanism akin to negative selection, potentially mediated by FEZF2, to eliminate autoreactive immune cells.
Conclusion
Our comprehensive analysis has revealed a multifaceted view of the regulation of TRA expression. Despite the absence of a recent common evolutionary origin in the mechanisms governing TRA regulation by FEZF2 and AIRE1, we have observed their co-expression in various contexts beyond the thymus, particularly in the germline. Both pathways appear to contribute to the process of autophagy, an ancient mechanism documented in organisms like Hydra, Drosophila, and Spiralia. Interestingly, while the AIRE1 pathway emerged in bony fish, the FEZF2-mediated pathway displays remarkable conservation in invertebrates. This suggests that Fezf2 may have been one of the ancient genes responsible for facilitating self-elimination, possibly by regulating gene expression related to autophagy. AIRE1, on the other hand, appears to have evolved to support this function in more complex vertebrate gonads and immune systems.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Abbas A, Lichtman A, Pillai S (2018) Cellular and molecular immunology, 9th edn. Elsevier
Adamson KA, Pearce SHS, Lamb JR, Seckl JR, Howie SEM (2004) A comparative study of MRNA and protein expression of the autoimmune regulator gene (Aire) in embryonic and adult murine tissues. Journal of Pathology. https://doi.org/10.1002/path.1493
Akiyama T, Shimo Y, Yanai H, Qin J, Ohshima D, Maruyama Y, Asaumi Y et al (2008) The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. https://doi.org/10.1016/j.immuni.2008.06.015
Albà M (2017) Zinc-finger domains in metazoans: evolution gone wild. Genome Biol. https://doi.org/10.1186/s13059-017-1307-y
Bajoghli B, Guo P, Aghaallaei N, Hirano M, Strohmeier C, McCurley N, Bockman DE, Schorpp M, Cooper MD, Boehm T (2011) A Thymus candidate in lampreys. Nature. https://doi.org/10.1038/nature09655
Beck G, Habicht GS (1996) Immunity and the Invertebrates. Sci Am. https://doi.org/10.1038/scientificamerican1196-60
Benlaribi R, Gou Q, Takaba H (2022) Thymic self-antigen expression for immune tolerance and surveillance. Inflamm Regen. https://doi.org/10.1186/s41232-022-00211-z
Bhaumik S, Łazarczyk M, Kubick N, Klimovich P, Gurba A, Paszkiewicz J, Teodorowicz P et al (2023) Investigation of the molecular evolution of Treg suppression mechanisms indicates a convergent origin. Curr Issues Mol Biol 45(1):628–648. https://doi.org/10.3390/cimb45010042
Buss LW, Moore JL, Green DR (1985) Autoreactivity and self-tolerance in an invertebrate. Nature. https://doi.org/10.1038/313400a0
Chera S, Buzgariu W, Ghila L, Galliot B (2009) Autophagy in hydra: a response to starvation and stress in early animal evolution. Biochim Biophys Acta—Mol Cell Res. https://doi.org/10.1016/j.bbamcr.2009.03.010
Copley RR (2005) The EH1 motif in metazoan transcription factors. BMC Genom. https://doi.org/10.1186/1471-2164-6-169
Dobosz E, Lorenz G, Ribeiro A, Wurf V, Wadowska M, Kotlinowski J, Schmaderer C et al (2021) Murine myeloid cell MCPIP1 suppresses autoimmunity by regulating b-cell expansion and differentiation. DMM Dis Models Mech. https://doi.org/10.1242/DMM.047589
Dragin N, Bismuth J, Cizeron-Clairac G, Biferi MG, Berthault C, Serraf A, Nottin R et al (2016) Estrogen-mediated downregulation of AIRE influences sexual dimorphism in autoimmune diseases. J Clin Investig. https://doi.org/10.1172/JCI81894
Forsdyke DR (2020) When few survive to tell the tale: thymus and gonad as auditioning organs: historical overview. Theory Biosci. https://doi.org/10.1007/s12064-019-00306-1
Gu X, Velden KV (2002) DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics 18(3):500–501. https://doi.org/10.1093/bioinformatics/18.3.500
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. https://doi.org/10.1093/molbev/msj030
Jean C, Oliveira NMM, Intarapat S, Fuet A, Mazoyer C, De Almeida I, Trevers K et al (2015) Transcriptome analysis of chicken ES, blastodermal and germ cells reveals that Chick ES cells are equivalent to mouse ES cells rather than EpiSC. Stem Cell Res. https://doi.org/10.1016/j.scr.2014.11.005
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Klein L, Kyewski B, Allen PM, Hogquist KA (2014) Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol. https://doi.org/10.1038/nri3667
Kubick N, Brösamle D, Mickael M-E (2018) Molecular evolution and functional divergence of the IgLON family. Evol Bioinf 20(Jan):1–10. https://doi.org/10.1177/1176934318775081
Kubick N, Klimovich P, Bieńkowska I, Poznanski P, Łazarczyk M, Sacharczuk M, Mickael M-E (2021a) Investigation of evolutionary history and origin of the Tre1 family suggests a role in regulating hemocytes cells infiltration of the blood-brain barrier. InSects 12(10):882. https://doi.org/10.3390/insects12100882
Kubick N, Klimovich P, Flournoy PH, Bieńkowska I, Łazarczyk M, Sacharczuk M, Bhaumik S, Mickael M-E, Basu R (2021b) Interleukins and interleukin receptors evolutionary history and origin in relation to CD4+ T cell evolution. Genes 12(6):813. https://doi.org/10.3390/genes12060813
Kulmanov M, Khan MA, Hoehndorf R (2018) DeepGO: predicting protein functions from sequence and interactions using a deep ontology-aware classifier. Edited by Jonathan Wiren. Bioinformatics 34(4):660–668. https://doi.org/10.1093/bioinformatics/btx624
Kumar M, Gouw M, Michael S, Sámano-Sánchez H, Pancsa R, Glavina J, Diakogianni A et al (2019) ELM—the eukaryotic linear motif resource in 2020. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz1030
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P et al (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz268
Mickael ME, Rajput A, Steyn J, Wiemerslage L, Bürglin T (2016) An optimised phylogenetic method sheds more light on the main branching events of rhodopsin-like superfamily. Comp Biochem Physiol D: Genomics Proteomics 20(Dec):85–94. https://doi.org/10.1016/j.cbd.2016.08.005
Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L (2008) Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. https://doi.org/10.1038/nature07208
Negi S, Imanishi M, Matsumoto M, Sugiura Y (2008) New redesigned zinc-finger proteins: design strategy and its application. Chem Eur J. https://doi.org/10.1002/chem.200701320
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. https://doi.org/10.1093/molbev/msu300
Peterson P, Org T, Rebane A (2008) Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nat Rev Immunol 8(12):948–957. https://doi.org/10.1038/nri2450
Pommier Y, Sun Y, Huang SYN, Nitiss JL (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. https://doi.org/10.1038/nrm.2016.111
Radhakrishnan K, Bhagya KP, Tr AK, Devi AN, Sengottaiyan J, Kumar PG (2016) Autoimmune regulator (AIRE) is expressed in spermatogenic cells, and it altered the expression of several nucleic-acid-binding and cytoskeletal proteins in germ cell 1 spermatogonial (GC1-Spg) cells. Mol Cell Proteomics. https://doi.org/10.1074/mcp.M115.052951
Rakhra G, Rakhra G (2021) Zinc finger proteins: insights into the transcriptional and post transcriptional regulation of immune response. Mol Biol Rep 48(7):5735–5743. https://doi.org/10.1007/s11033-021-06556-x
Sano H, Renault AD, Lehmann R (2005) Control of lateral migration and germ cell elimination by the drosophila melanogaster lipid phosphate phosphatases Wunen and Wunen 2. J Cell Biol. https://doi.org/10.1083/jcb.200506038
Shevyrev D, Tereshchenko V, Kozlov V, Sennikov S (2022) Phylogeny, structure, functions, and role of AIRE in the formation of T-cell subsets. Cells. https://doi.org/10.3390/cells11020194
Shimizu T, Nakazawa M, Kani S, Bae YK, Shimizu T, Kageyama R, Hibi M (2010) Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. https://doi.org/10.1242/dev.047167
Stern DL (2013) The genetic causes of convergent evolution. Nat Rev Genet. https://doi.org/10.1038/nrg3483
St-Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C (2015) Differential features of AIRE-Induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J Immunol. https://doi.org/10.4049/jimmunol.1500558
Takaba H, Takayanagi H (2017) The mechanisms of T cell selection in the thymus. Trends Immunol. https://doi.org/10.1016/j.it.2017.07.010
Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, Kodama T, Takayanagi H (2015) Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell. https://doi.org/10.1016/j.cell.2015.10.013
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Tomofuji Y, Takaba H, Suzuki HI, Benlaribi R, Martinez CDP, Abe Y, Morishita Y et al (2020) Chd4 choreographs self-antigen expression for central immune tolerance. Nat Immunol. https://doi.org/10.1038/s41590-020-0717-2
Törönen P, Medlar A, Holm L (2018) PANNZER2: a rapid functional annotation web server. Nucleic Acids Res 46(W1):W84-88. https://doi.org/10.1093/nar/gky350
Wiemerslage L, Gohel PA, Maestri G, Hilmarsson TG, Mickael M, Fredriksson R, Williams MJ, Schiöth HB (2016) The drosophila ortholog of TMEM18 regulates insulin and glucagon-like signaling. J Endocrinol 229(3):233–243. https://doi.org/10.1530/JOE-16-0040
Xing Y, Hogquist KA (2012) T-cell tolerance: central and peripheral. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a006957
Yadav PK, Tiwari M, Gupta A, Sharma A, Prasad S, Pandey AN, Chaube SK (2018) Germ cell depletion from mammalian ovary: possible involvement of apoptosis and autophagy. J Biomed Sci. https://doi.org/10.1186/s12929-018-0438-0
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. https://doi.org/10.1093/molbev/msm088
Yoshida H, Bansal K, Schaefer U, Chapman T, Rioja I, Proekt I, Anderson MS et al (2015) Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc Natl Acad Sci 112(32):E4448–E4457. https://doi.org/10.1073/pnas.1512081112
Acknowledgements
We would like to acknowledge the efforts of Macrious Abraham and Meriam Joachim for their informative discussions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Handling editor: Robert Noble.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mickael, M., Łazarczyk, M., Kubick, N. et al. FEZF2 and AIRE1: An Evolutionary Trade-off in the Elimination of Auto-reactive T Cells in the Thymus. J Mol Evol 92, 72–86 (2024). https://doi.org/10.1007/s00239-024-10157-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-024-10157-0