Abstract
When a dispensable gene is duplicated (referred to the ancestral dispensability denoted by O+), genetic buffering and duplicate compensation together maintain the duplicate redundancy, whereas duplicate compensation is the only mechanism when an essential gene is duplicated (referred to the ancestral essentiality denoted by O−). To investigate these evolutionary scenarios of genetic robustness, I formulated a simple mixture model for analyzing duplicate pairs with one of the following states: double dispensable (DD), semi-dispensable (one dispensable one essential, DE), or double essential (EE). This model was applied to the yeast duplicate pairs from a whole-genome duplication (WGD) occurred about 100 million years ago (mya), and the mouse duplicate pairs from a WGD occurred about more than 500 mya. Both case studies revealed that the proportion of essentiality for those duplicates with ancestral essentiality [PE(O−)] was much higher than that for those with ancestral dispensability [PE(O+)]. While it was negligible in the yeast duplicate pairs, PE(O+) (about 20%) was shown statistically significant in the mouse duplicate pairs. These findings, together, support the hypothesis that both sub-functionalization and neo-functionalization may play some roles after gene duplication, though the former may be much faster than the later.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The role of functional compensation by duplicate genes has been examined in diverse organisms by comparing the proportion (PE) of essential genes in duplicates to PE in singletons (Wagner 2000; Gu et al. 2003; Conant and Wagner 2004; Hanada et al. 2009). Technically, a gene is called ‘essential’ if the single-gene deletion phenotype is severe or lethal, or ‘dispensable’ if its deletion phenotype is normal or nearly normal (Ihmels et al. 2007; Hsiao and Vitkup 2008; Su et al. 2014; Kabir et al. 2017; Cacheiro et al. 2020). One may see Rancati et al. (2018) for a comprehensive review of gene essentiality. Due to different gene-silence/knockout technologies that are feasible, the criteria to determine gene essentiality or dispensability are usually not comparable between species such as yeasts and mice. The concept of gene essentiality is, therefore, theoretical, depending on different experimental conditions; it has been used as the first-order proxy to study the evolutionary pattern of genetic robustness: how an organism is resilient against the occurrence of null mutations.
Intuitively, one may speculate that if duplicates play a significant role in functional compensation, the PE for duplicates should be significantly lower than that of singletons. In other words, duplicate genes have major contributions to the genetic robustness at the organismal level. While this is indeed the case in yeasts, worms, and plants (Gu 2003; Kamath et al. 2003; Qian et al. 2010; Hanada et al. 2011), no significant difference in PE was found between mouse single-copy and duplicate genes (Liang and Li 2007; Liao and Zhang 2007). A number of explanations were proposed (Li et al. 2010; Makino and McLysaght 2010; Vandersluis et al. 2010; Mendonça et al. 2011; Plata and Vitkup 2014; Zhang et al. 2015). For instance, Su and Gu (2008) noticed that the effect of sampling bias: recently duplicated genes, e.g., after the mammalian radiation, are severely underrepresented in the current mouse knockout database. Because most of the mouse gene knockouts were generated by individual laboratories for finding knockout phenotypes, recently duplicated genes may have been purposely avoided to minimize the experimental cost due to negative-phenotype results. In other words, the age distribution of duplicates in the data sample is upwardly biased, resulting in underestimation of the overall duplicate effect on the genetic robustness. One may see Su et al. (2014) for a substantial follow-up analysis. Some studies showed that functional and protein connectivity bias between essential and dispensable duplicate genes may be the cause (Liang and Li 2009; Makino et al. 2009).
Although the pattern of duplicate compensation is universal, including essential genes in cancer cell lines (de Kegel and Ryan 2019), the pattern of duplicate compensation is complex (Szklarczyk et al. 2008; Hahn 2009; Chen et al. 2012; Keane et al. 2014; Saito et al. 2014; Diss et al. 2017; Teufel et al. 2018; Láruson et al. 2020; Mallik and Tawfik 2020). When an essential gene is duplicated (termed ancestral essentiality), duplicate compensation is the only mechanism to keep two resulting copies dispensable. On the other hand, when a dispensable gene is duplicated (termed ancestral dispensability), the ancient genetic buffering and duplicate compensation together keep both duplicate copies dispensable. Note that almost all previous PE-related analyses in the literature did not distinguish between these two possibilities. Indeed, duplication of dispensable genes virtually results in no change of PE, except for being essential by neo-functionalization. By contrast, after sufficiently long time, duplication of essential genes would be ultimately back to essentiality with no change of PE, except for the long-term functional compensation.
This paper will address this issue as follows. We first develop a statistical model to analyze duplicate pairs with three possible states: double dispensable (DD), semi-dispensable (one dispensable one essential, DE) or double essential (EE). Under some biologically reasonable assumptions, a probabilistic model is then developed to estimate the proportion of essential genes duplicated from essential genes or that from dispensable genes, respectively. Exemplified by the yeast and mouse duplicate pairs from their own whole-genome duplications (WGD), respectively, some new insights about the evolutionary pattern of genetic robustness after gene duplication are discussed.
Results
Genetic Robustness Between Duplicate Genes
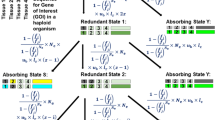
A gene is called ‘essential’ (denoted by d−) if the single-gene deletion phenotype is severe or lethal, or ‘dispensable’ (denoted by d+) if its deletion phenotype is normal or nearly normal (Ihmels et al. 2007; Hsiao and Vitkup 2008; Su et al. 2014; Kabir et al. 2017; Cacheiro et al. 2020). Consider two paralogous genes (A and B) duplicated from a common ancestor (O) t time units ago. There are four combined states, denoted by (dA, dB), representing double dispensable (d+, d+), semi-dispensable (d+, d−) or (d−, d+), and double essential (d−, d−), respectively.
We are interested in the derivation of Qt(dA, dB), the probability of any joint states (dA, dB) at time t since the duplication. To this end, one should distinguish between the duplication of an essential gene (ancestral essentiality, denoted by O−) and the duplication of a dispensable gene (ancestral dispensability, denoted by O+). Let Qt(dA, dB|O−) be the probability of being (dA, dB) after t time units since gene duplication, conditional of the ancestral essentiality (O−), and Qt(dA, dB|O+) be the probability conditional of the ancestral dispensability (O+). Since the ancestral state (dispensable or essential) for a duplicate pair is usually unknown, a mixture model is then implemented: let R0 = P(O+) be the probability of a gene pair duplicated from a dispensable gene, and 1 − R0 = P(O−) be that from an essential gene (Liang and Li 2007; Liao and Zhang 2007; Su and Gu 2008). Together, one can write
where (dA, dB) = (d+, d+), (d+, d−), (d−, d+) or (d−, d−), respectively (Table 1).
It should be noticed that, the process of non-functionalization of one duplicate copy was not conceptualized in the model, which is the most common fate of duplicated genes (Gu and Nei 1999). This treatment is rational under the assumption that the rate of non-functionalization was the same between dispensable and essential genes before duplication. Otherwise, Eq. (1) would be affected. One may see Stark et al. (2017) for a detailed discussion.
Duplication of Essential Gene: The Sub-functionalization
When an essential gene was duplicated, the process of sub-functionalization, probably driven by rapid regulatory motif divergence (Zhang et al. 2004), or trans-TF evolution (Zhou et al. 2014), or TATA Box changes (Zou et al. 2011) or histone modification changes (Zou et al. 2012), has been thought to be the major evolutionary mechanism for duplicate preservation (Force et al. 1999; Stoltzfus 1999; Prince and Pickett 2002; Innan and Kondrashov 2010; Stark et al. 2017). It would be worth mentioning that the sub-functionalization prior to duplication model has been described by Des Marais and Rausher (2008). As a result, both duplicate copies can be preserved without invoking positive selection. Suppose a duplicate pair has m independent functional components, each of which is either ‘active’ (denoted by ‘1’) or ‘inactive’ (denoted by ‘0’). Let U11 be the probability of a component being active in both genes; U01 (or U10) is that of being inactive in gene A but active in gene B (or active in A but inactive in B); and U00 is the probability of a component being inactive in both genes. Without loss of generality, it is assumed that U01 = U10. According to the not-all-inactive constraint, i.e., each component is functionally active at least in one duplicate copy, we claim U00 = 0, leading to U11 = 1-2U and U10 = U01 = U, respectively. That is, with a probability of 2U, a functional component is active in one duplicate but inactive in another one, and with a probability of 1-2U, a component is active in both duplicates.
If these functional components of a gene are statistically independent and identical, Qt(dA, dB|O−) can be derived in terms of the component parameter (U) and the number (m) of functional components, that is,
The rationale of Eq. (2) is follows. Under the m-component model (m > 1), two duplicate copies remain both dispensable only when each component is active in both duplicates (with a probability of 1 − 2U), which leads to the derivation of Qt(d+, d+|O−) directly. Next we consider the (marginal) probability of dispensability (d+) conditional of the ancestral essentiality (O−), denoted by Qt(d+|O−). It appears that Qt(d+|O−) = (1 − U)m because the probability of a component for being active in one duplicate is given by (1 − U). Since Qt(d+|O−) = Qt(d+, d+|O−) + Qt(d+, d−|O−), it is straightforward to obtain the second and third equations of Eq. (2). The last equation of Eq. (2) is derived by the sum of probabilities to be one.
Equation (2) implies a gradual process of state transition. The starting states are apparently double dispensable (d+, d+), most of which would be transformed to semi-dispensable (d+, d−) and further to double essential (d−, d−). That said, double essentiality can only be achieved after the occurrence of semi-dispensability after the gene duplication.
Duplication of Dispensable Gene: The Rare Neo-functionalization
When a dispensable gene was duplicated, gene dispensability can be maintained through ancient genetic buffering and/or duplicate compensation (Prince and Pickett 2002; Innan and Kondrashov 2010; Stark et al. 2017). As a result, sub-functionalization becomes an ineffective approach for the retention of duplicate gene, because the process of complementation between functional components (Force et al. 1999) is difficult to achieve. To explain this argument, one may consider a simple case: two duplicates A and B have two sub-functions (F1 and F2). After a complete sub-functionalization, duplicate A has functional F1 and nonfunctional F2, whereas duplicate B has nonfunctional F1 and functional F2. Since both sub-functions are required at the organismal level, duplicates A and B obviously become essential in the case of no genetic buffering. However, if duplicates A and B are from the duplication of a dispensable genes, the status of dispensability would not be altered.
While the neo-functionalization has been suggested for the duplicate preservation in the case of genetic buffering (Chen et al. 2010; Vankuren and Long 2018; Lee and Szymanski 2021), it is unlikely that both copes acquire new functions simultaneously. In this sense, one can assume that
This assumption holds well except for very ancient duplicates that may acquire new functions in the later stage. Although the link between molecular function and selection has not been explicitly formulated, one may reasonably argue that the retention of dispensable genes through neo-functionalization may be mainly driven by a positive selection.
Analysis of Genetic Robustness Model Between Duplicates
Model Formulation and Estimation
Together with Eqs. (2) and (3), the model of genetic robustness between duplicates formulated by Eq. (1) can be further specified as follows:
where Q(d−|O+) is the probability of an O+-duplicate being essential (d−); under Eq. (3), one can show Q(d−|O+) = Q(d−, d−|O+) + Q(d−, d+|O+) = Q(d−, d+|O+).
There are four unknown parameters, R0, U, m, and Q(d−|O+) in two independent equations (Table 1). A practically feasible approach is then implemented to solve this difficulty, as shown below.
-
(i)
Suppose we have a set (N) of duplicate pairs; all 2N genes have single-gene deletion phenotypes (dispensable or essential). Three types of duplicate pairs are considered, that is, DD for (d+, d+), DE for (d+, d−) or (d−, d+), and EE for (d−, d−), and their frequencies are denoted by fDD, fDE, and fEE, respectively.
-
(ii)
R0, the (prior) probability of a gene being dispensable before gene duplication can be replaced by the proportion of single-copy dispensable genes in the current genome as a proxy, under the assumption that R0 remained a rough constant during the long-term evolution (Su and Gu 2008).
-
(iii)
The parameter U can be estimated by replacing Qt(d−, d−) in the last equation of Eq. (4) by fEE, that is,
$$1 - 2\left( {1 - \widehat{U}} \right)^{m} + \left( {1 - 2\widehat{U}} \right)^{m} = \frac{{f_{EE} }}{{1 - R_{0} }}$$(5)where m, the number of functional components, is treated as a known integer, i.e., m=2, 3,….
-
(iv)
The proportion of essential O−-duplicates, i.e., those duplicated from essential genes, is given by Q(d−|O−) = Q(d−, d−|O−) + Q(d−, d+|O−). When U is estimated by Eq. (5) (for any fixed m), according to Eq. (2), one can estimate Q(d−|O−) by
$$\widehat{Q}\left( {d^{ - } \left| {O^{ - } } \right.} \right) = 1 - \left( {1 - \widehat{U}} \right)^{m}$$(6) -
(v)
After replacing Qt(d+, d+) in the first equation of Eq. (4) by fDD, one can show that the proportion of essential O+-duplicates is estimated by
$$\widehat{Q}\left( {d^{ - } \left| {O^{ + } } \right.} \right) = \frac{1}{2} - \frac{{f_{DD} - \left( {1 - R_{0} } \right)\left( {1 - 2\widehat{U}} \right)^{m} }}{{2R_{0} }}$$(7)
In short, from the observed frequencies fDD, fDE, and fEE with two degrees of freedom, we attempt to estimate two parameters Q(d−|O−) and Q(d−|O+) by Eqs.(5–7). To this end, we use the proportion of single-copy dispensable genes in the current genome as a proxy of R0, and m as a constant that may only affect our estimation marginally (see below).
Statistical Evaluation
The statistical property of two estimates, Q(d−|O−) and Q(d−|O+), can be evaluated by two approaches. First, their large-sample variances can be obtained by the delta-method under a multinomial model of fDD, fDE, and fEE. The analytical formulas can be approximately obtained though the algebra was tedious (not shown). Second, a bootstrapping approach was implemented to empirically determine the sampling variance, as well as the confidence internals of these estimates.
Effect of the Number of Functional Components (m)
By computer simulations, we examined how the number (m) of functional components may affect our analysis. Note that the model of sub-functionalization requires at least two functional components. Hughes and Liberles (2007) suggested that between m = 2 and m = 12 regulatory regions would be biologically realistic. By extensive simulation analysis, Stark et al. (2017) argued that it was unlikely that a gene would have in excess of m = 20 functional components. Our main results are follows: (i) the estimate of Q(d−|O−) tends to decrease slightly when m is increased from 2 to 5 (about 20%), whereas that of Q(d−|O+) tends to increase slightly; (ii) in both cases little difference was observed for m = 5 or more; and (iii) all estimates are virtually the same from m = 7 to m = ∞. In short, it seems that the effect of variable m is negligible as long as it is reasonably large, say, m = 5 or more.
The number of sub-functions (m) involved in the process of sub-functionalization after gene duplication actually represents a subset of sub-functions that are essential for the fitness of the organism. There are likely to have more non-essential sub-functions, for instance, some minor expression patterns in a tissue. In this case, it remains unclear whether a large m seems biologically plausible.
Prediction of Joint Conditional Probabilities
In practice, it is desirable to know two types of conditional probabilities, Qt(dA, dB|O−) and Qt(dA, dB|O+), based on the observed frequencies fDD, fDE, and fEE. According to Eq. (2), it is straightforward to calculate the conditional probabilities of (dA, dB) after duplication of an essential gene (O−) as follows:
where \(\widehat{U}\) is the positive solution of Eq. (5). Next, one can predict Q(d+, d+|O+) by equating Qt(d+, d+) with fDD in Eq. (1) in the case of dA = d+ and dB = d+, and replacing Q(d+, d+|O−) by its prediction given by the first equation of Eq. (8). They are, respectively, given by
As indicated before, for a set of duplicate pairs with observed fDD, fDE, and fEE, there are only two degrees of freedoms. Hence, the statistical procedure described above treated R0 and m as known constants and then estimated U and Q(d−|O+). In this sense, Eqs. (8) and (9) are not statistically well justified to be treated as ‘estimates’; instead, they should be regarded as predicted values.
Case Study: Duplicate Pairs from the Whole-Genome Duplication (WGD) in Yeast or Mouse
Data Availability
Due to different gene-silence/knockout technologies, the criteria to determine gene essentiality or dispensability are usually not comparable between species such as yeasts and mice. Because fitness phenotypes after single-gene deletions were identified under the experimental conditions, the population size under the natural condition would not affect the outcome.
In total, 325 yeast duplicate pairs were collected, which were from the yeast WGD (whole-genome duplication) about 100 million years ago (Kim and Yi 2006; Guan et al. 2007; Musso et al. 2008). According to the common practice in the yeast single-gene deletion genomics, the mean fitness of single-gene deletion for any yeast gene is measured by the growth rate of the strain with a single gene deleted relative to the average growth rate of wild strains under five growth media. Qualitatively, it can be further grouped into lethal, the strong effect, the moderate effect, and the very weak effect (Gu et al. 2003). From the evolutionary view, a yeast gene is then classified as d+ if it belongs to the very weak-effect group, or d− otherwise. Under this classification, the proportion of dispensable single-copy genes (0.605) from Gu et al. (2003) is used as a proxy of R0. One may wonder how the analysis would be affected by the binned fitness data. Actually, the fitness histogram showed a U-like pattern where the moderate-effect group is the least. In other words, our classification of yeast essential or dispensable gene should be robust against the bin cutoff.
The second dataset includes 217 mouse duplicate pairs from the WGD occurred (Makino and McLysaght 2010), about more than 500 million years ago (in the early stage of vertebrates)(Wang and Gu 2000). Each pair was assigned by the mouse knockout phenotypes as follows (Su and Gu 2008). First, mouse phenotype and genotype association file (MGI_PhenoGenoMP.rpt) were downloaded from Mouse Genome Informatics (ftp://ftp.informatics.jax.org). Here, an essential gene was defined as a gene of which knockout phenotype is annotated as lethality (including embryonic, prenatal, and postnatal lethality) or infertility. We excluded all the phenotypic annotations due to multiple gene knockout experiments, and only used those of null mutation homozygotes by target deletion or gene-trap technologies.
Analysis
Our analysis is focused on three variables: (i) PE is the observed proportion of essential duplicates; (ii) PE(O−) is the expected proportion of essential O−-duplicates, i.e., those duplicated from essential genes, as estimated by \(\widehat{Q}\)(d−|O−) in Eq. (6); and (iii) PE(O+) is the expected proportion of essential O+-duplicates, i.e., those duplicated from dispensable genes, as estimated by \(\widehat{Q}\)(d−|O+) in Eq. (7). Their relationship is simply given by
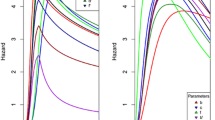
(see Table 1). The frequencies of duplicate pairs with DD (double dispensable), DE (dispensable essential) and EE (double essential) are presented in Figs. 1A (yeast) and 2A (mouse), respectively. While there is no empirical information about the number of functional components (m) for mouse and yeast genes, the robustness of the following analysis against various ms is important. Consistent with the simulation result, our analysis was generally not affected by m (the number of functional components); overall it revealed little difference among those cases of m = 3 or more. Our analysis of yeast WGD duplicate pairs is shown in Fig. 1B, and that of mouse in Fig. 2B (m = 6). Roughly speaking, yeast WGD pairs represent the case of recent WGD event, whereas mouse WGD pairs represent the ancient one.
Analysis of yeast 325 WGD pairs. A Frequencies of duplicate pairs with DD (double dispensable), DE (dispensable essential), and EE (double essential) are presented. B The proportion of essential duplicates (PE), the estimated PE in O−-duplicates (duplication of essential genes), PE(O−), and the estimated PE in O+-duplicates (duplication of dispensable genes), PE(O+), are presented. In the analysis, the number of functional components is set to be m = 6. For comparison, the proportion of essential genes in single-copy genes (1 − R0) is also presented
Analysis of mouse 217 WGD pairs. A Frequencies of duplicate pairs with DD (double dispensable), DE (dispensable essential), and EE (double essential) are presented. B The proportion of essential duplicates (PE), the estimated PE in O−-duplicates (duplication of essential genes), PE(O−), and the estimated PE in O+-duplicates (duplication of dispensable genes), PE(O+), are presented. In the analysis, the number of functional components is set to be m = 6. For comparison, the proportion of essential genes in single-copy genes (1 − R0) is also presented
In the case of yeast WGD pairs, the proportion of essential duplicates (PE = 10.3%) is significantly larger than zero (p-value < 10–6), yet it is much lower than that of single-copy yeast genes (PE,sin = 0.395). The new analysis showed that the PE in O−-duplicates (duplication of essential genes) was PE(O−) = 21.2%, significantly greater than zero (p < 0.001), whereas PE in O+-duplicates (duplication of dispensable genes) is PE(O+) = 3.0% that was not significant (p > 0.05). As expected, fEE (the proportion of double-essential duplicate pairs) is so small that the estimation of U is subject to a large sampling variance. At any rate, one should be cautious to draw any conclusion based on a non-significant result. Nevertheless, it appears that the increase of PE in the yeast WGD was mainly due to O−-duplicates, those duplicated from essential genes. Since the duplication time is the same for all duplicate pairs, one may predict that the rate of essentiality in O−-duplicates through sub-functionalization is about as sevenfold (21.2/3.0) as that in O+-duplicates through neo-functionalization.
In the case of mouse WGD pairs representing an ancient WGD, we observed PE = 62.2%, virtually the same as PE in single-copy genes (Liang and Li 2007; Liao and Zhang 2007; Su and Gu 2008). As expected, the estimate of PE(O−) = 86.0% indicated that the majority of O−-duplicates in mouse WGD pairs, i.e., those duplicated from essential genes, may have become essential. Interestingly, the estimate of PE(O+) = 21.2% was significantly greater than zero (p < 0.001). Indeed, a nontrivial portion of O+-duplicates in mice, i.e., those duplicated from dispensable genes, may be essential, which were subjected to neo-functionalization after the gene duplication (Chen et al. 2010; Vankuren and Long 2018; Lee and Szymanski 2021).
We observed that, strikingly, PE(O−) > PE(O+) significantly in both WGD duplicate pairs (p < 0.005), which can be tentatively interpreted as follows: after the occurrence of WGD, the proportion of essential duplicates (PE) increases with time t through two distinct evolutionary routes: a fast process of essentiality in O−-duplicates through sub-functionalization, and a slow process of essentiality in O+-duplicates through neo-functionalization; the difference is about fourfold (86.2/21.2). Finally, Fig. 3 shows the predicted conditional probabilities of yeast duplicate pairs: indeed, only marginal differences appeared when m = 2, and all estimates were virtually the same between m = 5 and m = ∞. It was, therefore, concluded that the effect of variable m is usually negligible.
Discussion
In this paper, we described a mixture model to study the pattern of genetic robustness after gene duplication, which made a distinction between two evolutionary scenarios: duplication of essential genes and duplication of dispensable genes. Case studies of yeast (Gu et al. 2003) and mouse (Makino and McLysaght 2010) WGD duplicate pairs provided some new insights about the evolution of genetic robustness, which can be further validated when more genome-wide gene deletion data are available in different organisms. While the mouse WGD is older in years than the yeast WGD, one should be cautious because yeasts have a much shorter generation time than mice. Hence, the evolutionary stage of yeast WGD may not necessarily be much younger than the mouse WGD. Further study is required to test whether the yeast WGD is ‘nearly resolved’ in the sense that further years of evolution will not allow further duplicate losses. Indeed, as both PE(O−) and PE(O+) are apparently time dependent, an interesting problem is to what extent PE(O−) and PE(O+) of yeast or mice WGD were close to the equilibrium. We shall address this issue when fitness phenotypes are available for the groups of duplicate pairs with different evolutionary ages.
It has been fully acknowledged that there are many factors at different levels that may affect the essentiality–dispensability evolution between duplicates. More explicit discussions are helpful although most of them cannot be embedded in the current model. For instance, it has been shown that highly pleiotropic genes evolves slowly (Gu 2007; Su et al. 2010; Zeng and Gu 2010).This so-called gene pleiotropy theory of molecular evolution (Su et al. 2010; Gu 2014) predicts that a highly pleiotropic gene tends to be essential. Another example is the tissue-driven hypothesis (Gu and Su 2007; Su et al. 2007), claiming that functionality of tissues in which the gene normally expresses may shape the evolutionary rate as well as the essentiality. How these genomic factors influence the functional divergence after gene duplication remains further study.
The new model for the evolution of genetic robustness is certainly oversimplified. It has been known that essentiality and dispensability are relative categories for genes. In yeast, Hillenmeyer et al. (2008) found that 97% of gene deletions exhibited a measurable growth phenotype, suggesting that nearly all genes are essential for optimal growth in at least one condition. Hence, the model of genetic robustness actually depends on a cutoff of fitness effect under a given environmental condition (Nowak et al. 1997; Visser et al. 2003; Flatt 2005). Indeed, dispensable genes in our case studies (yeast or mouse) should be interpreted as ‘nearly dispensable’ under ideal experimental conditions, whereas essential genes are likely to be truly ‘essential’ under the wild condition. One may speculate that natural selection may act on those dispensable genes that are only ‘essential’ under certain conditions, as illustrated by Hillenmeyer et al. (2008).
When an essential gene was duplicated, the current model assumed that two duplicate copies evolved under sub-functionalization, neglecting other possibilities such as neo-functionalization. Each functional component is assumed to undergo sub-functionalization independently, which is not biologically realistic (Szklarczyk et al. 2008; Hahn 2009; Chen et al. 2012; Keane et al. 2014; Saito et al. 2014; Diss et al. 2017; Teufel et al. 2018; Láruson et al. 2020; Mallik and Tawfik 2020). Meanwhile, after the duplication of a dispensable gene, interactions between ancestral genetic buffering, duplicate compensation, and neo-functionalization remain largely unknown. In addition, some attributes of genetic mechanisms have not been taken into accounts, such as the effect of dosage balance, or the later-stage functional divergence (Prince and Pickett 2002; Innan and Kondrashov 2010). For instance, a high dosage requirement for a duplicated gene pair could result in both being essential (since loss of expression from either copy would bring the expression below the required threshold). In particular, for WGD-produced duplicates, some evidence showed that much of the duplicate preservation is due to the need of dosage balance (Birchler and Veitia 2012). Indeed, duplicate genes that are subject to dosage selection and constraint tends to be essential, raising an important question how much the estimated neo-functionalization in mouse WGD pairs is actually due to the dosage constraints. Our future study will focus on the development of a more realistic model of gene duplication.
A key assumption in our analyses is Eq. (3), that is, after duplication of a dispensable gene (O+), the chance for both duplicate copies to be essential is negligible. While it is biologically intuitive, it may cause some biases, especially for some very ancient duplicate pairs. We conducted a simulation study to examine this effect by letting Q(d−, d+|O+) = q, where q is a small positive value. Our preliminary result showed that the estimation bias was usually marginal, except for an extremely long evolutionary span after gene duplication (not shown). In addition, the current model does not consider the neo-functionalization after the duplication of an essential gene if the acquired new function would not impair the current functions. Nevertheless, the neo-functionalization after sub-functionalization, or sub-neo-functionalization for short, would not change the status of essentiality.
Dean et al. (2008) demonstrated that yeast-duplicated genes can maintain substantial redundancy for extensive periods of time following duplication (over 100 million years). In another study, Vavouri et al. (2008) showed genetic redundancy was not just a transient consequence of gene duplication but is often an evolutionary stable state; that is why some genes have retained redundant functions since the divergence of the animal, plant, and fungi kingdoms (Gu 1997). Although the current study supported the basic idea provided by Vavouri et al. (2008) and Dean et al. (2008), a more careful analysis is required to clarify the difference in the evolutionary time scale.
For the purpose of biomedical science, a number of computational and experimental approaches were proposed to define human essential genes (Georgi et al. 2013; Wang et al. 2015; Chen et al. 2017; Fuller et al. 2019). One may also use mouse databases (Brown et al. 2018), for example, the international mouse phenotyping consortium (IMPC) (Muñoz-fuentes et al. 2018), or the mouse genome database (Smith et al. 2018), to predict the essentiality of human orthologous genes; see Brown et al. (2018) for a comprehensive review. It is, therefore, intriguing to ask whether our conclusion can be applied to the relationship between gene essentiality and human diseases (Fuller et al. 2019; Pengelly et al. 2019).
As the final comment, we notice that the effect of dominance has been neglected in this study, because the model implies that the genetic model is additive. Whether the gene is dominant or recessive will certainly contribute to the evolution of essentiality after the gene duplication. To take the dominance into account, we have to develop a population genetic model of gene duplication, which has been lacking (). We shall address the issue of whether the essential/dispensable genes are homozygous or heterozygous theoretically and experimentally.
Data Availability
In the current study, most work is theoretical, which did not include any original dataset. All datasets involved in the case analysis has been well cited in the text.
References
Birchler JA, Veitia RA (2012) Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA 109:14746–14753
Brown SDM, Holmes CC, Mallon AM et al (2018) High-throughput mouse phenomics for characterizing mammalian gene function. Nat Rev Genet 19:357–370
Cacheiro P, Muñoz-Fuentes V, Murray SA et al (2020) Human and mouse essentiality screens as a resource for disease gene discovery. Nat Commun. https://doi.org/10.1038/s41467-020-14284-2
Chen S, Zhang YE, Long M (2010) New genes in Drosophila quickly become essential. Science. https://doi.org/10.1126/science.1196380
Chen WH, Trachana K, Lercher MJ, Bork P (2012) Younger genes are less likely to be essential than older genes, and duplicates are less likely to be essential than singletons of the same age. Mol Biol Evol. https://doi.org/10.1093/molbev/mss014
Chen WH, Lu G, Chen X et al (2017) OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw1013
Conant GC, Wagner A (2004) Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc R Soc B Biol Sci. https://doi.org/10.1098/rspb.2003.2560
de Kegel B, Ryan CJ (2019) Paralog buffering contributes to the variable essentiality of genes in cancer cell lines. PLoS Genet. https://doi.org/10.1371/journal.pgen.1008466
Dean EJ, Davis JC, Davis RW, Petrov DA (2008) Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. https://doi.org/10.1371/journal.pgen.1000113
Des Marais DL, Rausher MD (2008) Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. https://doi.org/10.1038/nature07092
Diss G, Gagnon-Arsenault I, Dion-Coté AM et al (2017) Gene duplication can impart fragility, not robustness, in the yeast protein interaction network. Science. https://doi.org/10.1126/science.aai7685
Flatt T (2005) The evolutionary genetics of canalization. Q Rev Biol 80:287–316
Force A, Lynch M, Pickett FB et al (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics. https://doi.org/10.1093/genetics/151.4.1531
Fuller ZL, Berg JJ, Mostafavi H et al (2019) Measuring intolerance to mutation in human genetics. Nat Genet 51:772–776
Georgi B, Voight BF, Bućan M (2013) From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. https://doi.org/10.1371/journal.pgen.1003484
Gu X (1997) The age of the common ancestor of eukaryotes and prokaryotes: statistical inferences. Mol Biol Evol. https://doi.org/10.1093/oxfordjournals.molbev.a025827
Gu X (2003) Functional divergence in protein (family) sequence evolution. Genetica. https://doi.org/10.1023/A:1024197424306
Gu X (2007) Evolutionary framework for protein sequence evolution and gene pleiotropy. Genetics. https://doi.org/10.1534/genetics.106.066530
Gu X (2014) Pleiotropy can be effectively estimated without counting phenotypes through the rank of a genotype-phenotype map. Genetics. https://doi.org/10.1534/genetics.114.164673
Gu X, Nei M (1999) Locus specificity of polymorphic alleles and evolution by a birth-and death process in mammalian MHC genes. Mol Biol Evol. https://doi.org/10.1093/oxfordjournals.molbev.a026097
Gu X, Su Z (2007) Tissue-driven hypothesis of genomic evolution and sequence-expression correlations. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.0610797104
Gu Z, Steinmetz LM, Gu X et al (2003) Role of duplicate genes in genetic robustness against null mutations. Nature. https://doi.org/10.1038/nature01198
Guan Y, Dunham MJ, Troyanskaya OG (2007) Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. https://doi.org/10.1534/genetics.106.064329
Hahn MW (2009) Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered 100:604–617
Hanada K, Kuromori T, Myouga F et al (2009) Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis. Genome Biol Evol. https://doi.org/10.1093/gbe/evp043
Hanada K, Sawada Y, Kuromori T et al (2011) Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol Biol Evol 28:377–382. https://doi.org/10.1093/molbev/msq204
Hillenmeyer ME, Fung E, Wildenhain J et al (2008) The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. https://doi.org/10.1126/science.1150021
Hsiao T-L, Vitkup D (2008) Role of duplicate genes in robustness against deleterious human mutations. PLoS Genet 4:e1000014. https://doi.org/10.1371/journal.pgen.1000014
Hughes T, Liberles DA (2007) The pattern of evolution of smaller-scale gene duplicates in mammalian genomes is more consistent with neo- than subfunctionalisation. J Mol Evol. https://doi.org/10.1007/s00239-007-9041-9
Ihmels J, Collins SR, Schuldiner M et al (2007) Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol. https://doi.org/10.1038/msb4100127
Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11:97–108
Kabir M, Barradas A, Tzotzos GT et al (2017) Properties of genes essential for mouse development. PLoS ONE. https://doi.org/10.1371/journal.pone.0178273
Kamath RS, Fraser AG, Dong Y et al (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. https://doi.org/10.1038/nature01278
Keane OM, Toft C, Carretero-Paulet L et al (2014) Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae. Genome Res. https://doi.org/10.1101/gr.176792.114
Kim SH, Yi SV (2006) Correlated asymmetry of sequence and functional divergence between duplicate proteins of Saccharomyces cerevisiae. Mol Biol Evol. https://doi.org/10.1093/molbev/msj115
Láruson ÁJ, Yeaman S, Lotterhos KE (2020) The importance of genetic redundancy in evolution. Trends Ecol Evol 35:809–822
Lee Y, Szymanski DB (2021) Multimerization variants as potential drivers of neofunctionalization. Sci Adv. https://doi.org/10.1126/sciadv.abf0984
Li J, Yuan Z, Zhang Z (2010) The cellular robustness by genetic redundancy in budding yeast. PLoS Genet. https://doi.org/10.1371/journal.pgen.1001187
Liang H, Li WH (2007) Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet 23:375–378
Liang H, Li WH (2009) Functional compensation by duplicated genes in mouse. Trends Genet. https://doi.org/10.1016/j.tig.2009.08.001
Liao BY, Zhang J (2007) Mouse duplicate genes are as essential as singletons. Trends Genet 23:378–381
Makino T, McLysaght A (2010) Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.0914697107
Makino T, Hokamp K, McLysaght A (2009) The complex relationship of gene duplication and essentiality. Trends Genet 25:152–155
Mallik S, Tawfik DS (2020) Determining the interaction status and evolutionary fate of duplicated homomeric proteins. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1008145
Mendonça AG, Alves RJ, Pereira-Leal JB (2011) Loss of genetic redundancy in reductive genome evolution. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1001082
Muñoz-fuentes V, Cacheiro P, Meehan TF et al (2018) The International Mouse Phenotyping Consortium (IMPC): a functional catalogue of the mammalian genome that informs conservation. Conserv Genet. https://doi.org/10.1007/s10592-018-1072-9
Musso G, Costanzo M, Huangfu MQ et al (2008) The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res. https://doi.org/10.1101/gr.076174.108
Nowak MA, Boerlijst MC, Cooke J, Smith JM (1997) Evolution of genetic redundancy. Nature. https://doi.org/10.1038/40618
Pengelly RJ, Vergara-Lope A, Alyousfi D et al (2019) Understanding the disease genome: gene essentiality and the interplay of selection, recombination and mutation. Brief Bioinform. https://doi.org/10.1093/bib/bbx110
Plata G, Vitkup D (2014) Genetic robustness and functional evolution of gene duplicates. Nucleic Acids Res. https://doi.org/10.1093/nar/gkt1200
Prince VE, Pickett FB (2002) Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet 3:827–837
Qian W, Liao BY, Chang AYF, Zhang J (2010) Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. https://doi.org/10.1016/j.tig.2010.07.002
Rancati G, Moffat J, Typas A, Pavelka N (2018) Emerging and evolving concepts in gene essentiality. Nat Rev Genet 19:34–49
Saito N, Ishihara S, Kaneko K (2014) Evolution of genetic redundancy: the relevance of complexity in genotype-phenotype mapping. New J Phys. https://doi.org/10.1088/1367-2630/16/6/063013
Smith CL, Blake JA, Kadin JA et al (2018) Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx1006
Stark TL, Liberles DA, Holland BR, O’Reilly MM (2017) Analysis of a mechanistic Markov model for gene duplicates evolving under subfunctionalization. BMC Evol Biol. https://doi.org/10.1186/s12862-016-0848-0
Stoltzfus A (1999) On the possibility of constructive neutral evolution. J Mol Evol. https://doi.org/10.1007/PL00006540
Su Z, Gu X (2008) Predicting the proportion of essential genes in mouse duplicates based on biased mouse knockout genes. J Mol Evol. https://doi.org/10.1007/s00239-008-9170-9
Su Z, Huang Y, Gu X (2007) Tissue-driven hypothesis with Gene Ontology (GO) analysis. Ann Biomed Eng. https://doi.org/10.1007/s10439-007-9269-y
Su Z, Zeng Y, Gu X (2010) A preliminary analysis of gene pleiotropy estimated from protein sequences. J Exp Zool B Mol Dev Evol. https://doi.org/10.1002/jez.b.21315
Su Z, Wang J, Gu X (2014) Effect of duplicate genes on mouse genetic robustness: an update. Biomed Res Int. https://doi.org/10.1155/2014/758672
Szklarczyk R, Huynen MA, Snel B (2008) Complex fate of paralogs. BMC Evol Biol. https://doi.org/10.1186/1471-2148-8-337
Teufel AI, Johnson MM, Laurent JM et al (2018) Withdrawn as duplicate: the many nuanced evolutionary consequences of duplicated genes. Mol Biol Evol 35:e1. https://doi.org/10.1093/molbev/msy216
Vandersluis B, Bellay J, Musso G et al (2010) Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol Syst Biol. https://doi.org/10.1038/msb.2010.82
Vankuren NW, Long M (2018) Gene duplicates resolving sexual conflict rapidly evolved essential gametogenesis functions. Nat Ecol Evol. https://doi.org/10.1038/s41559-018-0471-0
Vavouri T, Semple JI, Lehner B (2008) Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. https://doi.org/10.1016/j.tig.2008.08.005
Visser JAGM, Hermisson J, Wagner GP et al (2003) Perspective: evolution and detection of genetic robustness. Evolution. https://doi.org/10.1111/j.0014-3820.2003.tb00377.x
Wagner A (2000) Robustness against mutations in genetic networks of yeast. Nat Genet. https://doi.org/10.1038/74174
Wang Y, Gu X (2000) Evolutionary patterns of gene families generated in the early stage of vertebrates. J Mol Evol. https://doi.org/10.1007/s002390010159
Wang T, Birsoy K, Hughes NW et al (2015) Identification and characterization of essential genes in the human genome. Science. https://doi.org/10.1126/science.aac7041
Zeng Y, Gu X (2010) Genome factor and gene pleiotropy hypotheses in protein evolution. Biol Direct. https://doi.org/10.1186/1745-6150-5-37
Zhang Z, Gu J, Gu X (2004) How much expression divergence after yeast gene duplication could be explained by regulatory motif evolution? Trends Genet. https://doi.org/10.1016/j.tig.2004.07.006
Zhang W, Landback P, Gschwend AR et al (2015) New genes drive the evolution of gene interaction networks in the human and mouse genomes. Genome Biol. https://doi.org/10.1186/s13059-015-0772-4
Zhou Z, Zhou J, Su Z, Gu X (2014) Asymmetric evolution of human transcription factor regulatory networks. Mol Biol Evol. https://doi.org/10.1093/molbev/msu163
Zou Y, Huang W, Gu Z, Gu X (2011) Predominant gain of promoter TATA box after gene duplication associated with stress responses. Mol Biol Evol. https://doi.org/10.1093/molbev/msr116
Zou Y, Su Z, Huang W, Gu X (2012) Histone modification pattern evolution after yeast gene duplication. BMC Evol Biol. https://doi.org/10.1186/1471-2148-12-111
Acknowledgements
The author is grateful to all members of the research group for constructive comments in the early version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: David Liberles.
Rights and permissions
About this article
Cite this article
Gu, X. A Simple Evolutionary Model of Genetic Robustness After Gene Duplication. J Mol Evol 90, 352–361 (2022). https://doi.org/10.1007/s00239-022-10065-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-022-10065-1