Abstract
This article is part of an anniversary issue of Journal of Molecular Evolution, commenting on a paper published on 1999 by the Nobel laureate Frances Arnold and her colleague Kentaro Miyazaki. The paper by Miyazaki and Arnold presented saturation mutagenesis as an alternative method to random mutagenesis for obtaining enzymes with increasing stability. Both techniques were conceived to accomplish directed evolution, an approach honoured by the Nobel Prize of Chemistry 2018. Here, I am commenting on the pros and cons of random and saturation mutagenesis, while also discussing important results from directed evolution. I conclude that molecular evolution is finding new applications in science and it is definitely an integral part of the genomic era’s revolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For more than 100 years, evolutionary biology was viewed exclusively as a theoretical field devoted to understanding how biological diversity originates and is maintained. Today, it is widely acknowledged that evolutionary theory is a unifying principle in biology with many scientific applications (Liberles et al. 2020), especially in conservation biology, agriculture, industrial chemistry, pharmacology, and biomedicine (Colautti et al. 2017; Stearns 2020). Antibiotic resistance and cancer development are two great examples where deeper understanding of these conditions came through evolutionary models (Christaki et al. 2020; Zahir et al. 2020). Another great practical example is the long-term evolutionary experiment by Prof. Richard Lenski. His team has been re-culturing 12 initially identical populations of asexual Escherichia coli bacteria since 24 February 1988, tracking genetic changes affecting fitness (Lenski et al. 1991; Lenski 2017). This is one of the longest lab experiments in the history of science.
It was a matter of time for evolutionary theory to become more applied. Using the power of evolution, researchers started to experimentally obtain modified enzymes with improved functions in the 1990s. This field was rapidly expanded by protein engineers and new methods were developed, finding applications in industry and medicine. Due to the great success of these applications, the Nobel Prize in Chemistry of 2018 was awarded to the pioneers of this field, Frances Arnold, for the accelerated evolution of enzymes, and George Smith and Gregory Winter for phage display of modified proteins. Prof. Frances Arnold mentioned in her Nobel award lecture (Arnold 2019) that the influential paper entitled “Natural Selection and the Concept of a Protein Space” (Maynard Smith 1970) by the famous evolutionary biologist John Maynard Smith that was published in Nature on 1970, inspired her to engineer proteins in the lab using evolutionary procedures.
Directed Evolution
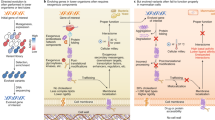
There are three main steps in directed evolution: mutagenesis, selection/screening of the desired enzyme property, and decision of which parental genes will proceed to the next round of evolution. The experiments can be performed in vivo, inside cells (bacteria or yeast), or through in vitro transcription–translation systems. In the classical way, the researcher begins with the gene of a parent protein (Bloom and Arnold 2009; Zeymer and Hilvert 2018). Using error-prone PCR or a similar method, a library of thousands of mutant clones of the gene is produced. Each clone is transferred through a plasmid inside a cell (bacterial or yeast) to express the mutant protein. Then a high-throughput selection/screening method is followed. The mutant enzyme can be immobilized on a solid surface for testing its properties. Alternatively, a massive screening of the enzymatic activity or stability can be performed, usually using 96-well plates (Bloom and Arnold 2009). The decision about the next round of mutagenesis/evolution is the final step. Gene clones that expressed a protein with an improved property, are selected for further mutagenesis, going again though the whole procedure. This can be repeated tens or hundreds of times until the evolved protein reaches a desired level of performance (Bloom and Arnold 2009; Arnold 2019). Of course, these experiments do not always end by producing a successful outcome. Directed evolution is highly unpredictable, just as evolution in nature is. Enzymes can be improved but not always at the level that it is intended. In such cases, complementary methods can be helpful, like saturation mutagenesis, which will be explained below.
Theoretical Background
The Kimura neutral theory of molecular evolution (Kimura 1968) provides the theoretical background to experiments of directed evolution. According to empirical observations, 30–50% of random mutations are strongly deleterious, 50–70% are neutral or slightly affect enzyme function, and only 0.5–0.01% are beneficial (Bloom and Arnold 2009). In this context, “beneficial” usually means an increase of enzyme activity or stability. The experimenter plays the role of natural selection, trying to find the beneficially mutated gene, in order to proceed to further mutagenesis. Other kinds of mutations may play a role in directed evolution, as well. For example, compensatory mutations may appear in proteins. These mutations correct the deleterious effect of other mutations and tend to occur in certain regions of the proteins (Davis et al. 2009).
Prof. F. Arnold describes directed evolution as a 'fitness landscape' containing multiple peaks (Arnold 2019). Every peak represents a desired property of the enzyme, for example increased activity for its substrate, increased stability, activity for another substrate, etc. In each round of selection, the researcher tries to climb up a hill of the 'fitness landscape'. Repeated rounds of mutagenesis/selection can permit reaching a desired level of function (“peak”) (Renata et al. 2015). The final result may not be always the ideal one. Mutations may stop being additive or an early saturation level of improvement can be reached (Sayous et al. 2020). In those cases, other methods can be combined with random mutagenesis, like saturation mutagenesis.
The Cytochrome P450 Example
The cytochrome P450 family of enzymes is vital for many forms of life in Earth. Genes encoding these enzymes have evolved through gene duplication (Omura 2013). Members of this enzyme family transfer an oxygen atom to organic molecules to make specific hydroxylated compounds or epoxides, oxidize heteroatoms, nitrate aromatics, and many more. Prof. F. Arnold and her team used directed evolution trying to give new functions to these enzymes. One of their achievements was the creation of cyclopropane stereoisomers by P450 (Coelho et al. 2013), that is now used by pharmaceutical companies for drug development and production (Wang et al. 2014). Another discovery was that P450 enzymes can perform nitrene chemistry directing the nitrene to C–H bonds for C–H amination (McIntosh et al. 2013). This and other new functions of P450 can contribute to the development of new products in industry. P450 is just an example. Directed evolution has contributed to many other achievements, like the creation of new catalytic sites in enzymes and the catalysis of new covalent bonds like C–Si (Kan et al. 2016).
Saturation Mutagenesis and the Paper by Miyazaki and Arnold 1999, in JME
Saturation mutagenesis is a complementary method to random mutagenesis. Sometimes, through random mutagenesis, a fraction of the protein is not altered at all or is altered with only a few mutants. Saturation mutagenesis can be an alternative strategy (Sayous et al. 2020). Researchers can use site-directed mutagenesis through synthetic oligos in order to create all the possible versions of a protein for a specific amino-acid position (Gupta and Varadarajan 2018). Miyazaki and Arnold published a paper in Journal of Molecular Evolution in 1999 entitled: “Exploring Nonnatural Evolutionary Pathways by Saturation Mutagenesis: Rapid Improvement of Protein Function” (Miyazaki and Arnold 1999). This paper has 256 citations in Google Scholar as of 14/09/2020, an important contribution to the field.
Miyazaki and Arnold (1999) used this method for the protein psychrophilic protease subtilisin S4 in order to investigate the thermostability effect of different amino acids at positions 211 (natively Lys) and 212 (natively Arg). These positions seem to be related with the thermostability of the protein but are not easily accessible by random point mutagenesis. Miyazaki and Arnold found a number of amino acid combinations for the 211/212 positions (Pro/Ala, Pro/Val, Leu/Val, and Trp/Ser) that increase protein’s thermostability. The authors state in their paper that, “These nonconservative replacements, accessible only by multiple (two to three) base substitutions in a single codon, would be extremely rare in a point mutation library. Such replacements are also extremely rare in natural evolution. Saturation mutagenesis may be used advantageously during directed evolution to explore nonnatural evolution pathways and enable rapid improvement in protein traits”. This work shows that saturation mutagenesis can be used successfully in accordance with random mutagenesis, for increasing the stability of enzymes.
Conclusion
Directed evolution is not equivalent to evolution we observe in nature. Many protein versions produced by directed evolution could be incompatible with life but probably useful in industry. On the other hand, some protein versions may not appear at all in the lab due to molecular obstacles. Saturation mutagenesis can help to produce protein forms that rarely appear in the lab or to study all the possible amino acids for a certain residue of the protein.
I consider that the most important achievement of directed evolution is transforming evolutionary theory into a valuable application in the laboratory, with industrial extensions, showing for one more time that basic and theoretical science feeds applied science. Evolutionary theory will continue to feed other areas of science in the near future. We are currently living in the genomic era, where massive genomic data are produced daily and frequently are analysed under an evolutionary lens. This gives a powerful perspective to biomedical sciences. Molecular evolution has undoubtedly become a valuable field for data interpretation and for possible scientific applications, in keeping with the heritage of the great evolutionary biologists of the past century.
References
Arnold FH (2019) Innovation by evolution: bringing new chemistry to life (Nobel lecture). Angew Chem Int Ed 58:14420–14426
Bloom JD, Arnold FH (2009) In the light of directed evolution: pathways of adaptive protein evolution. Proc Natl Acad Sci USA 106(Suppl):9995–10000
Christaki E, Marcou M, Tofarides A (2020) Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol 88:26–40
Coelho PS, Brustad EM, Kannan A, Arnold FH (2013) Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes. Science 339:307–310. https://doi.org/10.1126/science.1231434
Colautti RI, Alexander JM, Dlugosch KM et al (2017) Invasions and extinctions through the looking glass of evolutionary ecology. Philos Trans R Soc B 372:20160031
Davis BH, Poon AFY, Whitlock MC (2009) Compensatory mutations are repeatable and clustered within proteins. Proc R Soc B 276:1823–1827. https://doi.org/10.1098/rspb.2008.1846
Gupta K, Varadarajan R (2018) Insights into protein structure, stability and function from saturation mutagenesis. Curr Opin Struct Biol 50:117–125
Kan SBJ, Lewis RD, Chen K, Arnold FH (2016) Directed evolution of cytochrome c for carbon-silicon bond formation: bringing silicon to life. Science 354:1048–1051. https://doi.org/10.1126/science.aah6219
Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626. https://doi.org/10.1038/217624a0
Lenski RE (2017) Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J 11:2181–2194
Lenski RE, Rose MR, Simpson SC, Tadler SC (1991) Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. Am Nat 138:1315–1341. https://doi.org/10.1086/285289
Liberles DA, Chang B, Geiler-Samerotte K et al (2020) Emerging frontiers in the study of molecular evolution. J Mol Evol 88:211–226
Maynard Smith J (1970) Natural selection and the concept of a protein space. Nature 225:563–564. https://doi.org/10.1038/225563a0
McIntosh JA, Coelho PS, Farwell CC et al (2013) Enantioselective intramolecular C-H amination catalyzed by engineered cytochrome P450 enzymes in vitro and in vivo. Angew Chem Int Ed 52:9309–9312. https://doi.org/10.1002/anie.201304401
Miyazaki K, Arnold FH (1999) Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J Mol Evol 49:716–720. https://doi.org/10.1007/PL00006593
Omura T (2013) Contribution of cytochrome P450 to the diversification of eukaryotic organisms. Biotechnol Appl Biochem 60:4–8
Renata H, Wang ZJ, Arnold FH (2015) Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution. Angew Chem Int Ed 54:3351–3367
Sayous V, Lubrano P, Li Y, Acevedo-Rocha CG (2020) Unbiased libraries in protein directed evolution. Biochim Biophys Acta 1868:140321
Stearns SC (2020) Frontiers in molecular evolutionary medicine. J Mol Evol 88:3–11
Wang ZJ, Renata H, Peck NE et al (2014) Improved cyclopropanation activity of histidine-ligated cytochromeP450 enables the enantioselective formal synthesis of levomilnacipran. Angew Chem Int Ed 53:6810–6813. https://doi.org/10.1002/anie.201402809
Zahir N, Sun R, Gallahan D et al (2020) Characterizing the ecological and evolutionary dynamics of cancer. Nat Genet 52:759–767. https://doi.org/10.1038/s41588-020-0668-4
Zeymer C, Hilvert D (2018) Directed evolution of protein catalysts. Annu Rev Biochem 87:131–157
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Not any.
Additional information
Handling editor: Aaron Goldman.
Rights and permissions
About this article
Cite this article
Voskarides, K. Directed Evolution. The Legacy of a Nobel Prize. J Mol Evol 89, 189–191 (2021). https://doi.org/10.1007/s00239-020-09972-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-020-09972-y