Abstract
Kunitz-type domains are ubiquitously found in natural systems as serine protease inhibitors or animal toxins in venomous animals. Kunitz motif is a cysteine-rich peptide chain of ~ 60 amino acid residues with alpha and beta fold, stabilized by three conserved disulfide bridges. An extensive dataset of amino acid variations is found on sequence analysis of various Kunitz peptides. Kunitz peptides show diverse biological activities like inhibition of proteases of other classes and/or adopting a new function of blocking or modulating the ion channels. Based on the amino acid residues at the functional site of various Kunitz-type inhibitors, it is inferred that this ‘flexibility within the structural rigidity’ is responsible for multiple biological activities. Accelerated evolution of functional sites in response to the co-evolving molecular targets of the hosts of venomous animals or parasites, gene sharing, and gene duplication have been discussed as the most likely mechanisms responsible for the functional heterogeneity of Kunitz-domain inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction: Elucidating the Structural and Functional Biodiversity of Kunitz Domain
Peptidases are indispensable for the survival of all kinds of organisms as they break down substrate proteins but their activities need to be kept under strict control. Inhibitors of peptidases, the protease inhibitors play crucial roles in natural systems by tightly regulating the protease activity and acting as a switch in many signaling pathways (Laskowski and Kato 1980; Rawlings et al. 2004a). Several inherited diseases have been attributed to the abnormalities in the functioning of proteases and their inhibitors (Molinari et al. 2003; Fregonese and Stalk 2008; Cleynen et al. 2011; Ketterer et al. 2016). Protease inhibitors have been classified either by their mechanism of action or by the type of protease they inhibit: aspartic, cysteine, metallo, serine, and threonine inhibitors (Laskowski and Kato 1980). They can be also classified into families/superfamilies based on the similarities at the amino acid sequence level and/or tertiary structure (Rawlings et al. 2004a). Similarities in primary structure and tertiary structure have supported the common ancestry of many inhibitor families. The research on protease inhibitors has always been in attention owing to their potential applications in medicine, agriculture, and biotechnology (Sabotič and Kos 2012; Cotabarren et al. 2020).
Kunitz-type serine protease inhibitors are found in several organisms including animals, plants, and microbes (Rawlings et al. 2004a). Kunitz-domain inhibitors known from animal sources are classified under the inhibitor family I2, Clan IB according to the MEROPS database (Rawlings et al. 2004a, b). They are typically of 50–70 amino acids in length and adopt a conserved structural fold with two antiparallel β-sheets and one or two helical regions that are stabilized with three disulfide bridges with the bonding pattern of 1–6, 2–4, 3–5 (Rawlings et al. 2004b; Buchanan and Revell 2015) (Fig. 1a, b). The disulfide bridges maintain the structural integrity of the inhibitor and also present the protease-binding loop at its surface. A highly exposed P1 active site residue at position 15 is usually arginine (Arg) or lysine (Lys) inserts into the S1 site of the cognate protease and is the primary determinant of the specificity of serine protease inhibition. This motif was first identified in the bovine pancreatic trypsin inhibitor (BPTI)-like protease inhibitors, which are strong inhibitors of serine proteases like trypsin and chymotrypsin (Vincent and Lazdunski 1973; Deisenhofer and Steigemann 1975). An overall high content of basic amino acids and exceptionally high basicity of the guanidine moiety on the side chain of arginine imparts the inhibitor with a basic isoelectric point. These inhibitors are found in many tissues throughout the body and inhibit several serine proteases (Ascenzi et al. 2003). The solvent-exposed loop (residue 8 to 19) is highly complementary to the enzyme active site (S1 pocket) with the P1 residue (Lys15 in BPTI) interacting with the Asp189 at the bottom of the S1 pocket of trypsin (Fig. 1c). Kunitz-domain can function as a standalone protease inhibitor (~ 6 kDa) in its free form by recognizing specific protein structures. Kunitz inhibitors may have a single inhibitory domain or even more forming a multi-domain, single-chain inhibitor.
Structure and activity of Kunitz-domain inhibitors. a Schematic representation of Kunitz-domain inhibitor showing the disulfide bonding pattern of 1–6, 2–4, 3–5. Conserved cysteine residues are marked with yellow and the P1 active site residue, the primary determinant of the specificity of serine protease inhibition is marked with a blue asterisk. b Crystal structure of BPTI (PDB: 4Y0Y) showing three disulfide bonds (in yellow), the solvent-exposed loop (P8 to I19) highly complementary to the enzyme active site, and the P1 residue (K15). c Trypsin in complex with BPTI (PDB: 4Y0Y) showing P1 residue (K15 in BPTI) interacting with the Asp189 at the bottom of the S1 pocket of Trypsin. d Amino acid sequence alignment of Kunitz-domain inhibitors from different source organisms: BPTI from Bos taurus (P00974); Delta-dendrotoxin from Dendroaspis angusticeps (P00982); KappaPI-theraphotoxin from Haplopelma schmidti (P68425); KappaPI-stichotoxin from Stichodactyla haddoni (B1B5I8); Kunitz domain from human (Homo sapiens) amyloid β-protein precursor (P05067); Kunitz domain from human (Homo sapiens) collagen alpha-3(VI) chain (P12111); Kunitz domain from human (Homo sapiens) Tissue factor pathway inhibitor 2 (P48307). Conserved residues are highlighted in red and the P1 residue (K/R) is marked with an asterisk. Kunitz family signature sequence (F*Y*GC****N*F*****C) is shown in a box (Color figure online)
Kunitz-type domains exist in multiple forms in numerous tissues imparting proteins with specific (serine) protease inhibitory function: the Kunitz-type toxin in venomous animals like snakes, spiders, and scorpions (Fry et al. 2005); mammalian inter-alpha-trypsin inhibitors (Fries and Kaczmarczyk 2003); domain found in Alzheimer's amyloid β-protein in humans (Hynes et al. 1990); domains at the C-termini of the alpha-1 and alpha-3 chains of type VI and type VII collagen (Chen et al. 2013a, b) and tissue factor pathway inhibitor (TFPI) (Broze and Girard 2012) (Fig. 1d). It is worth giving mention to the Kunitz-soybean trypsin inhibitor (STI) family found in legume seeds. Inhibitors of Kunitz-STI family are about ~ 20 kDa in size and characterized by ‘β-trefoil fold’ and two disulfide bridges (Azarkan et al. 2011; Oliva et al. 2011). They constitute the inhibitor family I3A, Clan IC according to the MEROPS database (Rawlings et al. 2004a, b). Therefore, Kunitz-STI family of plants is quite unrelated in its primary as well as tertiary structure from Kunitz-domain inhibitors and thus beyond the scope of this discussion. I2 and I3 inhibitor families are also known as the ‘Kunitz-A’ and ‘Kunitz-P’ families, respectively, because members are known from animals and plants.
Functionally, Kunitz-domain inhibitors are known to be involved in various physiological processes such as host defense against microbial infection, blood coagulation, fibrinolysis, and inflammation by exhibiting inhibition of serine proteases (viz. trypsin/chymotrypsin/elastase/kallikrein) (Ranasinghe and McManus 2013; Wan et al. 2013a, b). However, few inhibitors of cysteine and aspartic proteases belonging to the Kunitz family are also known (Rawlings et al. 2004a). Kunitz-domain inhibitors are also known as frequent components of venoms from poisonous animals acting as ion channel blockers (Yuan et al. 2008; Peigneur et al. 2011). The present review aims at introspecting at the various physiological targets of these inhibitors, the structural basis of these multifunctional proteins, and the evolutionary aspects of the progression of Kunitz domain towards multiple target recognition.
Kunitz Trypsin Inhibitor Domains in Mammals
Kunitz-type serine protease inhibitors are found in blood plasma, saliva, and several tissues in mammals. Several examples of mammalian proteins have made evident that Kunitz-type inhibitor domains are often present as ‘insertion’ in various proteins most likely as a result of exon shuffling, thereby incorporating domains from different evolutionary precursors (Ikeo et al. 1992).
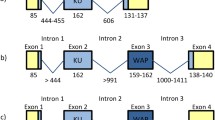
A Kunitz-domain sequence is found in the precursor amyloid β-protein (APPI) which accumulates in the neuritic plaques and cerebrovascular deposits of patients with Alzheimer’s disease, the most common neurodegenerative disorder (Kitaguchi et al. 1988, 1990; Ikeo et al. 1992) (Fig. 2a). The APPI precursor protein has three alternatively spliced versions of which two have an insertion sequence of the Kunitz-type trypsin inhibitor domain. Studies have indicated that the overproduction of inhibitor-containing species can be associated with the progression of disease in Alzheimer’s patients (Johnson et al. 1989, 1990; Ben Khalifa et al. 2012). Specifically, a major role of Kunitz domain in APPI dimerization leading to the formation of β-amyloid plaques has been found (Ben Khalifa et al. 2012). The Kunitz domain of APPI shows 43% sequence identity with the BPTI. The structural superimposition of APPI Kunitz domain with BPTI has shown identical backbone conformation except for residues 39–41 (Hynes et al. 1990). A Gly-Gly-Asn sequence is found in APPI whereas BPTI has an Arg-Ala-Lys sequence. This substitution results in a significant alteration in the backbone fold most likely Gly-40 and Asn-41 influencing the target specificity by altering the geometry of residue 39. Few other amino acid substitutions in the protease-binding loops of APPI result in significant changes in affinity and specificity of target protease (Hynes et al. 1990). The conformation of side chains, three disulfide bridges, the hydrophobic core, and the hydrogen bonding network due to internal waters remain conserved in APPI identical to BPTI. APPI inhibits trypsin and also inhibits chymotrypsin.
Schematic representation of the Kunitz domain present as ‘insertions’ in various proteins in mammals. a Amyloid β-protein precursor (100–135 kDa) is an integral membrane glycoprotein that contains the Kunitz domain as an insertion between the acidic domain and the glycosylated domain. Alternative splicing results in overproduction of Kunitz-inhibitor-containing species in Alzheimer’s patients. b Inter-α-trypsin inhibitor consists of three polypeptides: 2 homologous heavy chains (~ 75 kDa) and a light chain known as bikunin (~ 16 kDa). Bikunin contains two Kunitz domains with protease inhibitory function. c Collagen VI is a major extracellular matrix protein distributed in various tissues. α3 chain of collagen VI is large (250–300 kDa) with several splice variants produced under different conditions. Kunitz domain is present towards the C-terminal end and is known to be released by proteolytic processing after collagen VI secretions
The inter-alpha-trypsin inhibitor (IαTI), an acid-labile plasma glycoprotein, is one of the most fascinating proteins in which the Kunitz-type inhibitor domain is present at the NH2-terminal as a distinct structural and functional domain called bikunin (Fig. 2b). Bikunin moiety comprises of two Kunitz-type inhibitory domains. Bikunin is found in most of the IαTI family inhibitors and also exists in a free state in plasma, urine, and hepatocyte cultures (Salier 1990; Salier et al. 1996; Pugia et al. 2007). The Kunitz domains in bikunin show high homology with BPTI and are targeted towards trypsin, chymotrypsin, cathepsin G, elastase, thrombin, and plasmin (Fioretti et al. 1987; Salier et al. 1996; Zhuo et al. 2004). These inhibitors are proposed to be involved in the control of plasminogen activation by regulating the activity of plasminogen-activators like kallikrein and other serine proteases. Plasminogen activation leads to the production of plasmin which functions in the fibrinolytic mechanism by degrading the fibrin blood clots (Chapin and Hajjar 2015). Thus, the inhibitory activity of Kunitz domain towards serine proteases including plasmin and kallikrein suggests their regulatory role in blood clotting and fibrinolysis. Bikunin is also known to effectively inhibit cell surface plasmins of cancer cells which are involved in metastasis of tumor cells (Fries and Blom 2000). Their presence in human sera in free form has proposed further physiological roles of Kunitz inhibitors such as in inflammation and modulation of cell growth (Fries and Blom 2000). Bikunin is one of the main anti-inflammatory response mediators whose high levels are observed in plasma and urine in case of infection, cancer, tissue injury, or vascular diseases (Pugia et al. 2007). Bikunin-free members of the IαTI family are known for their role in extracellular matrix biology because of their hyaluronic acid-binding capacity (Fries and Kaczmarczyk 2003).
The α3 chain of collagen type VI (α3-VI), a cell-binding protein identified in humans, chicken, mouse, guinea pig, and rat also has an insertion of a Kunitz domain towards the C-termini (Fig. 2c) (Bonaldo and Colombatti 1989). The most C-terminal subdomain of α3-VI also referred to as the ‘C5 domain’ has high amino acid sequence similarity to Kunitz family serine protease inhibitors yet it lacks trypsin inhibition activity (Zweckstetter et al. 1996). Collagen VI is widely distributed in several tissues and contributes to the properties of the extracellular matrix also regulating several cell and tissue processes like autophagy, apoptosis, proliferation, and inflammation (Chen et al. 2013a, b; Cescon et al. 2015). Formation of collagen VI extracellular microfibrils is a complex process that involves the assembly of heterotrimeric monomers into dimers and tetramers followed by their end-to-end joining. The C-terminal domain C5 is critical for interactions between tetramers and thus efficient microfibril formation (Lamande et al. 2006). Detection of the C5 domain of α3-VI in the extracellular matrix of normal human fibroblasts as well as in immediate pericellular matrix of articular cartilage indicates that Kunitz domains might also be playing role in growth and remodeling of connective tissues (Lamande et al. 2006). Therefore, the ubiquitous presence of Kunitz domains within several mosaic gene architectures suggests its multiple functions in various tissues over an array of organisms.
Functional Variants of Kunitz-Domain Inhibitor in Parasites
Parasites secrete a plentiful of proteases and protease inhibitors to invade the host and support their survival within the host. Protease inhibitors play multiple roles including immune evasion, parasite development, and protein regulation (Jefferies et al. 2001; Robinson et al. 2009; De Magalhaes et al. 2018). Kunitz-type protease inhibitors have been identified in several parasitic organisms, though a few have been characterized functionally against trypsin, chymotrypsin, neutrophil elastase, and plasma kallikrein (Ranasinghe and McManus 2013; Ranasinghe et al. 2015a, b; Morais et al. 2018).
A protein (FhKT1) with homology to Kunitz domain have been identified in the gut and parenchymal tissues of the infective juvenile stage of Fasciola hepatica (a helminth parasite) which exhibits no inhibitory activity toward serine proteases but is a potent inhibitor of major cathepsin cysteine proteases (Smith et al. 2016). FhKT1 shares only 23% sequence identity with BPTI but possesses the characteristic Kunitz-type fold because of the six conserved cysteine residues that form three disulfide bonds. This implies that although sequence variations are there, the structural integrity defining Kunitz domain inhibitors remains intact. The P1 residue in the reactive site loop of FhKT1 is leucine (Leu), which is important in docking into the S2 active site of Cathepsin L- like cysteine proteases. Leu15 of the protease-binding loop of FhKT1 forms strong hydrophobic interactions with residues Leu176, Met177, and Val267 of the S2 pocket of Fasciola cathepsin L. Therefore, the hydrophilic S1 pocket of serine proteases is unable to form a protein–protein complex with FhKT1. However, it was observed that upon substitution of unusual P1Leu15 for more common Arg, its cysteine protease inhibition profile was modestly reduced and bestowed the protein with trypsin-inhibitory activity. Therefore, FhKT1 presents an adequate example of Kunitz-domain inhibitor variant targeting proteases of different mechanistic classes which may be important in parasitic or other infectious diseases.
Kunitz-Type Toxins (KTTs): Kunitz-Domain Inhibitors in Venomous Animals
Kunitz-type toxins (KTTs) have been identified from many blood-sucking and venomous animals, such as snakes, spiders, ticks, scorpions, cone snails, and sea anemones (Chang et al. 2001; Zupunski et al. 2003; Yuan et al. 2008; Zhao et al. 2011; Peigneur et al. 2011). The first KTT in animal venom was isolated from snake in the 70′s with a serine protease inhibition function (Strydom 1973, 1977). The physiological role of these inhibitors is to resist prey proteases from degrading their venom protein toxins (Zupunski et al. 2003). Since then, various KTTs with diverse pathophysiological significance have been identified (Mukherjee and Mackessy 2014; Thakur and Mukherjee 2017; Martins et al. 2020). Some of them have shown inhibitory potential against proteases of other classes or ability to block voltage-gated potassium (Kv) ion channels, which are crucial regulators of various physiological processes such as host defense, blood coagulation, platelet modulation, fibrinolysis, and action potential transduction (Masci et al. 2000; Lu et al. 2008; Flight et al. 2009; Wan et al. 2013a, b; Kalita et al. 2019). KTTs have been categorized into five groups: body trypsin inhibitors, chymotrypsin inhibitors in venom, trypsin inhibitors in venom, dual function toxins, and potassium ion channel blockers (Yuan et al. 2008).
Inhibitors with Multiple Protease Recognition
Several KTTs have been characterized with further new functions in addition to the typical inhibition of serine proteases. AvKTI is a Kunitz-type serine protease inhibitor characterized from spider (Araneus ventricosus) that exhibits inhibitory activity against trypsin, chymotrypsin, plasmin, and neutrophil elastase (Wan et al. 2013a, b). AvKTI 57-amino acid mature peptide displays typical Kunitz-type inhibitor scaffold including six conserved cysteine residues and P1-P1′ (Lys-Ala) residues like BPTI. Novel functions of AvKTI against plasmin and neutrophil elastase signify their regulatory role in modulation of thrombosis and fibrinolysis and the regulation of inflammatory signaling. Interestingly, its expression observed only in the epidermis (not in venom gland) has posed new questions on its physiological target and roles in spiders.
Similarly, ShPI-1 is a Kunitz-type inhibitor from the Carribean Sea anemone (Stichodactyla helianthus) which displays inhibitory activity against not only serine but also cysteine and aspartic proteases (Fig. 3a) (Garcia-Fernandez et al. 2012). The unusual multifunctional activity of this inhibitor is intriguing and suggests exceptional structural and functional properties of protease inhibitors from marine invertebrates. The interaction maps of the ShPI-1: trypsin and BPTI: trypsin complexes were found almost identical at positions P2, P1, and P1′ forming primary direct interactions with the S1 pocket of enzyme and found completely buried after complex formation. Recently, its pseudo-wild-type variant, rShPI-1A was identified which also exhibits tight-binding potency against different Kv ion channels (Garcia-Fernandez et al. 2016). However, to understand the cross-reactivity with proteases of other classes and identifying key residues for Kv ion channel inhibiting activity, further design of ShPI-1 mutants and their structural validation is necessary.
Functional diversification of Kunitz-type toxins on conserved Kunitz fold. a Amino acid sequence alignment of Kunitz-type toxin representatives of the three functional subtypes: Inhibitors with multiple protease recognition (P31713; ShPI-1 from the sea anemone Stichodactyla helianthus), dual function toxins (P68425; HWTX-XI from Chinese bird spider Ornithoctonus huwena or Haplopelma schmidti) and ion channel inhibitors or neurotoxins (P00982; DTX-K from mamba snakes Dendroaspis angusticeps). The alignment shows the conserved cysteine residues bonded with disulfide bridges and the conserved P1 residue (K) marked with a dotted arrow. An enrichment of basic residues (in blue) in the N-terminal half has been proposed to have an important role in binding to the potassium ion channels and adopting the new function as ion channel blockers among KTTs. b Electrostatic surface representations (PDB: 1shp, 2jot, 1dtk) show the structural similarities but different electrostatic surfaces for protease inhibitors and the ion channel blockers (red: negative; blue: positive; gray: neutral). The phylogram of KTTs of the three functional subtypes shows their common evolutionary origin. KTTs show a shift of losing their ability to function as serine protease inhibitors and instead adopt a new function to block or modulate ion channels, effectively becoming more toxic (Color figure online)
Dual Function Toxins
Dual function toxins have developed the ability to block ion channels besides their original function of serine protease inhibition. HWTX-XI is a representative dual function toxin from the distinct superfamily of KTTs in spiders (Tarantulas: Ornithoctonus huwena) (Fig. 3a). It is a potent trypsin inhibitor (30-fold stronger than BPTI) as well as a weak Kv ion channel blocker (Yuan et al. 2008). The solution structure of HWTX-XI resembles a typical Kunitz-type fold very similar to BPTI. Structural analysis together with functional studies on 18 expressed mutants of this toxin reveals that there are two separate sites located on the two ends of the molecule, responsible for the two types of activities exhibited. Mutation in K14 residue in the loop region led to a 105-fold reduction in inhibitory potency against trypsin while it did not influence its inhibitory potency toward Kv1.1 channels. Similarly, residue Leu6 seems to be critical for the Kv1.1 channel blockage having no effect on the trypsin inhibition activity. These results suggest that these two sites are not only distinct but also functionally independent of each other. LmKTT-1a is another KTT homolog found in scorpion which has been functionally characterized for both protease and potassium ion channel inhibition (Chen et al. 2013a, b). Interestingly, the amino acid sequence homology of LmKTT-1a with classical Kunitz-type peptides is low yet it adopts a conserved Kunitz-type fold very similar to other KTTs like HWTX-XI from the spider.
Neurotoxins or Ion Channel Blockers
Dendrotoxins (DTX) are representative neurotoxins found in mamba snakes (Dendroaspis) that block Kv ion channels in neurons by slowing down the channel activation and inactivation kinetics thereby modulating the neuronal activity (Robertson et al. 1996; Harvey 2001; Báez et al. 2015). These peptide toxins are quite large molecules (~ 2.9 nm) as compared to Kv ion channels. They can affect potassium ion channels by two mechanisms: First, by directly occluding the channel pore and second, by causing sufficient steric hindrance to slow down the conformational changes necessary for channel opening and closing (Wulff et al. 2009). Potassium ion channels are crucial for many aspects of cellular regulation and signal transduction as they regulate ion flux across biological membranes. These neurotoxins are KTT homologs as they possess a conserved Kunitz-type structural scaffold with one α-helix, two β-sheets, and a conserved disulfide bonding pattern. They have lost their protease inhibitory activity, even though they have characteristic residues such as Lys or Tyr at the reactive sites in the protease contact regions (Fig. 3a). The implication being that the region important for the neuromuscular activity of snake toxins is different than the reactive sites. However, this does not mean that the reactive sites (for contacts with protease) are useless for activity as a toxin as they may be playing a regulatory role in enzymatic activities on the presynaptic membrane (Ikeo et al. 1992).
Dendrotoxins have served as useful pharmacological tools in designing candidate drugs targeting various potassium ion channels which have an important regulatory role in neurons of the mammalian central nervous systems (Wulff et al. 2009). Thus, the development of neurotoxic functions on the KTT scaffold in spiders, scorpions, and snakes reflects on the molecular diversity of Kunitz-type fold while still maintaining the structural conservation.
Functional Diversification of the KTT Subfamily
The addition of new protein functions to KTTs has proven beneficial by generating more effective toxins capable of paralyzing the prey/predator. Specifically, elucidation of the molecular basis for the activity of these multifunctional inhibitors against structurally unrelated proteases/other targets has been of interest. Thus, the origin and evolution of toxins have always been very interesting to study and also under debate. It has been hypothesized that the recruitment of Kunitz-type scaffold (i.e., BPTI) by the venomous animals to produce dual function toxins viz. ion channel blockers or neurotoxins is the effect of Darwinian selection pressure on the evolution of KTTs (Fry et al. 2005). However, fundamental questions about how stable are these newly grafted functions on to the ‘old’ scaffold remain perplexing due to limited correlating sequence and 3D-structure data for KTTs. Alone, the sequence data from various KTTs have reflected clear patterns of amino acid substitutions to discard the inhibition function when dual function toxin gets finally transformed into an ion channel blocker (Fig. 3a, b). Exclusive replacement of amino acids in the loop regions on the surface leading to an overall change of charge or electrostatic potential of the Kunitz proteins is observed (Župunski and Kordiš 2016). An enrichment of basic residues is found in key sites responsible for interacting with the ion channel. Several mutational studies which have tried to identify crucial residues for blocking the activity of DTX have proposed that positively charged lysines in the N-terminal half, especially Lys5 plays an important role in DTX binding to potassium ion channels (Fig. 3a, b) (Harvey 1997; Wang et al. 1999; Garcia-Fernandez et al. 2016). Nevertheless, additional residues might play important roles in deciding the specificity of DTX to different subtypes of Kv ion channels (Harvey and Robertson 2004).
KTTs themselves comprise a multi-gene subfamily within the Kunitz-type superfamily as evident by several functional variants within this subgroup. Development of a multi-gene family is a complex process that involves a series of gene duplication events that also go on accumulating a series of mutations over time resulting in sequence divergence and acquiring new functions (Kordis and Gubensek 2000; Walsh and Stephan 2001) (Fig. 4). While the original function is conserved, the duplicated copies come under selection constraints for further changes. The conservation between various KTT proteins and peptides at the primary and tertiary structure level provides compelling evidence for their common evolutionary origins from an ancestral gene. The evolution of toxins from the BPTI scaffold seems to be a transformation of protease inhibitor function (e.g., AvKTI) into ion channel blockers (e.g., DTX-K) via dual function toxins (e.g., HWTx-XI) as intermediates (Fig. 3b). However, this transformation seems to be prominent in KTTs from snakes and spider venoms where Kv ion channel-blocking activity could be important for paralyzing their prey in the shortest time (Kordis and Gubensek 2000; Yuan et al. 2008; Schwarz et al. 2014). This brings in to picture the role of a natural selection pressure which is favoring the ion channel-blocking activity within these venomous animals (Župunski and Kordiš 2016). KTTs in venomous animals are secretory proteins and thus have more interaction with the environment as compared to body proteins (like BPTI) which are involved in physiological functions within the body (Fry et al. 2005; Yuan et al. 2008). Therefore, they may have a higher evolutionary rate based on the positive Darwinian selection of traits which might be beneficial to kill or immobilize the prey. Phylogenetic analyses based on comparison of mature KTT sequences from several taxonomic groups implies that the KTTs may have evolved through grafting of new function (i.e., ion channel blocking) onto an old Kunitz protein scaffold (S1-protease inhibitors) by gene duplication, mutation and driving force from the selection pressure (Fig. 3b) (Yuan et al. 2008). However, the presence of dual function toxins with weak Kv ion channel blocking activity in spiders and scorpions yet raises more questions on our full understanding of KTT evolution. Development of strong and specific Kv ion channel blockers mostly in snakes reflects on further underlying intricacies in the process of KTT evolution which may vary in different organisms. A full understanding of the KTT family evolution process needs yet more data related to their DNA and protein sequence, 3D structure, and functions. Nonetheless, the evolution of novel functions on a conserved structural scaffold brings in to spotlight the convergent evolution of animal toxins and their evolutionary diversity.
Target-oriented evolution of Kunitz domain functions. Ancestral Kunitz domain diversified its functions by duplications and chance mutations resulting in variants with novel functions and newer targets. Co-evolution of the host molecular targets viz. enzymes involved in physiological processes, agonists of hemostasis & inflammation, platelets, neuronal ion channels (prey and predators in this context) is the driving force towards the selection of these variants in the population based on their functional essentiality. Once selected, these functional variants are reinforced by gene duplication and gene sharing mechanisms within the population
Ecological Significance of the Functional Evolution of Kunitz-Domain Inhibitors
Kunitz-type domains are ubiquitously found in natural systems, either acting as serine protease inhibitors and/or toxins in animal venoms. Inhibitors having Lys or Arg at the reactive site ‘P1 residue’ inhibit trypsin-like enzymes, those with Tyr, Phe, Trp, Leu, Met at P1 site presumably inhibit chymotrypsin-like enzymes and those having Ala/Ser inhibit elastase-like enzymes. On the other hand, KTTs having neuromuscular activity have lost their protease inhibitory potential and acquired the new function of blocking Kv ion channels. The phylogenetic analysis of all Kunitz-type inhibitor sequences has indicated that the ancestral gene of the Kunitz-type inhibitor must have appeared about 500 million years ago. After that, gene duplicated itself many times and some of the duplicates got inserted into other protein-coding genes (Ikeo et al. 1992). In mammals, it is present as an insertion in various protein-coding genes while the majority of the venom proteins are almost completely composed of this domain (Figs. 2, 3). Analysis of several Kunitz-type inhibitors offers an extensive database of amino acid sequence variations (Fig. 1d). Single Kunitz-domain peptides are highly represented but several tick salivary Kunitz-domain proteins possessing multiple domains (1–7) have been also characterized as serine protease inhibitors (Schwarz et al. 2014). In some instances, Kunitz peptides vary in their cysteine motifs (more or less than 6 conserved Cys residues) therefore lacking the typical disulfide bonds probably leading to more flexible fold capable of further diversifying its inhibitory spectrum (example, LmKTT-1a). Kunitz-type inhibitors have evolved so much that modern members of Kunitz superfamily can be distinguished only by their conserved disulfide bonding structure.
Based on the residues at the functional site of various Kunitz-type inhibitors, it is inferred that the ‘flexibility within the rigidity’ and variations of amino acid residues in loop regions and β-turns are responsible for multiple biological functions. Mostly, amino acid substitutions in the reactive-center regions have resulted in inhibitors with varying specificities. It has been observed in several inhibitor families that sites which are in contact with the enzyme show greater interspecific heterogeneity than the other sites which are not in contact with the target molecule (Laskowski et al. 1987a, b; Graur and Li 1988). BPTI or Kunitz-domain family is one of the inhibitor families showing accelerated evolution at functional sites brought about by positive Darwinian selection (Župunski and Kordiš 2016). Thus, the changes that favorably affect the activity of the protein are fixed more rapidly in a population as compared to the neutral mutations (Hill and Hastie 1987).
In natural systems, protease inhibitors have a general defensive role against prey and predators. Venomous animals have evolved with several morphological and physiological adaptations to counteract the host’s immune response. They naturally produce chemical toxins with a primary function either to kill or paralyze the prey or defense against predators. To counter host defense mechanisms, their salivary glands produce venoms that are composed of pharmacological proteins which are injected into the host during blood feeding (Andersen 2010; Schwarz et al. 2014). Thus, venom is under constant selection by nature (Fig. 4). Kunitz-domain toxins identified in several venomous and blood-sucking animals performing multiple functions (protease inhibition to ion channel blockers) endorse that they are important elements in the arms race with the host/predators. Several Kunitz peptides in venoms show a shift of losing their ability to function as serine protease inhibitors and instead adopt a new function to block or modulate ion channels, effectively becoming more toxic (Fig. 3). The main evolutionary pressure driving these variations could be the necessity of functional diversity against the host molecular targets which also keep co-evolving rapidly during the arms race (Andersen 2010; Schwarz et al. 2014) (Fig. 4). This is perhaps ‘target-oriented evolution’ in the benefit of venomous animals to prolong their feeding on the host thereby increasing their chances of survival. Once any protein variant becomes functionally essential as a venom/toxin, this adaptation is reinforced by gene duplication and gene sharing mechanisms in the population (Schwarz et al. 2014).
Perspectives and Concluding Remarks
Kunitz-domain inhibitors are found in a variety of organisms with involvement in various physiological roles. More insights on their functional aspects are required for direct information on their cellular role in various biological systems. KTTs evolved with dual functions and toxins have attracted huge interest not only for evolutionary research but are prospective candidates for pharmacological research as well (Shigetomi et al. 2010; Thakur and Mukherjee 2017). Promising pharmacological potential of Kunitz-domain inhibitors has been demonstrated in murine renal cell carcinoma model (De Souza et al. 2016) and human glioblastoma cells for anti-tumor effects (Morjen et al. 2013). It is recommended that a classification of Kunitz inhibitors based on their inhibition profile: serine-specific, cysteine-specific, serine/cysteine and serine/cysteine/aspartic protease inhibitors can help in designing specific inhibitors against proteases of various mechanistic classes by making defined alterations in the ~ 60 amino acid scaffold of the Kunitz inhibitor (Smith et al. 2016). This has been possible because residues in the loops can be substituted or grafted into desired ones without destabilizing the basic structural framework. Kunitz-domain scaffold is also present in human proteins and thus may possess very low immunogenic potential when used in therapeutics.
Nature-inspired structural leads have always been potential candidates for drug development and therapeutics. Kunitz domain has acquired proximate attention in protein engineering efforts to create specific protease inhibitors relevant for therapeutic applications (Sheffield et al. 2018; Ding et al. 2018; Simeon and Chen 2018). A few Kunitz-domain-derived inhibitors have been successfully approved by the Food and Drug Administration. Ecallantide (DX-88), a substitute kallikrein inhibitor is approved for the treatment of hereditary angioedema, a rare autosomal inherited blood disorder caused by malfunction of plasma C1 kallikrein inhibitor (Lehmann 2008). DX-88, engineered to bind kallikrein with picomolar affinity was derived from the Kunitz domain of lipoprotein-associated coagulation inhibitor (Williams and Baird 2003; Simeon and Chen 2018). In a much recent effort, the Kunitz domain was stabilized against proteolysis by mesotrypsin through disulfide engineering. Mesotrypsin, a human protease is an atypical form of trypsin that exhibits resistance to biological trypsin inhibitors. Kunitz domain in several human protease inhibitors has shown specific substrate like kinetic profiles with mesotrypsin (Pendlebury et al. 2014). Addition of a new disulfide bond (Cys17-Cys34) within the APPI Kunitz domain improves its stability against proteolysis by mesotrypsin by 74-fold (Cohen et al. 2019). Mesotrypsin has been implicated in promoting tumor progression. Therefore, the Kunitz domain can serve as an important scaffold for therapeutics and drug development. The functional evolution of Kunitz-domain inhibitors not only offers a debate on molecular evolution but also provides a tremendous opportunity for application in therapeutics.
References
Andersen JF (2010) Structure and mechanism in salivary proteins from blood-feeding arthropods. Toxicon 56(7):1120–1129
Ascenzi P, Bocedi A, Bolognesi M et al (2003) The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Curr Protein Pept Sci 4(3):231–251
Azarkan M, Martinez-Rodriguez S, Buts L, Baeyens-Volant D, Garcia-Pino A (2011) The plasticity of the β-trefoil fold constitutes an evolutionary platform for protease inhibition. J Biol Chem 286(51):43726–43734
Báez A, Salceda E, Fló M et al (2015) α-Dendrotoxin inhibits the ASIC current in dorsal root ganglion neurons from rat. Neurosci Lett 606:42–47
Ben Khalifa N, Tyteca D, Courtoy PJ et al (2012) Contribution of Kunitz protease inhibitor and transmembrane domains to amyloid precursor protein homodimerization. Neurodegener Dis 10(1–4):92–95
Bonaldo P, Colombatti A (1989) The carboxyl terminus of the chicken α3 chain of collagen VI is a unique mosaic structure with glycoprotein Ib-like, fibronectin type III, and Kunitz modules. J Bio Cem 264(34):20235–20239
Broze GJ Jr, Girard TJ (2012) Tissue factor pathway inhibitor: structure-function. Front Biosci (Landmark Ed) 17:262–280
Buchanan A, Revell JD (2015) Novel therapeutic proteins and peptides. In: Manmohan S, Maya S (eds) Novel approaches and strategies for biologics, vaccines and cancer therapies. Academic Press, Cambridge, ISBN 9780124166035, pp. 483–509.
Cescon M, Gattazzo F, Chen P, Bonaldo P (2015) Collagen VI at a glance. J Cell Sci 128:3525–3531
Chang LS, Chung C, Huang HB, Lin SR (2001) Purification and characterization of a chymotrypsin inhibitor from the venom of Ophiophagus hannah (king cobra). Biochem Biophys Res Commun 283:862–867
Chapin JC, Hajjar KA (2015) Fibrinolysis and the control of blood coagulation. Blood Rev 29(1):17–24
Chen P, Cescon M, Bonaldo P (2013a) Collagen VI in cancer and its biological mechanisms. Trends in Mol Med 19(7):410–417
Chen Z, Luo F, Feng J, Yang W et al (2013b) Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins. PLoS ONE 8(4):e60201
Cleynen I, Juni P, Bekkering GE, Nuesch E et al (2011) Genetic evidence supporting the association of protease and protease inhibitor genes with inflammatory bowel disease: a systematic review. PLoS ONE 6(9):e24106
Cohen I, Coban M, Shahar A, Sankaran B et al (2019) Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin. J Biol Chem 294(13):5105–5120
Cotabarren J, Lufrano D, Graciela Parisi M et al (2020) Biotechnological, biomedical and agronomical applications of plant protease inhibitors with high stability: a systematic review. Plant Sci 292:110398
De Magalhães MTQ, Mambelli FS, Santos BPO, Morais SB, Oliveira SC (2018) Serine protease inhibitors containing a Kunitz domain: their role in modulation of host inflammatory responses and parasite survival. Microbes Infect 20(9–10):606–609
De Souza JG, Morais KL, Anglés-Cano E et al (2016) Promising pharmacological profile of a Kunitz-type inhibitor in murine renal cell carcinoma model. Oncotarget 7(38):62255–62266
Deisenhofer J, Steigemann W (1975) Crystallographic refinement of the structure of bovine pancreatic trypsin inhibitor at l.5 Å resolution. Acta Cryst B31:238–250
Ding L, Hao J, Luo X, Chen Z (2018) Engineering varied serine protease inhibitors by converting P1 site of BF9, a weakly active Kunitz-type animal toxin. Int J Biol Macromol 120(Pt A):1190–1197
Fioretti E, Angeletti M, Citro G, Barra D, Ascoli F (1987) Kunitz-type inhibitors in human serum. Identification and characterization. J Biol Chem 262(8):3586–3589
Flight SM, Johnson LA, Du QS, Warner RL et al (2009) Textilinin-1, an alternative anti-bleeding agent to aprotinin: importance of plasmin inhibition in controlling blood loss. Br J Hematol 145:207–211
Fregonese L, Stolk J (2008) Hereditary alpha-I-antitrypsin deficiency and its clinical consequences. Orphanet J Rare Dis 3:16
Fries E, Blom AM (2000) Bikunin-not just a plasma proteinase inhibitor. Int J Biochem Cell Biol 32(2):125–137
Fries E, Kaczmarczyk A (2003) Inter-α-inhibitor, hyaluronan and inflammation. Acta Biochim Polon 50(3):735–742
Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA (2005) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet 10(1):483–511
Garcia-Fernandez R, Pons T, Meyer A, Perbandt M et al (2012) Structure of the recombinant BPTI/Kunitz-type inhibitor rShPI-1A from the marine invertebrate Stichodactyla helianthus. Acta Crystallogr. Sect F Struct Biol Cryst Commun 68:1289–1293
Garcia-Fernandez R, Peigneur S, Pons T, Alvarez C et al (2016) The Kunitz-type protein ShPI-1 inhibits serine proteases and voltage-gated potassium channels. Toxins 8:110
Graur D, Li WH (1988) Evolution of protein inhibitors of serine proteinases: positive Darwinian selection or compositional effects? J Mol Evol 28:131–135
Harvey AL (1997) Recent studies on dendrotoxins and potassium ion channels. Gen Pharmacol Vasc Syst 28(1):7–12
Harvey AL (2001) Twenty years of dendrotoxins. Toxicon 39(1):15–26
Harvey AL, Robertson B (2004) Dendrotoxins: structure-activity relationships and effects on potassium ion channels. Curr Med Chem 11(23):3065–3072
Hill RE, Hastie ND (1987) Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature 326:96–99
Hynes TR, Randal M, Kennedy LA, Eigenbrot C, Kossiakoff AA (1990) X-ray crystal structure of the protease inhibitor domain of Alzheimer’s amyloid β-precursor protein. Biochemistry 29(43):10018–10022
Ikeo K, Takahashi K, Gojobori T (1992) Evolutionary origin of a Kunitz-type trypsin inhibitor domain inserted in the amyloid β precursor protein of Alzheimer’s disease. J Mol Evol 34:536–543
Jefferies JR, Campbell AM, Van Rossum AJ, Barrett J, Brophy PM (2001) Proteomic analysis of Fasciola hepatica excretory-secretory products. Proteomics 1:1128–1132
Johnson SA, Rogers J, Finch CE (1989) APP-695 transcript prevalence is selectively reduced during Alzheimer’s disease in cortex and hippocampus but not in cerebellum. Neurobiol Aging 10:267–272
Johnson SA, Mc Neill T, Cordell B, Finch CE (1990) Relation of neuronal APP-751/APP-695 mRNA ratio and neuritic plaque density in Alzheimer’s disease. Science 248:854–857
Kalita B, Dutta S, Mukherjee AK (2019) RGD-independent binding of Russell’s viper venom kunitz-type protease inhibitors to platelet GPIIb/IIIa receptor. Sci Rep 9:8316
Ketterer S, Gomez-Auli A, Hillebrand LE, Petrera A et al (2016) Inherited diseases caused by mutations in cathepsin protease genes. FEBS J 284:1437–1454
Kitaguchi N, Takahashi Y, Tokushima Y, Shiojiri S, Ito H (1988) Novel precursor of Alzheimer’s disease amyloid protein shows protease inhibitor activity. Nature 331:530–532
Kitaguchi N, Takahashi Y, Oishi K, Shiojiri S et al (1990) Enzyme specificity of proteinase inhibitor region in amyloid precursor protein of Alzheimer’s disease: different properties compared with protease nexin I. Biochim Biophys Acta 1038:105–113
Kordis D, Gubensek F (2000) Adaptive evolution of animal toxin multigene families. Gene 261(1):43–52
Lamande SR, Morgelin M, Adams NE et al (2006) The C5 domain of the collagen VI α3(VI) chain is critical for extracellular microfibril formation and is present in the extracellular matrix of cultured cells. J Bio Chem 281(24):16607–16614
Laskowski M, Kato I (1980) Protein inhibitors of proteinases. Annu Rev Biochem 49:593–626
Laskowski M, Kato I, Ardelt W, Cook J, Denton A et al (1987a) Ovomucoid third domains from 100 avian species: isolation, sequences and hypervariability of enzyme-inhibitor contact residues. Biochemistry 26:202–221
Laskowski M, Kato I, Kohr WJ, Park SJ, Tashiro M, Whatley HE (1987b) Positive Darwinian selection in evolution of protein inhibitors of serine proteinases. Cold Spring Harbor Symp Quant Biol 52:545–553
Lehmann A (2008) Ecallantide (DX-88), a plasma kallikrein inhibitor for the treatment of hereditary angioedema and the prevention of blood loss in on-pump cardiothoracic surgery. Expert Opin Biol Ther 8(8):1187–1199
Lu J, Yang H, Yu H et al (2008) A novel serine protease inhibitor from Bungarus fasciatus venom. Peptides 29(3):369–374
Martins LA, Kotál J, Bensaoud C et al (2020) Small protease inhibitors in tick saliva and salivary glands and their role in tick-host-pathogen interactions. Biochim Biophys Acta Proteins Proteom 1868(2):140336
Masci PP, Whitaker AN, Sparrow LG, Jersey JD, Winzor DJ et al (2000) Textilinins from Pseudonaja textilis textilis. Characterization of two plasmin inhibitors that reduce bleeding in an animal model. Blood Coagul Fibrinolysis 11:385–393
Molinari F, Meskanaite V, Munnich A, Sonderegger P, Colleaux L (2003) Extracellular proteases and their inhibitors in genetic diseases of the central nervous system. Hum Mol Genet 12(2):R195–R200
Morais SB, Figueiredo BC, Assis NRG et al (2018) Schistosoma mansoni SmKI-1 serine protease inhibitor binds to elastase and impairs neutrophil function and inflammation. PLoS Pathog 14(2):e1006870
Morjen M, Kallech-Ziri O, Bazaa A et al (2013) PIVL, a new serine protease inhibitor from Macrovipera lebetina transmediterranea venom, impairs motility of human glioblastoma cells. Matrix Biol 32(1):52–62
Mukherjee AK, Mackessy SP (2014) Pharmacological properties and pathophysiological significance of a Kunitz-type protease inhibitor (Rusvikunin-II) and its protein complex (Rusvikunin complex) purified from Daboia russelii russelii venom. Toxicon 89:55–66
Oliva ML, Ferreira Rda S, Ferreira JG, de Paula CA et al (2011) Structural and functional properties of kunitz proteinase inhibitors from leguminosae: a mini review. Curr Protein Pept Sci 12(5):348–357
Peigneur S, Billen B, Derua R, Waelkens E et al (2011) A bifunctional sea anemone peptide with Kunitz type protease and potassium channel inhibiting properties. Biochem Pharmacol 82:81–90
Pendlebury D, Wang R, Henin RD et al (2014) Sequence and conformational specificity in substrate recognition: several human Kunitz protease inhibitor domains are specific substrates of mesotrypsin. J Biol Chem 289(47):32783–32797
Pugia MJ, Valdes R, Jortani SA (2007) Bikunin (urinary trypsin inhibitor): structure, biological relevance, and measurement. Adv In Clin Chem 44:223–245
Ranasinghe S, McManus DP (2013) Structure and function of invertebrate Kunitz serine protease inhibitors. Dev Com Immuno 39:219–227
Ranasinghe SL, Fischer K, Gobert GN et al (2015a) Functional expression of a novel Kunitz type protease inhibitor from the human blood fluke Schistosoma mansoni. Parasites Vectors 8:408
Ranasinghe SL, Fischer K, Gobert GN, McManus DP (2015b) A novel coagulation inhibitor from Schistosoma japonicum. Parasitology 142(14):1663–1672
Rawlings ND, Tolle DP, Barrett AJ (2004a) Evolutionary families of peptidase inhibitors. Biochem J 378:705–716
Rawlings ND, Tolle DP, Barrett AJ (2004b) MEROPS: the peptidase database. Nucleic acids Res 32:160–164
Robertson B, Owen D, Stow J, Butler C, Newland C (1996) Novel effects of dendrotoxin homologues on subtypes of mammalian Kv1 potassium channels expressed in Xenopus oocytes. FEBS Lett 383:26–30
Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S (2009) An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics 8:1891–1907
Sabotič J, Kos J (2012) Microbial and fungal protease inhibitors—current and potential applications. Appl Microbiol Biotechnol 93:1351–1375
Salier JP (1990) Inter-α-trypsin inhibitor: emergence of a family within the Kunitz-type protease inhibitor superfamily. Trends in Biochem Sci 15:435–439
Salier JP, Rouet P, Raguenez G, Daveau M (1996) The inter-α-inhibitor family: from structure to regulation. Biochem J 315:1–9
Schwarz A, Cabezas-Cruz A, Kopecky J, Valdes JJ (2014) Understanding the evolutionary structural variability and target specificity of tick salivary Kunitz peptides using next generation transcriptome data. BMC Evol Biol 14:4
Sheffield WP, Eltringham-Smith LJ, Bhakta V (2018) Fusion to human serum albumin extends the circulatory half-life and duration of antithrombotic action of the kunitz protease inhibitor domain of protease nexin 2. Cell Physiol Biochem 45(2):772–782
Shigetomi H, Onogi A, Kajiwara H et al (2010) Anti-inflammatory actions of serine protease inhibitors containing the Kunitz domain. Inflamm Res 59(9):679–687
Simeon R, Chen Z (2018) Invitro-engineered non-antibody protein therapeutics. Protein Cell 9(1):3–14
Smith D, Tikhonova IG, Jewhurst HL, Drysdale OC et al (2016) Unexpected activity of a novel Kunitz-type inhibitor: inhibition of cysteine proteases but not serine proteases. J Biol Chem 291(37):19220–19234
Strydom DJ (1973) Protease inhibitors as snake venom toxins. Nat New Biol 243:88–89
Strydom DJ (1977) Snake venom toxins. The amino acid sequence of toxin Vi2, a homologue of pancreatic trypsin inhibitor, from Dendroaspis polylepis polylepis (black mamba) venom. Biochim Biophys Acta 491:361–369
Thakur R, Mukherjee AK (2017) Pathophysiological significance and therapeutic applications of snake venom protease inhibitors. Toxicon 131:37–47
Vincent JP, Lazdunski M (1973) The interaction between alpha-chymotrypsin and pancreatic trypsin inhibitor (Kunitz inhibitor). Kinetic and thermodynamic properties. Eur J Biochem 38:365–372
Walsh JB, Stephan W (2001) Multigene families: evolution. Encycl Life Sci 1–6:673
Wan H, Lee KS, Kim BY, Zou FM, Yoon HJ et al (2013a) A spider-derived kunitz-type serine protease inhibitor that acts as a plasmin inhibitor and an elastase inhibitor. PLoS ONE 8(1):e53343
Wan H, Lee KS, Kim BY, Zou FM et al (2013b) A spider-derived Kunitz-type serine protease inhibitor that acts as a plasmin inhibitor and an elastase inhibitor. PLoS ONE 8(1):e53343
Wang FC, Bell N, Reid P, Smith LA et al (1999) Identification of residues in dendrotoxin K responsible for its discrimination between neuronal K+ channels containing Kv1.1 and 1.2a subunit. Eur J Biochem 263:222–229
Williams A, Baird LG (2003) DX-88 and HAE: a developmental perspective. Transfus Apher Sci 29(3):255–258
Wulff H, Castle NA, Pardo LA (2009) Voltage-gated potassium channels as therapeutic tragets. Nat Rev Drug Discov 8(12):982–1001
Yuan CH, He QY, Peng K, Diao JB, Tang X et al (2008) Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS ONE 3:e3414
Zhao R, Dai H, Qiu S, Li T, He Y et al (2011) SdPI, the first functionally characterized Kunitz-type trypsin inhibitor from scorpion venom. PLoS ONE 6:27548
Zhuo L, Hascall VC, Kimata K (2004) Inter-α-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J Biol Chem 279(37):38079–38082
Župunski V, Kordiš D (2016) Strong and widespread action of site-specific positive selection in the snake venom Kunitz/BPTI protein family. Sci Rep 6:37054
Zupunski V, Kordis D, Gubensek F (2003) Adaptive evolution in the snake venom Kunitz/BPTI protein family. FEBS Lett 547:131–136
Zweckstetter M, Czisch M, Mayer U et al (1996) Structure and multiple conformations of the Kunitz-type domain from human type VI collagen α3(VI) chain in solution. Structure 4:195–209
Acknowledgements
MM is a recipient of DST-INSPIRE Faculty fellowship award from Department of Science & Technology (DST) and Indo-Australian Career Boosting Gold Fellowship (IACBGF) from Department of Biotechnology (DBT), Government of India, New Delhi. Support from Shiv Nadar University and mentorship of Dr. Shailja Singh is sincerely acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Erich Bornberg-Bauer.
Rights and permissions
About this article
Cite this article
Mishra, M. Evolutionary Aspects of the Structural Convergence and Functional Diversification of Kunitz-Domain Inhibitors. J Mol Evol 88, 537–548 (2020). https://doi.org/10.1007/s00239-020-09959-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-020-09959-9